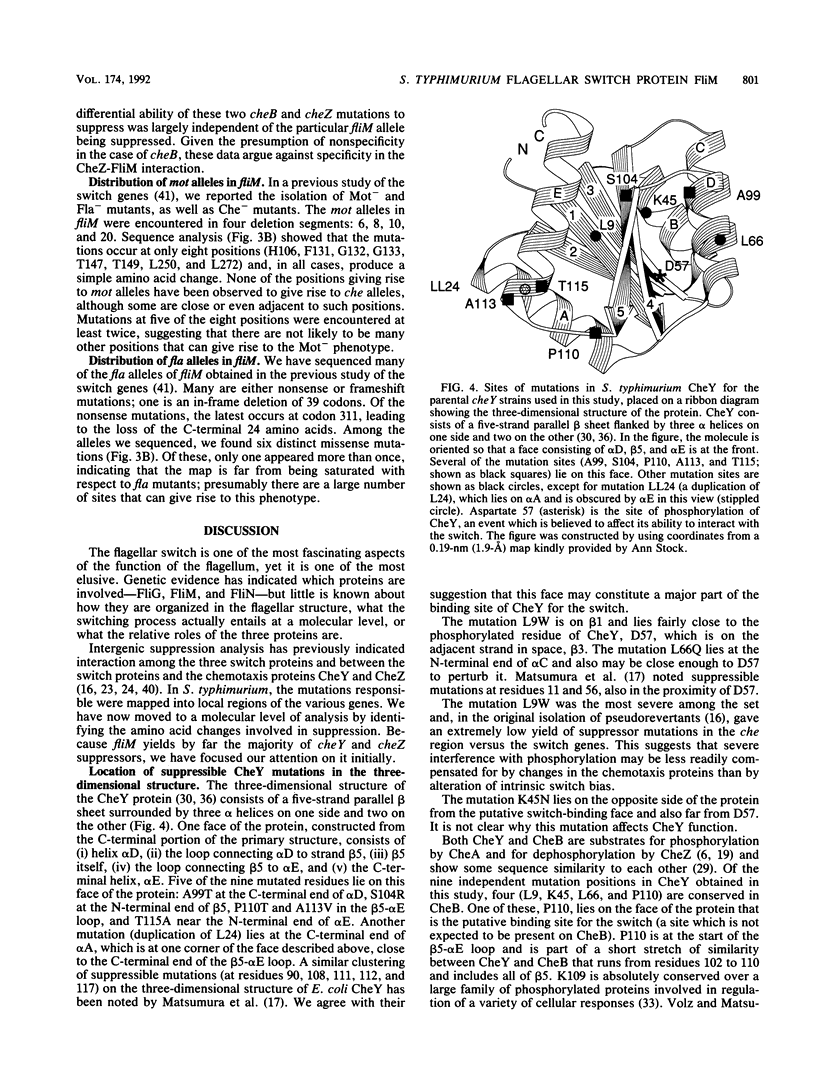
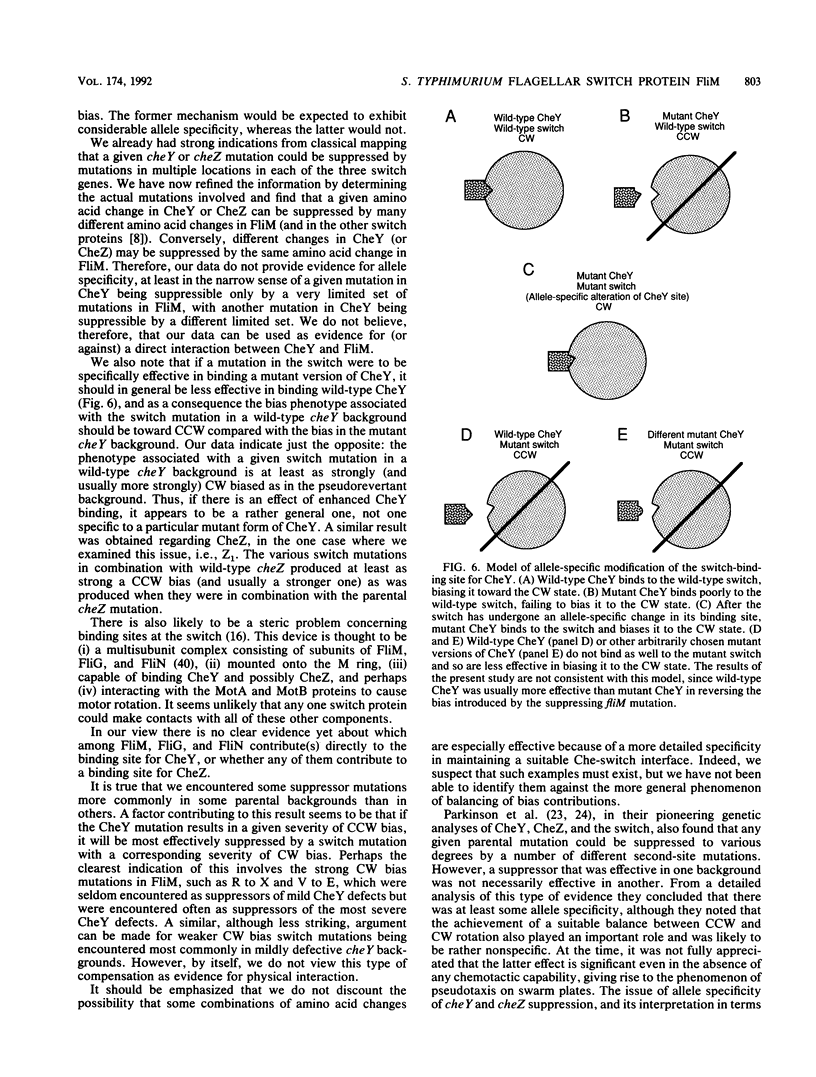
