Abstract
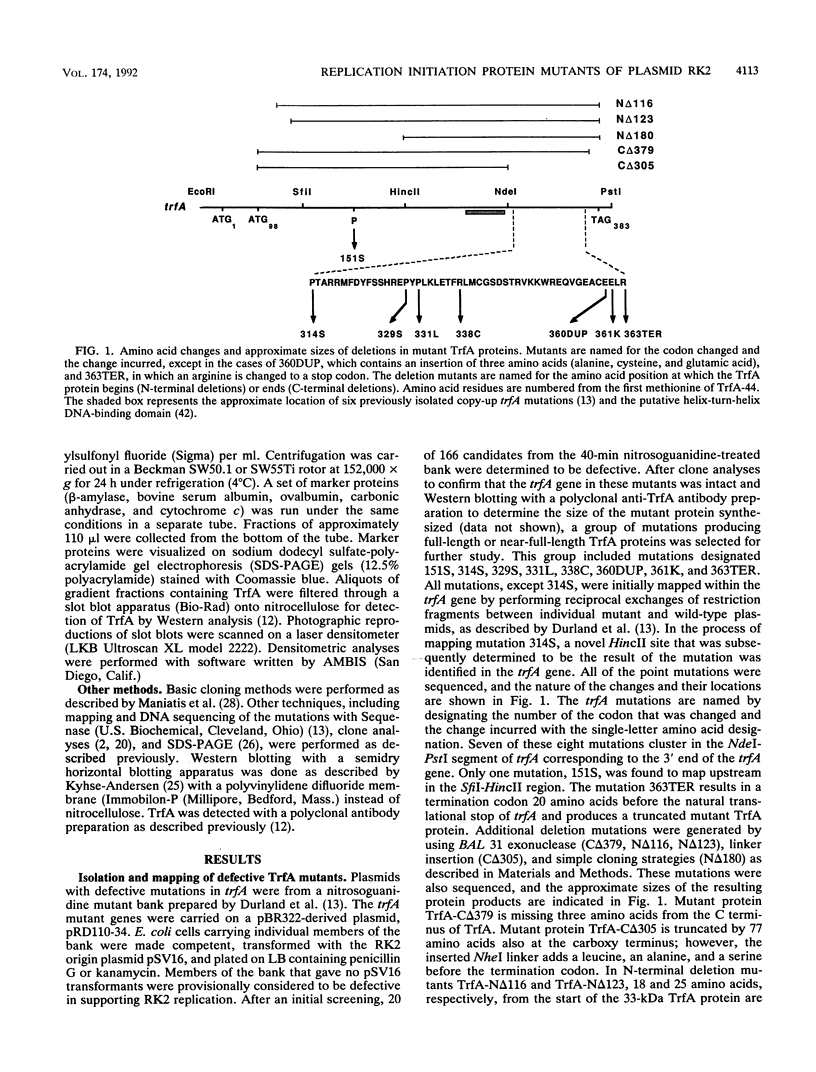
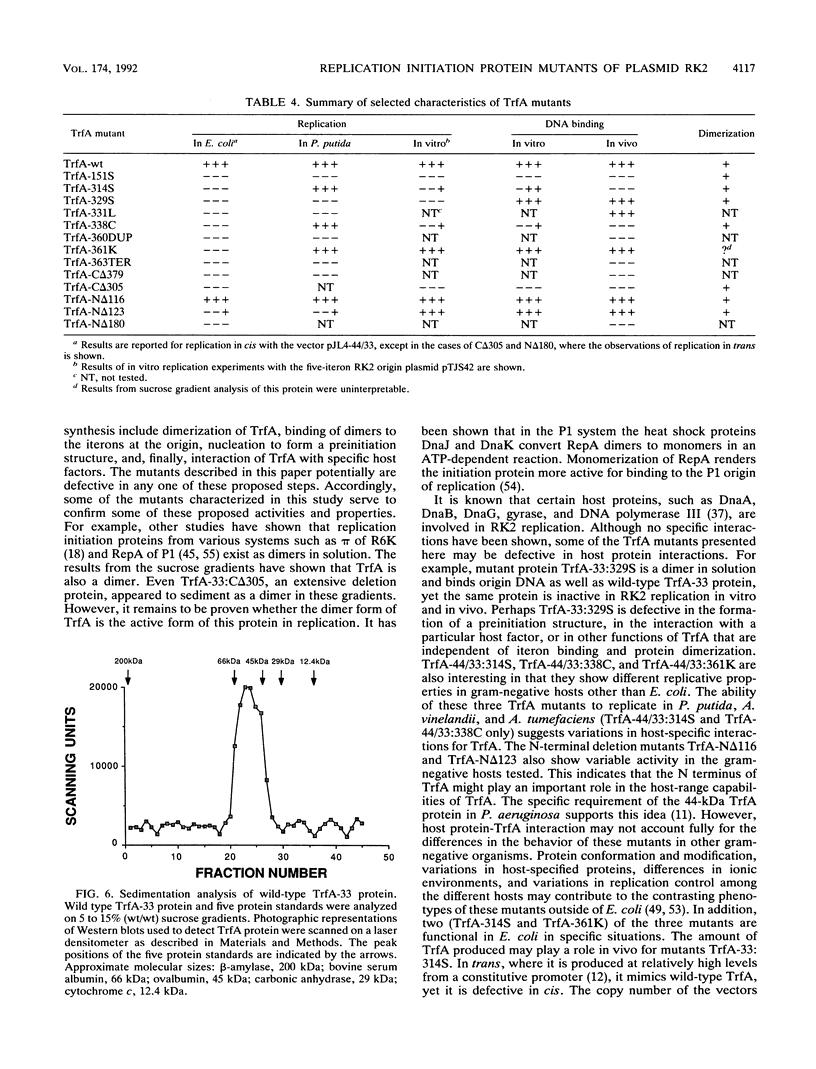
Plasmids with mutations in trfA, the gene encoding the replication initiation protein of the broad-host-range plasmid RK2, were isolated and characterized. Mutants identified from a nitrosoguanidine bank were defective in supporting the replication of a wild-type RK2 origin in Escherichia coli. Most of the mutations were clustered in a region of trfA corresponding to the carboxy-terminal quarter of the TrfA protein. 5' and 3' deletion mutants of trfA were also constructed. A C-terminal deletion of three amino acids of the Tr A protein was completely nonfunctional for RK2 replication. However, a deletion of 25 amino acids from the start of the 33-kDa TrfA protein was still competent for replication. Further characterization of the point and deletion trfA mutants in vivo revealed that a subset was capable of supporting RK2 replication in other gram-negative bacteria, including Pseudomonas putida, Agrobacterium tumefaciens, and Azotobacter vinelandii. Selected mutant TrfA proteins were partially purified and characterized in vitro. Velocity sedimentation analysis of these partially purified TrfA proteins indicated that the wild-type protein and all mutant TrfA proteins examined exist as dimers in solution. Results from in vitro replication assays corroborated the experimental findings in vivo. Gel retardation results clearly indicated that the point mutant TrfA-33:151S, which was completely defective in replication of an RK2 origin in all of the bacterial hosts tested in vivo, and a carboxy-terminal deletion mutant, TrfA-33:C delta 305, were not able to bind iterons in vitro. In addition to the partially defective or could not be distinguished from the wild-type protein in binding to the origin region. The mutant proteins with apparently normal DNA-binding activity in vitro either were inactive in all four gram-negative bacteria tested or exhibited differences in functionality depending on the host organism. These mutant TrfA proteins may be altered in the ability to interact with the replication proteins of the specific host bacterium.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin T. O., Berends T., Bunch T. A., Holzman T. F., Rausch S. K., Shamansky L., Treat M. L., Ziegler M. M. Cloning of the luciferase structural genes from Vibrio harveyi and expression of bioluminescence in Escherichia coli. Biochemistry. 1984 Jul 31;23(16):3663–3667. doi: 10.1021/bi00311a014. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop P. E., Brill W. J. Genetic analysis of Azotobacter vinelandii mutant strains unable to fix nitrogen. J Bacteriol. 1977 May;130(2):954–956. doi: 10.1128/jb.130.2.954-956.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Host ranges of R factors. J Gen Microbiol. 1972 May;70(3):453–460. doi: 10.1099/00221287-70-3-453. [DOI] [PubMed] [Google Scholar]
