Abstract
The PRO1 gene of Saccharomyces cerevisiae encodes the 428-amino-acid protein gamma-glutamyl kinase (ATP:L-glutamate 5-phosphotransferase, EC 2.7.2.11), which catalyzes the first step in proline biosynthesis. Amino acid sequence comparison revealed significant homology between the yeast and Escherichia coli gamma-glutamyl kinases throughout their lengths. Four close matches to the consensus sequence for GCN4 protein binding and one close match to the RAP1 protein-binding site were found in the PRO1 upstream region. The response of the PRO1 gene to changes in the growth medium was analyzed by measurement of steady-state mRNA levels and of beta-galactosidase activity encoded by a PRO1-lacZ gene fusion. PRO1 expression was not repressed by exogenous proline and was not induced by the presence of glutamate in the growth medium. Although expression of the PRO1 gene did not change in response to histidine starvation, both steady-state PRO1 mRNA levels and beta-galactosidase activities were elevated in a gcd1 strain and reduced in a gcn4 strain. In addition, a pro1 bradytrophic strain became completely auxotrophic for proline in a gcn4 strain background. These results indicate that PRO1 is regulated by the general amino acid control system.
Full text
PDF




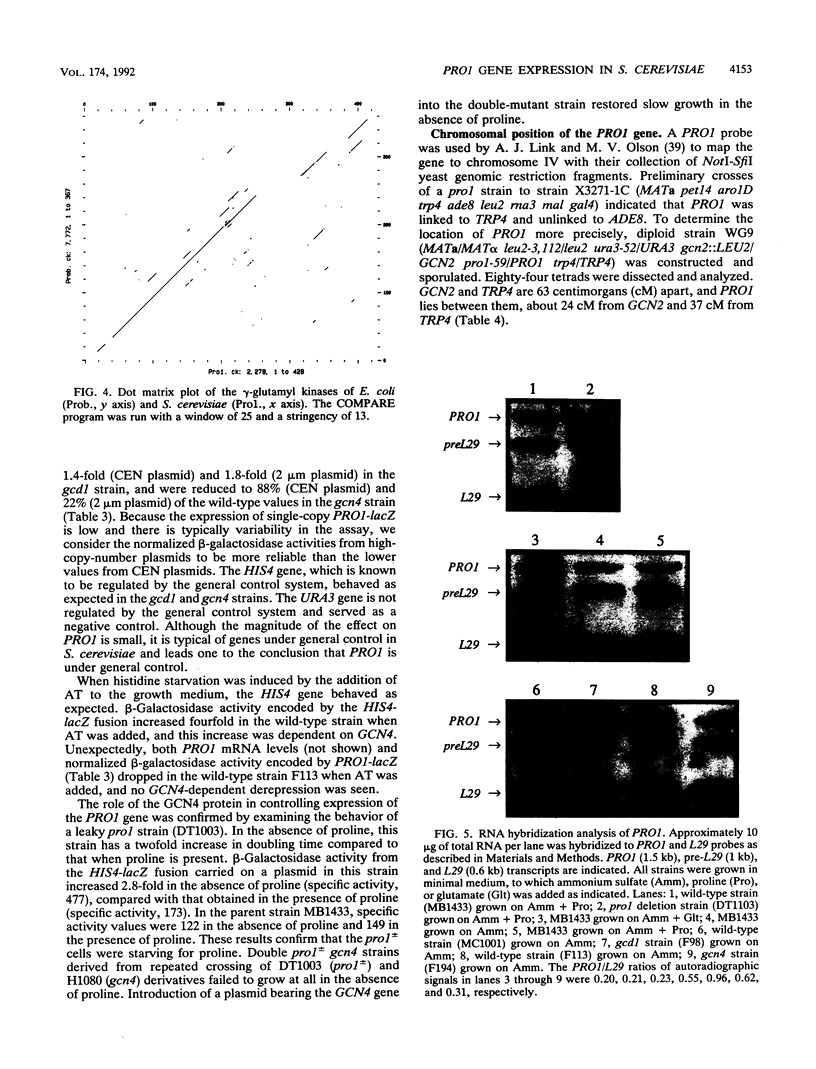
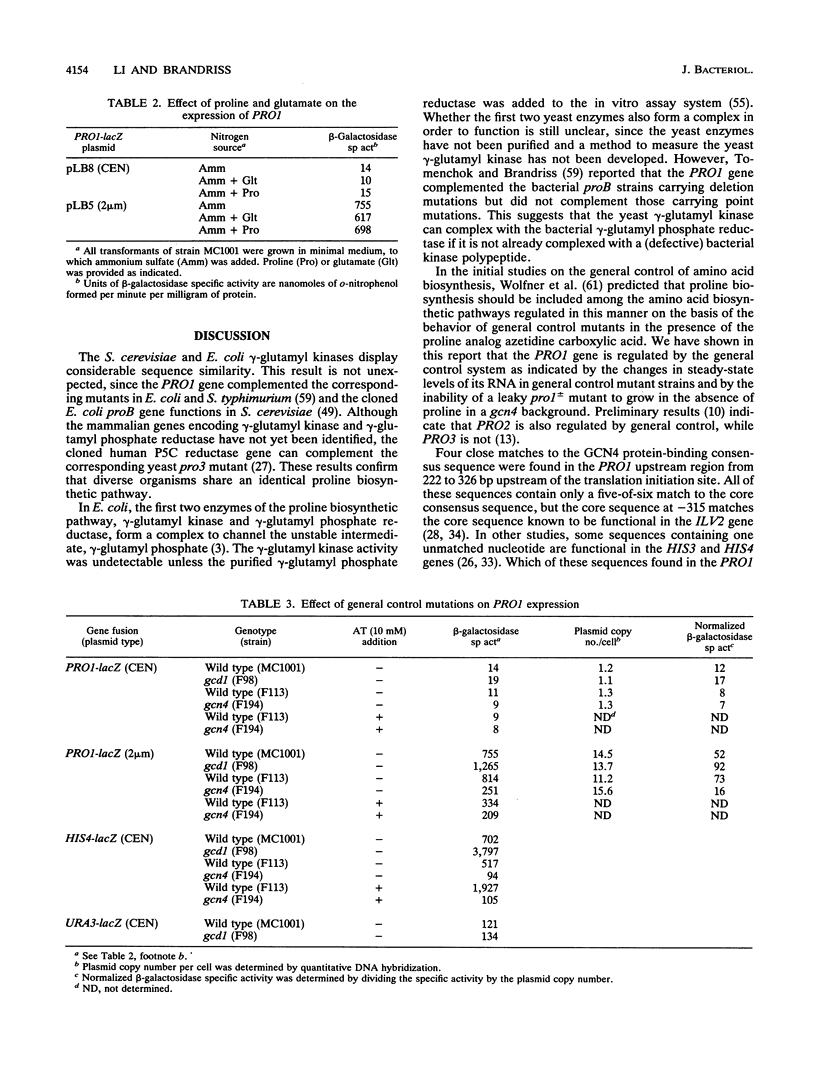


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Erickson B. W. Optimal sequence alignment using affine gap costs. Bull Math Biol. 1986;48(5-6):603–616. doi: 10.1007/BF02462326. [DOI] [PubMed] [Google Scholar]
- Andreadis A., Hsu Y. P., Hermodson M., Kohlhaw G., Schimmel P. Yeast LEU2. Repression of mRNA levels by leucine and primary structure of the gene product. J Biol Chem. 1984 Jul 10;259(13):8059–8062. [PubMed] [Google Scholar]
- Baich A. Proline synthesis in Escherichia coli. A proline-inhibitable glutamic acid kinase. Biochim Biophys Acta. 1969 Dec 30;192(3):462–467. doi: 10.1016/0304-4165(69)90395-x. [DOI] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Beltzer J. P., Chang L. F., Hinkkanen A. E., Kohlhaw G. B. Structure of yeast LEU4. The 5' flanking region contains features that predict two modes of control and two productive translation starts. J Biol Chem. 1986 Apr 15;261(11):5160–5167. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C., Falvey D. A. Proline biosynthesis in Saccharomyces cerevisiae: analysis of the PRO3 gene, which encodes delta 1-pyrroline-5-carboxylate reductase. J Bacteriol. 1992 Jun;174(11):3782–3788. doi: 10.1128/jb.174.11.3782-3788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C. Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J Bacteriol. 1979 Jun;138(3):816–822. doi: 10.1128/jb.138.3.816-822.