Abstract
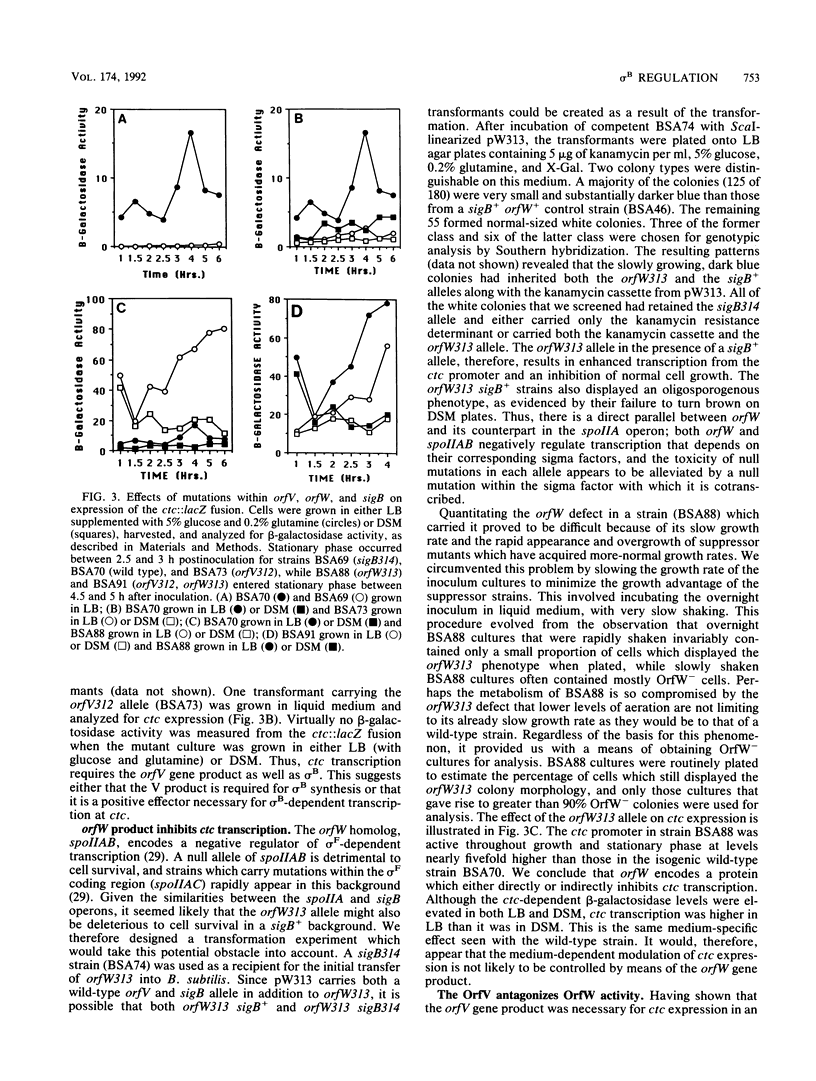
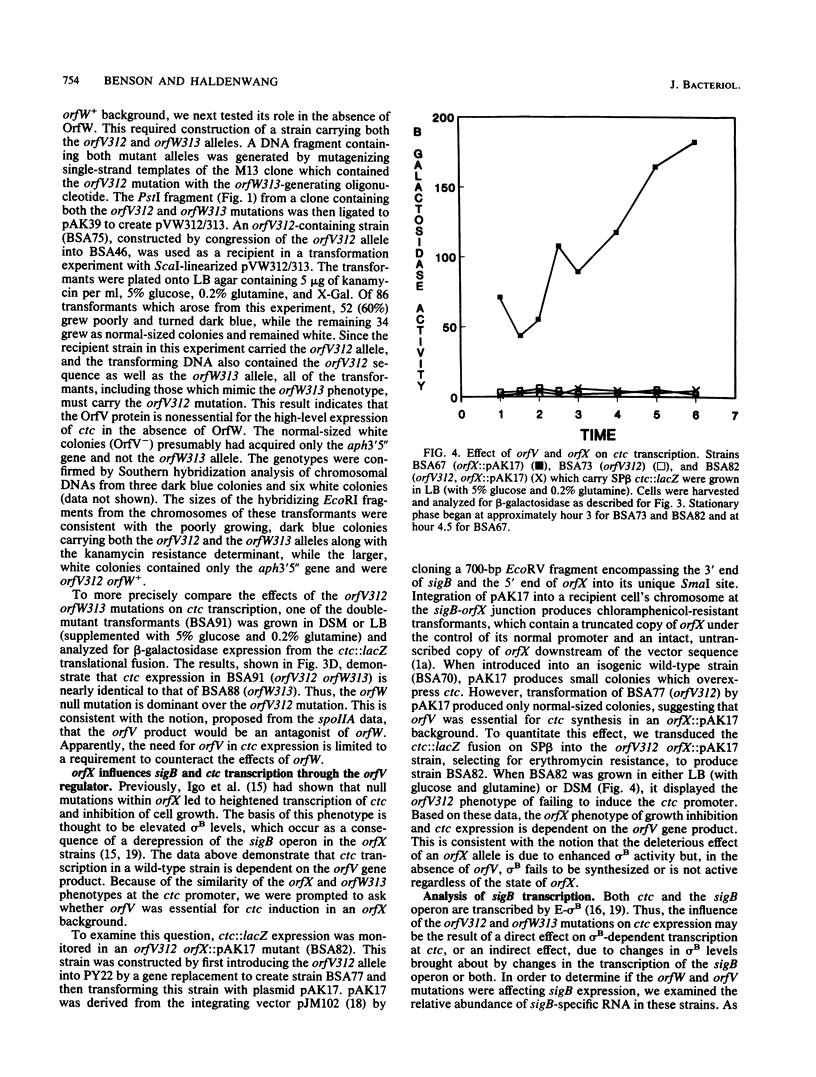
The sigB operon of Bacillus subtilis encodes sigma B and three additional open reading frames (orfV, orfW, and orfX). Having previously mapped several mutations that alter the induction pattern of a sigma B-dependent promoter (ctc) to regions of cloned B. subtilis DNA which contain these three open reading frames, we directly tested the regulatory potential of orfV, orfW, and orfX by creating null alleles of each of these genes and examining the effects of the mutations, either singly or in pairs, on transcription of ctc and the sigB operon. Using lacZ reporter gene fusions and Northern (RNA) blot analyses, we have determined that all three genes modulate the activation of the sigma B-dependent promoters at both the sigB operon and ctc. Our data are consistent with the three gene products participating in a single pathway of negative control. orfW and orfX single-mutant strains have high levels of sigB and ctc transcription. sigB and ctc transcription in an orfV strain is similar to that found in mutant strains which lack sigma B itself. The orfV mutation is dominant to orfX but recessive to orfW. These results suggest that OrfW is the primary inhibitor of sigma B-dependent transcription and that OrfV is capable of counteracting the negative control of OrfW but is prevented from doing this by the orfX gene product.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Hahn J., Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnie C., Lampe M., Losick R. Gene encoding the sigma 37 species of RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dubnau E., Weir J., Nair G., Carter L., 3rd, Moran C., Jr, Smith I. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol. 1988 Mar;170(3):1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M. L., Kalman S. S., Thomas S. M., Price C. W. Gene encoding the 37,000-dalton minor sigma factor of Bacillus subtilis RNA polymerase: isolation, nucleotide sequence, chromosomal locus, and cryptic function. J Bacteriol. 1987 Feb;169(2):771–778. doi: 10.1128/jb.169.2.771-778.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J., Mandelstam J. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):2967–2976. doi: 10.1099/00221287-132-11-2967. [DOI] [PubMed] [Google Scholar]
