Abstract
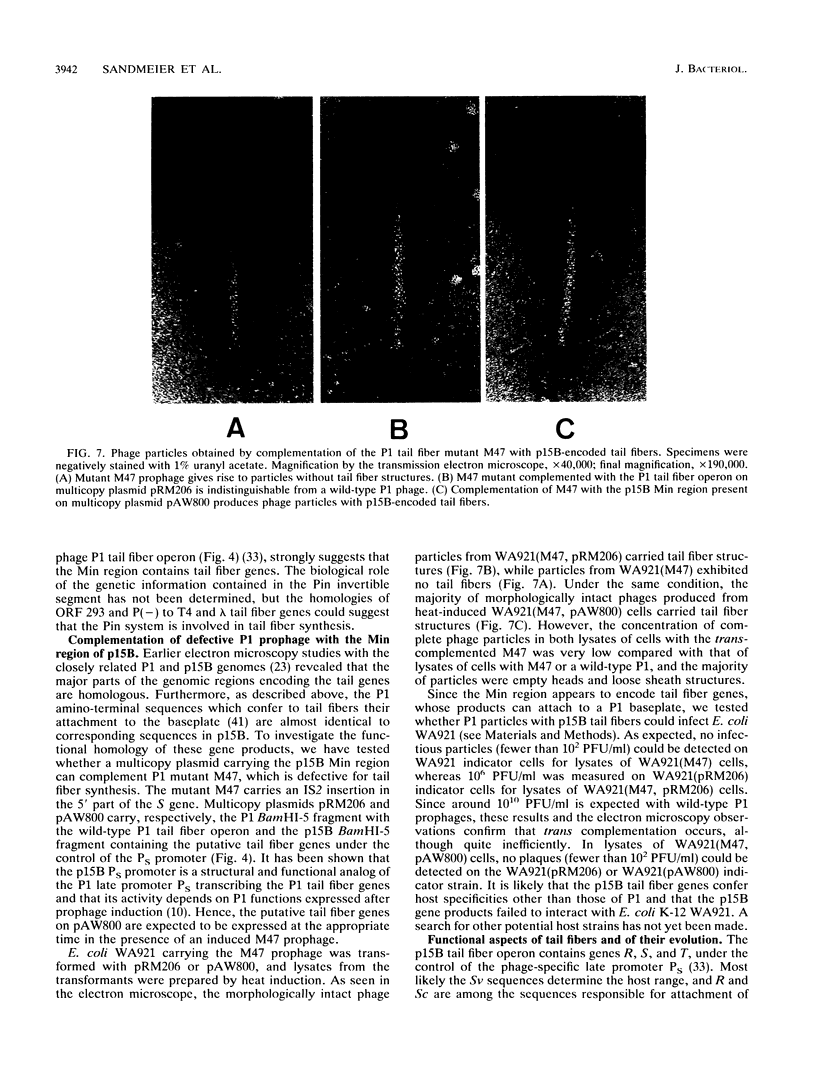
Plasmid p15B and the genome of bacteriophage P1 are closely related, but their site-specific DNA inversion systems, Min and Cin, respectively, do not have strict structural homology. Rather, the complex Min system represents a substitution of a Cin-like system into an ancestral p15B genome. The substituting sequences of both the min recombinase gene and the multiple invertible DNA segments of p15B are, respectively, homologous to the pin recombinase gene and to part of the invertible DNA of the Pin system on the defective viral element e14 of Escherichia coli K-12. To map the sites of this substitution, the DNA sequence of a segment adjacent to the invertible segment in the P1 genome was determined. This, together with already available sequence data, indicated that both P1 and p15B had suffered various sequence acquisitions or deletions and sequence amplifications giving rise to mosaics of partially related repeated elements. Data base searches revealed segments of homology in the DNA inversion regions of p15B, e14, and P1 and in tail fiber genes of phages Mu, T4, P2, and lambda. This result suggest that the evolution of phage tail fiber genes involves horizontal gene transfer and that the Min and Pin regions encode tail fiber genes. A functional test proved that the p15B Min region carries a tail fiber operon and suggests that the alternative expression of six different gene variants by Min inversion offers extensive host range variation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brody H., Greener A., Hill C. W. Excision and reintegration of the Escherichia coli K-12 chromosomal element e14. J Bacteriol. 1985 Mar;161(3):1112–1117. doi: 10.1128/jb.161.3.1112-1117.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George D. G., Yeh L. S., Barker W. C. Unexpected relationships between bacteriophage lambda hypothetical proteins and bacteriophage T4 tail-fiber proteins. Biochem Biophys Res Commun. 1983 Sep 30;115(3):1061–1068. doi: 10.1016/s0006-291x(83)80043-6. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin A., Zingg J. M., Arber W. Organization of the bacteriophage P1 tail-fibre operon. Gene. 1989;76(2):239–243. doi: 10.1016/0378-1119(89)90164-9. [DOI] [PubMed] [Google Scholar]
- Haggård-Ljungquist E., Halling C., Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992 Mar;174(5):1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S. Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology. 1984 Apr 30;134(2):421–434. doi: 10.1016/0042-6822(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Iida S., Hiestand-Nauer R. Localized conversion at the crossover sequences in the site-specific DNA inversion system of bacteriophage P1. Cell. 1986 Apr 11;45(1):71–79. doi: 10.1016/0092-8674(86)90539-8. [DOI] [PubMed] [Google Scholar]
- Iida S., Hiestand-Nauer R. Role of the central dinucleotide at the crossover sites for the selection of quasi sites in DNA inversion mediated by the site-specific Cin recombinase of phage P1. Mol Gen Genet. 1987 Jul;208(3):464–468. doi: 10.1007/BF00328140. [DOI] [PubMed] [Google Scholar]
- Iida S., Huber H., Hiestand-Nauer R., Meyer J., Bickle T. A., Arber W. The bacteriophage P1 site-specific recombinase cin: recombination events and DNA recognition sequences. Cold Spring Harb Symp Quant Biol. 1984;49:769–777. doi: 10.1101/sqb.1984.049.01.087. [DOI] [PubMed] [Google Scholar]
- Iida S., Sandmeier H., Hübner P., Hiestand-Nauer R., Schneitz K., Arber W. The Min DNA inversion enzyme of plasmid p15B of Escherichia coli 15T-: a new member of the Din family of site-specific recombinases. Mol Microbiol. 1990 Jun;4(6):991–997. doi: 10.1111/j.1365-2958.1990.tb00671.x. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Inuzuka M., Tomizawa J. I. P1-like plasmid in Escherichia coli 15. J Mol Biol. 1970 Jun 14;50(2):457–470. doi: 10.1016/0022-2836(70)90204-4. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kardas E., Ritthaler W., Sandulache R., Schmucker R., Stern B. Comparative analysis of invertible DNA in phage genomes. Cold Spring Harb Symp Quant Biol. 1984;49:301–311. doi: 10.1101/sqb.1984.049.01.036. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Nakao T., Iino T. A gene for DNA invertase and an invertible DNA in Escherichia coli K-12. Gene. 1985;34(2-3):343–350. doi: 10.1016/0378-1119(85)90143-x. [DOI] [PubMed] [Google Scholar]