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Krzywicki K. A. Amino-terminal fragments of delta 1-pyrroline-5-carboxylate dehydrogenase direct beta-galactosidase to the mitochondrial matrix in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Oct;6(10):3502–3512. doi: 10.1128/mcb.6.10.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol. 1979 Nov;140(2):498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol Cell Biol. 1983 Oct;3(10):1846–1856. doi: 10.1128/mcb.3.10.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W. E., Magasanik B. Ammonia regulation of amino acid permeases in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Apr;3(4):672–683. doi: 10.1128/mcb.3.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Huygen R., Verschueren K., Messenguy F., Tinel K., Cunin R., Glansdorff N. General amino acid control and specific arginine repression in Saccharomyces cerevisiae: physical study of the bifunctional regulatory region of the ARG3 gene. Mol Cell Biol. 1985 Nov;5(11):3139–3148. doi: 10.1128/mcb.5.11.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney A. J., Verma D. P. A soybean gene encoding delta 1-pyrroline-5-carboxylate reductase was isolated by functional complementation in Escherichia coli and is found to be osmoregulated. Mol Gen Genet. 1990 May;221(3):299–305. doi: 10.1007/BF00259392. [DOI] [PubMed] [Google Scholar]
- Deutch A. H., Rushlow K. E., Smith C. J. Analysis of the Escherichia coli proBA locus by DNA and protein sequencing. Nucleic Acids Res. 1984 Aug 10;12(15):6337–6355. doi: 10.1093/nar/12.15.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutch A. H., Smith C. J., Rushlow K. E., Kretschmer P. J. Escherichia coli delta 1-pyrroline-5-carboxylate reductase: gene sequence, protein overproduction and purification. Nucleic Acids Res. 1982 Dec 11;10(23):7701–7714. doi: 10.1093/nar/10.23.7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin C., Tice-Baldwin K., Shore D., Arndt K. T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991 Jul;11(7):3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Dougherty K. M., Brandriss M. C., Valle D. Cloning human pyrroline-5-carboxylate reductase cDNA by complementation in Saccharomyces cerevisiae. J Biol Chem. 1992 Jan 15;267(2):871–875. [PubMed] [Google Scholar]
- Falco S. C., Dumas K. S., Livak K. J. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 1985 Jun 11;13(11):4011–4027. doi: 10.1093/nar/13.11.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Hamilton P. T., Reeve J. N. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol Gen Genet. 1985;200(1):47–59. doi: 10.1007/BF00383311. [DOI] [PubMed] [Google Scholar]