- Durland R. H., Helinski D. R. Replication of the broad-host-range plasmid RK2: direct measurement of intracellular concentrations of the essential TrfA replication proteins and their effect on plasmid copy number. J Bacteriol. 1990 Jul;172(7):3849–3858. doi: 10.1128/jb.172.7.3849-3858.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland R. H., Helinski D. R. The sequence encoding the 43-kilodalton trfA protein is required for efficient replication or maintenance of minimal RK2 replicons in Pseudomonas aeruginosa. Plasmid. 1987 Sep;18(2):164–169. doi: 10.1016/0147-619x(87)90044-8. [DOI] [PubMed] [Google Scholar]
- Durland R. H., Toukdarian A., Fang F., Helinski D. R. Mutations in the trfA replication gene of the broad-host-range plasmid RK2 result in elevated plasmid copy numbers. J Bacteriol. 1990 Jul;172(7):3859–3867. doi: 10.1128/jb.172.7.3859-3867.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Sugiono P., Guarente L., Davis R. W. Genetic selection for genes encoding sequence-specific DNA-binding proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3689–3693. doi: 10.1073/pnas.86.10.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F. C., Helinski D. R. Broad-host-range properties of plasmid RK2: importance of overlapping genes encoding the plasmid replication initiation protein TrfA. J Bacteriol. 1991 Sep;173(18):5861–5868. doi: 10.1128/jb.173.18.5861-5868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Meyer R. J., Helinski D. R. Suppression of Co1E1 replication properties by the Inc P-1 plasmid RK2 in hybrid plasmids constructed in vitro. J Mol Biol. 1979 Sep 25;133(3):295–318. doi: 10.1016/0022-2836(79)90395-4. [DOI] [PubMed] [Google Scholar]
- Filutowicz M., Uhlenhopp E., Helinski D. R. Binding of purified wild-type and mutant pi initiation proteins to a replication origin region of plasmid R6K. J Mol Biol. 1986 Jan 20;187(2):225–239. doi: 10.1016/0022-2836(86)90230-5. [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., Simpson R. B., Ream L. W., White F. F., Gordon M. P., Nester E. W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981 Nov;27(1 Pt 2):143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ingram L. C., Richmond M. H., Sykes R. B. Molecular characterization of the R factors implicated in the carbenicillin resistance of a sequence of Pseudomonas aeruginosa strains isolated from burns. Antimicrob Agents Chemother. 1973 Feb;3(2):279–288. doi: 10.1128/aac.3.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M., Ow D., Sauer B., Rabinowitz A., Calendar R. Genetic analysis of bacteriophage P4 using P4-plasmid ColE1 hybrids. Mol Gen Genet. 1980 Feb;177(3):399–412. doi: 10.1007/BF00271478. [DOI] [PubMed] [Google Scholar]