- Meyer J., Stålhammar-Carlemalm M., Streiff M., Iida S., Arber W. Sequence relations among the IncY plasmid p15B, P1, and P7 prophages. Plasmid. 1986 Sep;16(2):81–89. doi: 10.1016/0147-619x(86)90066-1. [DOI] [PubMed] [Google Scholar]
- Michel C. J., Jacq B., Arquès D. G., Bickle T. A. A remarkable amino acid sequence homology between a phage T4 tail fibre protein and ORF314 of phage lambda located in the tail operon. Gene. 1986;44(1):147–150. doi: 10.1016/0378-1119(86)90055-7. [DOI] [PubMed] [Google Scholar]
- Montag D., Henning U. An open reading frame in the Escherichia coli bacteriophage lambda genome encodes a protein that functions in assembly of the long tail fibers of bacteriophage T4. J Bacteriol. 1987 Dec;169(12):5884–5886. doi: 10.1128/jb.169.12.5884-5886.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag D., Riede I., Eschbach M. L., Degen M., Henning U. Receptor-recognizing proteins of T-even type bacteriophages. Constant and hypervariable regions and an unusual case of evolution. J Mol Biol. 1987 Jul 5;196(1):165–174. doi: 10.1016/0022-2836(87)90519-5. [DOI] [PubMed] [Google Scholar]
- Montag D., Schwarz H., Henning U. A component of the side tail fiber of Escherichia coli bacteriophage lambda can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J Bacteriol. 1989 Aug;171(8):4378–4384. doi: 10.1128/jb.171.8.4378-4384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural R. J., Chesney R. H., Vapnek D., Kropf M. M., Scott J. R. Isolation and characterization of cloned fragments of bacteriophage P1 DNA. Virology. 1979 Mar;93(2):387–397. doi: 10.1016/0042-6822(79)90243-5. [DOI] [PubMed] [Google Scholar]
- Oliver D. B., Crowther R. A. DNA sequence of the tail fibre genes 36 and 37 of bacteriophage T4. J Mol Biol. 1981 Dec 15;153(3):545–568. doi: 10.1016/0022-2836(81)90407-1. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985 Jan;4(1):237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H., Iida S., Hübner P., Hiestand-Nauer R., Arber W. Gene organization in the multiple DNA inversion region min of plasmid p15B of E.coli 15T-: assemblage of a variable gene. Nucleic Acids Res. 1991 Nov 11;19(21):5831–5838. doi: 10.1093/nar/19.21.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H., Iida S., Meyer J., Hiestand-Nauer R., Arber W. Site-specific DNA recombination system Min of plasmid p15B: a cluster of overlapping invertible DNA segments. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1109–1113. doi: 10.1073/pnas.87.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selivanov N. A., Prilipov A. G., Mesyanzhinov V. V. Nucleotide and deduced amino acid sequence of bacteriophage T4 gene 12. Nucleic Acids Res. 1988 Mar 25;16(5):2334–2334. doi: 10.1093/nar/16.5.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C., Arber W. IS2 insertion is a major cause of spontaneous mutagenesis of the bacteriophage P1: non-random distribution of target sites. EMBO J. 1983;2(1):67–71. doi: 10.1002/j.1460-2075.1983.tb01382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C., Shepherd J. C., Arber W. The sequence of the bacteriophage P1 genome region serving as hot target for IS2 insertion. EMBO J. 1983;2(10):1777–1781. doi: 10.1002/j.1460-2075.1983.tb01657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga A., Ikemizu S., Enomoto M. Site-specific recombinase genes in three Shigella subgroups and nucleotide sequences of a pinB gene and an invertible B segment from Shigella boydii. J Bacteriol. 1991 Jul;173(13):4079–4087. doi: 10.1128/jb.173.13.4079-4087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]