- Fort P., Piggot P. J. Nucleotide sequence of sporulation locus spoIIA in Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2147–2153. doi: 10.1099/00221287-130-8-2147. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Lang N., Losick R. A sporulation-induced sigma-like regulatory protein from B. subtilis. Cell. 1981 Feb;23(2):615–624. doi: 10.1016/0092-8674(81)90157-4. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Losick R. A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature. 1979 Nov 15;282(5736):256–260. doi: 10.1038/282256a0. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Losick R. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7000–7004. doi: 10.1073/pnas.77.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Márquez L. M., Chamberlin M. J. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J Bacteriol. 1988 Apr;170(4):1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986 Oct 20;191(4):615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Igo M., Lampe M., Ray C., Schafer W., Moran C. P., Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987 Aug;169(8):3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Jonas R. M., Peters H. K., 3rd, Haldenwang W. G. Phenotypes of Bacillus subtilis mutants altered in the precursor-specific region of sigma E. J Bacteriol. 1990 Aug;172(8):4178–4186. doi: 10.1128/jb.172.8.4178-4186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman S., Duncan M. L., Thomas S. M., Price C. W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990 Oct;172(10):5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmazyn-Campelli C., Bonamy C., Savelli B., Stragier P. Tandem genes encoding sigma-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 1989 Feb;3(2):150–157. doi: 10.1101/gad.3.2.150. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Inamine J. M., Lee L. N. Broad geographical distribution of homologous erythromycin, kanamycin, and streptomycin resistance determinants among group D streptococci of human and animal origin. Antimicrob Agents Chemother. 1986 Apr;29(4):549–555. doi: 10.1128/aac.29.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda E. S., Anaguchi H., Yamada K., Kobayashi Y. Two developmental genes encoding sigma factor homologs are arranged in tandem in Bacillus subtilis. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7637–7641. doi: 10.1073/pnas.85.20.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel D. B., Chamberlin M. J. The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase. J Bacteriol. 1989 Jun;171(6):3095–3101. doi: 10.1128/jb.171.6.3095-3101.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Foulger D., Errington J. The role of sigma F in prespore-specific transcription in Bacillus subtilis. Mol Microbiol. 1991 Mar;5(3):757–767. doi: 10.1111/j.1365-2958.1991.tb00746.x. [DOI] [PubMed] [Google Scholar]