- Kittell B. L., Helinski D. R. Iteron inhibition of plasmid RK2 replication in vitro: evidence for intermolecular coupling of replication origins as a mechanism for RK2 replication control. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1389–1393. doi: 10.1073/pnas.88.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornacki J. A., West A. H., Firshein W. Proteins encoded by the trans-acting replication and maintenance regions of broad host range plasmid RK2. Plasmid. 1984 Jan;11(1):48–57. doi: 10.1016/0147-619x(84)90006-4. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984 Dec;10(3-4):203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Fürste J. P. Molecular cloning and mapping of SphI restriction fragments of plasmid RP4. Plasmid. 1983 Nov;10(3):303–307. doi: 10.1016/0147-619x(83)90047-1. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Bott M. A., Tooker P. A., Helinski D. R. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J., Engelberg A., Bairoch A. Using PC/GENE for protein and nucleic acid analysis. Biotechniques. 1988 Jun;6(6):566–572. [PubMed] [Google Scholar]
- NEWTON J. W., WILSON P. W., BURRIS R. H. Direct demonstration of ammonia as an intermediate in nitrogen fixation by Azotobacter. J Biol Chem. 1953 Sep;204(1):445–451. [PubMed] [Google Scholar]
- O'neill E. A., Berlinberg C., Bender R. A. Activity of Plasmid Replicons in CAULOBACTER CRESCENTUS : Rp4 and Cole1. Genetics. 1983 Apr;103(4):593–604. doi: 10.1093/genetics/103.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Shipley P. Host range and properties of the Pseudomonas aeruginosa R factor R1822. J Bacteriol. 1973 Feb;113(2):772–780. doi: 10.1128/jb.113.2.772-780.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. K., Chattoraj D. K. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol. 1988 Aug;170(8):3554–3560. doi: 10.1128/jb.170.8.3554-3560.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri S., Helinski D. R., Toukdarian A. Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J Biol Chem. 1991 Jul 5;266(19):12536–12543. [PubMed] [Google Scholar]
- Pinkney M., Diaz R., Lanka E., Thomas C. M. Replication of mini RK2 plasmid in extracts of Escherichia coli requires plasmid-encoded protein TrfA and host-encoded proteins DnaA, B, G DNA gyrase and DNA polymerase III. J Mol Biol. 1988 Oct 20;203(4):927–938. doi: 10.1016/0022-2836(88)90118-0. [DOI] [PubMed] [Google Scholar]
- Roberts R. C., Burioni R., Helinski D. R. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J Bacteriol. 1990 Nov;172(11):6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Filutowicz M., Helinski D. R. Replication of derivatives of the broad host range plasmid RK2 in two distantly related bacteria. Plasmid. 1983 May;9(3):325–330. doi: 10.1016/0147-619x(83)90010-0. [DOI] [PubMed] [Google Scholar]
