Abstract
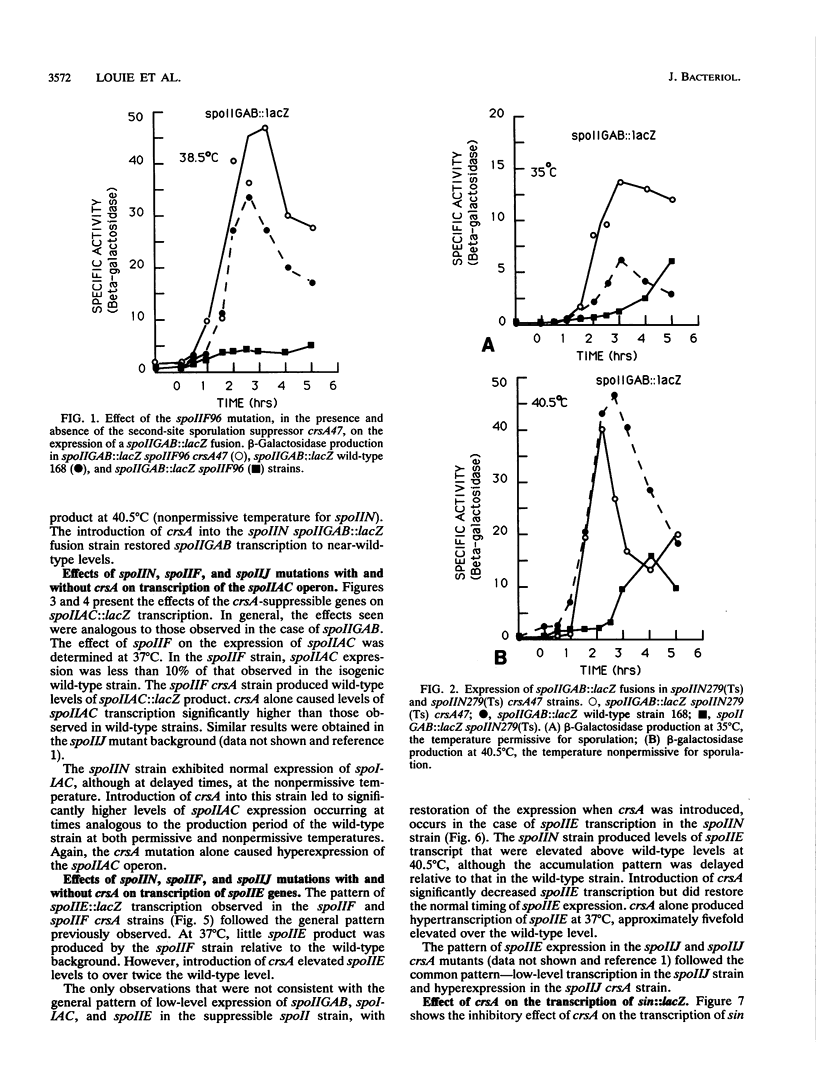
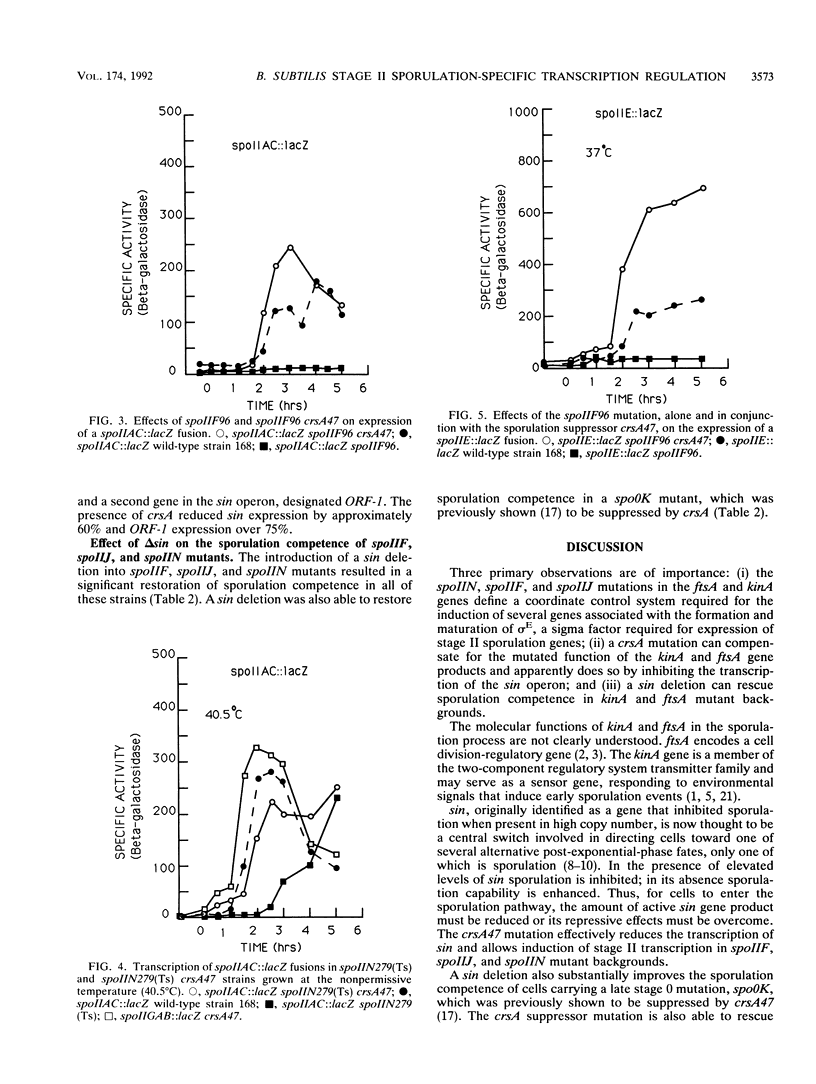
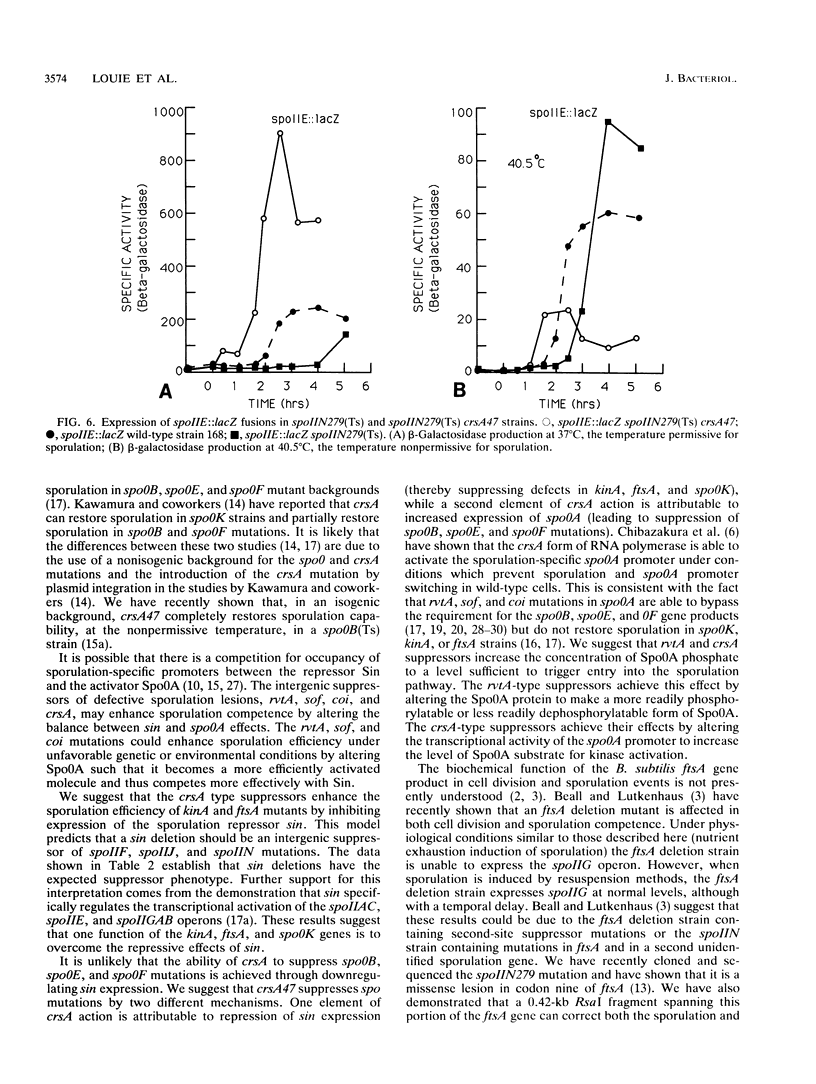
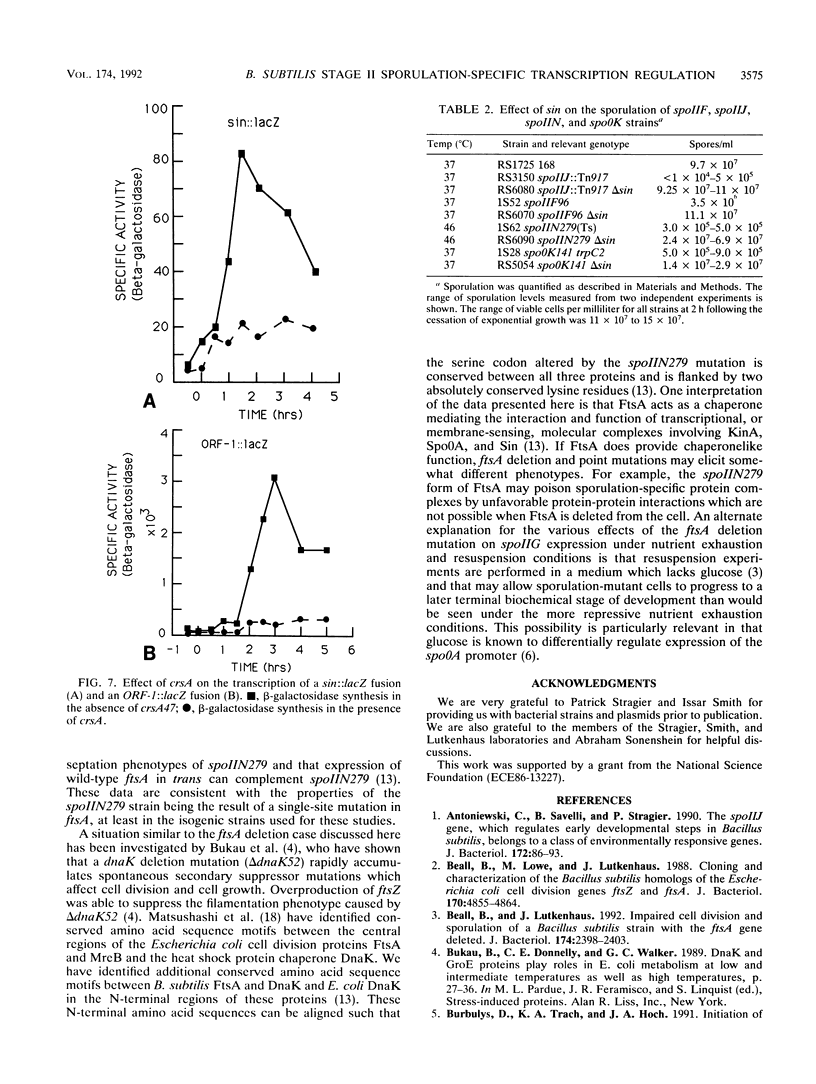
Bacillus subtilis strains containing defects in the sporulation gene spoIIF (kinA), spoIIJ (kinA), or spoIIN (ftsA) cannot transcribe the sigma E-dependent gene spoIID. Results presented here and by other workers demonstrate that the spoIIF, spoIIJ, and spoIIN gene products control spoIID transcription indirectly by coordinating the induction of the spoIIGAB, spoIIE, and spoIIAC operons, which are required for sigma E synthesis and processing. Sporulation competence and spoIIGAB, spoIIE, and spoIIAC transcription were restored in spoIIF, spoIIJ, and spoIIN mutants by introduction of crsA47, a mutation in the major vegetative sigma factor sigma A. crsA mutations are known to restore sporulation in certain spo0 mutants. crsA suppression of kinA and ftsA mutations was achieved through inhibition of the transcription of sin, a gene involved in the selection between several post-exponential-phase cell states. A deletion of sin restored sporulation competence in spoIIF, spoIIJ, or spoIIN mutant strains. A sin deletion was also able to restore sporulation competence in the crsA suppressible stage 0 mutant spo0K141.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antoniewski C., Savelli B., Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990 Jan;172(1):86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B., Lowe M., Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsZ and ftsA. J Bacteriol. 1988 Oct;170(10):4855–4864. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992 Apr;174(7):2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibazakura T., Kawamura F., Takahashi H. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J Bacteriol. 1991 Apr;173(8):2625–2632. doi: 10.1128/jb.173.8.2625-2632.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Cabane K., Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988 Mar;170(3):1046–1053. doi: 10.1128/jb.170.3.1046-1053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Dubnau E., Smith I. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J Bacteriol. 1986 Nov;168(2):860–869. doi: 10.1128/jb.168.2.860-869.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Oppenheim J., Smith I. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J Bacteriol. 1991 Jan;173(2):678–686. doi: 10.1128/jb.173.2.678-686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. D. Integration of developmental signals and the initiation of sporulation in B. subtilis. Cell. 1991 Apr 5;65(1):5–8. doi: 10.1016/0092-8674(91)90353-z. [DOI] [PubMed] [Google Scholar]
- Jonas R. M., Weaver E. A., Kenney T. J., Moran C. P., Jr, Haldenwang W. G. The Bacillus subtilis spoIIG operon encodes both sigma E and a gene necessary for sigma E activation. J Bacteriol. 1988 Feb;170(2):507–511. doi: 10.1128/jb.170.2.507-511.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura F., Wang L. F., Doi R. H. Catabolite-resistant sporulation (crsA) mutations in the Bacillus subtilis RNA polymerase sigma 43 gene (rpoD) can suppress and be suppressed by mutations in spo0 genes. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8124–8128. doi: 10.1073/pnas.82.23.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney T. J., York K., Youngman P., Moran C. P., Jr Genetic evidence that RNA polymerase associated with sigma A factor uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M., Wachi M., Ishino F. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res Microbiol. 1990 Jan;141(1):89–103. doi: 10.1016/0923-2508(90)90101-u. [DOI] [PubMed] [Google Scholar]
- Ng C., Buchanan C., Leung A., Ginther C., Leighton T. Suppression of defective-sporulation phenotypes by mutations in transcription factor genes of Bacillus subtilis. Biochimie. 1991 Jul-Aug;73(7-8):1163–1170. doi: 10.1016/0300-9084(91)90161-s. [DOI] [PubMed] [Google Scholar]
- Olmedo G., Ninfa E. G., Stock J., Youngman P. Novel mutations that alter the regulation of sporulation in Bacillus subtilis. Evidence that phosphorylation of regulatory protein SpoOA controls the initiation of sporulation. J Mol Biol. 1990 Oct 5;215(3):359–372. doi: 10.1016/s0022-2836(05)80357-2. [DOI] [PubMed] [Google Scholar]
- Perego M., Cole S. P., Burbulys D., Trach K., Hoch J. A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Hoch J. A. Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. J Bacteriol. 1991 Apr;173(8):2514–2520. doi: 10.1128/jb.173.8.2514-2520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Hoch J. A. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J Bacteriol. 1988 Jun;170(6):2560–2567. doi: 10.1128/jb.170.6.2560-2567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Spiegelman G. B., Hoch J. A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988 Nov;2(6):689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Perego M., Wu J. J., Spiegelman G. B., Hoch J. A. Mutational dissociation of the positive and negative regulatory properties of the Spo0A sporulation transcription factor of Bacillus subtilis. Gene. 1991 Apr;100:207–212. doi: 10.1016/0378-1119(91)90368-l. [DOI] [PubMed] [Google Scholar]