- Harbury P. A., Struhl K. Functional distinctions between yeast TATA elements. Mol Cell Biol. 1989 Dec;9(12):5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hsu Y. P., Schimmel P. Yeast LEU1. Repression of mRNA levels by leucine and relationship of 5'-noncoding region to that of LEU2. J Biol Chem. 1984 Mar 25;259(6):3714–3719. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Boonchird C. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol Cell Biol. 1991 May;11(5):2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45(3):299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Nam H. G., Fried H. M. Effects of progressive depletion of TCM1 or CYH2 mRNA on Saccharomyces cerevisiae ribosomal protein accumulation. Mol Cell Biol. 1986 May;6(5):1535–1544. doi: 10.1128/mcb.6.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman R. B., Kaback D. B., Dubin R. A., Perkins E. L., Rosenberg N. G., Sutherland K. A., Forrest D. B., Michels C. A. MAL6 of Saccharomyces: a complex genetic locus containing three genes required for maltose fermentation. Proc Natl Acad Sci U S A. 1984 May;81(9):2811–2815. doi: 10.1073/pnas.81.9.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Orser C. S., Goodner B. W., Johnston M., Gelvin S. B., Csonka L. N. The Escherichia coli proB gene corrects the proline auxotrophy of Saccharomyces cerevisiae pro1 mutants. Mol Gen Genet. 1988 Apr;212(1):124–128. doi: 10.1007/BF00322454. [DOI] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savioz A., Jeenes D. J., Kocher H. P., Haas D. Comparison of proC and other housekeeping genes of Pseudomonas aeruginosa with their counterparts in Escherichia coli. Gene. 1990 Jan 31;86(1):107–111. doi: 10.1016/0378-1119(90)90121-7. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol Cell Biol. 1988 Nov;8(11):4634–4641. doi: 10.1128/mcb.8.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Deutch A. H., Rushlow K. E. Purification and characteristics of a gamma-glutamyl kinase involved in Escherichia coli proline biosynthesis. J Bacteriol. 1984 Feb;157(2):545–551. doi: 10.1128/jb.157.2.545-551.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. J., Downing S. J., Phang J. M., Lodato R. F., Aoki T. T. Pyrroline-5-carboxylate synthase activity in mammalian cells. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5221–5225. doi: 10.1073/pnas.77.9.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomenchok D. M., Brandriss M. C. Gene-enzyme relationships in the proline biosynthetic pathway of Saccharomyces cerevisiae. J Bacteriol. 1987 Dec;169(12):5364–5372. doi: 10.1128/jb.169.12.5364-5372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y., Jones M. E. Pyrroline-5-carboxylate synthesis from glutamate by rat intestinal mucosa. J Biol Chem. 1983 Mar 25;258(6):3865–3872. [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. Mutationally altered 3' ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J Mol Biol. 1984 Jul 25;177(1):107–135. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]