- Schmidhauser T. J., Helinski D. R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985 Oct;164(1):446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingler V., Thomas C. M. Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J Mol Biol. 1984 May 25;175(3):229–249. doi: 10.1016/0022-2836(84)90346-2. [DOI] [PubMed] [Google Scholar]
- Shingler V., Thomas C. M. Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J Mol Biol. 1984 May 25;175(3):229–249. doi: 10.1016/0022-2836(84)90346-2. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Thomas C. M., Helinski D. R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Swack J. A., Pal S. K., Mason R. J., Abeles A. L., Chattoraj D. K. P1 plasmid replication: measurement of initiator protein concentration in vivo. J Bacteriol. 1987 Aug;169(8):3737–3742. doi: 10.1128/jb.169.8.3737-3742.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terawaki Y., Hong Z., Itoh Y., Kamio Y. Importance of the C terminus of plasmid Rts1 RepA protein for replication and incompatibility of the plasmid. J Bacteriol. 1988 Mar;170(3):1261–1267. doi: 10.1128/jb.170.3.1261-1267.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. Complementation analysis of replication and maintenance functions of broad host range plasmids RK2 and RP1. Plasmid. 1981 May;5(3):277–291. doi: 10.1016/0147-619x(81)90005-6. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Hussain A. A. The korB gene of broad host range plasmid RK2 is a major copy number control element which may act together with trfB by limiting trfA expression. EMBO J. 1984 Jul;3(7):1513–1519. doi: 10.1002/j.1460-2075.1984.tb02004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Meyer R., Helinski D. R. Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J Bacteriol. 1980 Jan;141(1):213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Stalker D. M., Helinski D. R. Replication and incompatibility properties of segments of the origin region of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):1–7. doi: 10.1007/BF00338996. [DOI] [PubMed] [Google Scholar]
- Valla S., Haugan K., Durland R., Helinski D. R. Isolation and properties of temperature-sensitive mutants of the trfA gene of the broad host range plasmid RK2. Plasmid. 1991 Mar;25(2):131–136. doi: 10.1016/0147-619x(91)90025-r. [DOI] [PubMed] [Google Scholar]
- Wickner S. H. Three Escherichia coli heat shock proteins are required for P1 plasmid DNA replication: formation of an active complex between E. coli DnaJ protein and the P1 initiator protein. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2690–2694. doi: 10.1073/pnas.87.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H., Zahn K. Characterization of the DNA binding domain of bacteriophage lambda O protein. J Biol Chem. 1986 Jun 5;261(16):7537–7543. [PubMed] [Google Scholar]
- Wickner S., Hoskins J., McKenney K. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]