- Price C. W., Doi R. H. Genetic mapping of rpoD implicates the major sigma factor of Bacillus subtilis RNA polymerase in sporulation initiation. Mol Gen Genet. 1985;201(1):88–95. doi: 10.1007/BF00397991. [DOI] [PubMed] [Google Scholar]
- Satola S., Kirchman P. A., Moran C. P., Jr Spo0A binds to a promoter used by sigma A RNA polymerase during sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4533–4537. doi: 10.1073/pnas.88.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R. A., Leighton T. Intergenic suppressors of temperature-sensitive sporulation in Bacillus subtilis are allele non-specific. Mol Gen Genet. 1981;183(3):532–537. doi: 10.1007/BF00268777. [DOI] [PubMed] [Google Scholar]
- Spiegelman G., Van Hoy B., Perego M., Day J., Trach K., Hoch J. A. Structural alterations in the Bacillus subtilis Spo0A regulatory protein which suppress mutations at several spo0 loci. J Bacteriol. 1990 Sep;172(9):5011–5019. doi: 10.1128/jb.172.9.5011-5019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Bonamy C., Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988 Mar 11;52(5):697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Trempy J. E., Bonamy C., Szulmajster J., Haldenwang W. G. Bacillus subtilis sigma factor sigma 29 is the product of the sporulation-essential gene spoIIG. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4189–4192. doi: 10.1073/pnas.82.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Penchev R., Dubnau E., Smith I., Dubnau D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990 May;4(5):860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]