- Rather P. N., Coppolecchia R., DeGrazia H., Moran C. P., Jr Negative regulator of sigma G-controlled gene expression in stationary-phase Bacillus subtilis. J Bacteriol. 1990 Feb;172(2):709–715. doi: 10.1128/jb.172.2.709-715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Margolis P., Duncan L., Coppolecchia R., Moran C. P., Jr, Losick R. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9221–9225. doi: 10.1073/pnas.87.23.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Kunkel B., Kroos L., Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989 Jan 27;243(4890):507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- Sun D. X., Stragier P., Setlow P. Identification of a new sigma-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 1989 Feb;3(2):141–149. doi: 10.1101/gad.3.2.141. [DOI] [PubMed] [Google Scholar]
- Trempy J. E., Bonamy C., Szulmajster J., Haldenwang W. G. Bacillus subtilis sigma factor sigma 29 is the product of the sporulation-essential gene spoIIG. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4189–4192. doi: 10.1073/pnas.82.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempy J. E., Morrison-Plummer J., Haldenwang W. G. Synthesis of sigma 29, an RNA polymerase specificity determinant, is a developmentally regulated event in Bacillus subtilis. J Bacteriol. 1985 Jan;161(1):340–346. doi: 10.1128/jb.161.1.340-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt C. L., Ray G. L., Trempy J. E., Da-Jian Z., Haldenwang W. G. Isolation of Bacillus subtilis mutants altered in expression of a gene transcribed in vitro by a minor form of RNA polymerase (E-sigma 37). J Bacteriol. 1985 Feb;161(2):515–522. doi: 10.1128/jb.161.2.515-522.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Wilson G. A., Young F. E. Transformation and transfection in lysogenic strains of Bacillus subtilis 168. J Bacteriol. 1973 Feb;113(2):540–548. doi: 10.1128/jb.113.2.540-548.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkin M. D. Structure and function in a Bacillus subtilis sporulation-specific sigma factor: molecular nature of mutations in spoIIAC. J Gen Microbiol. 1987 Mar;133(3):475–481. doi: 10.1099/00221287-133-3-475. [DOI] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]