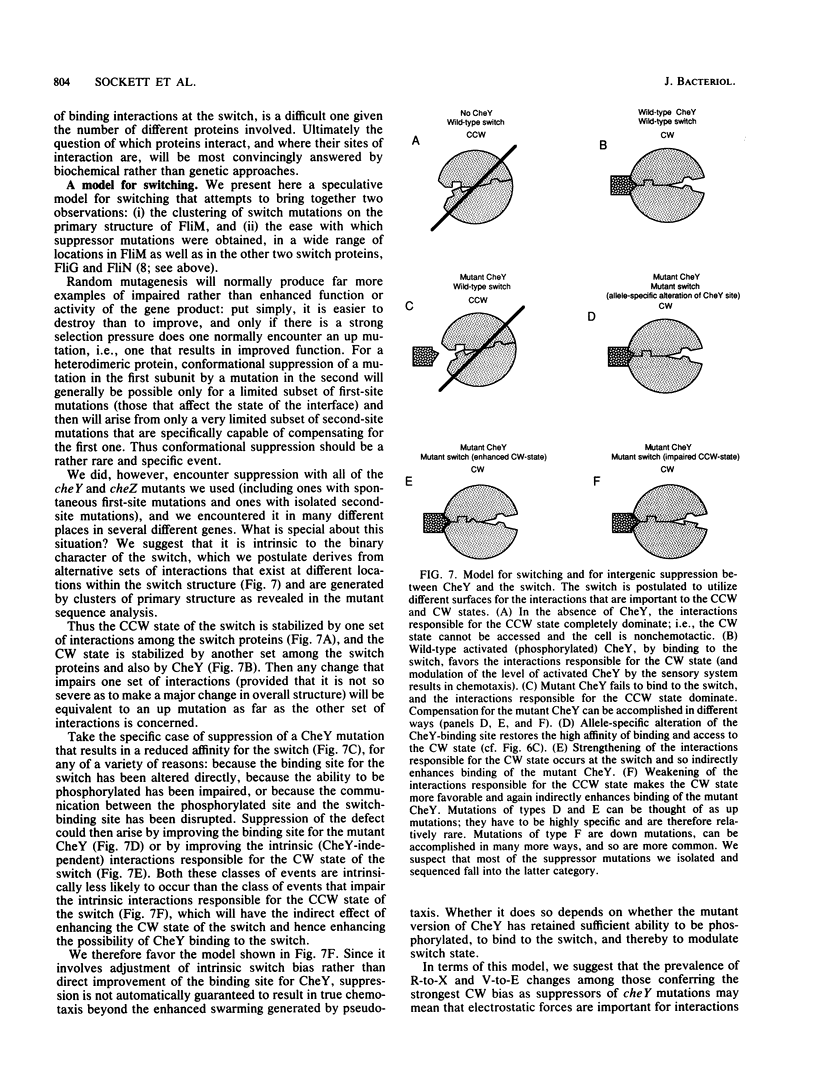
Abstract
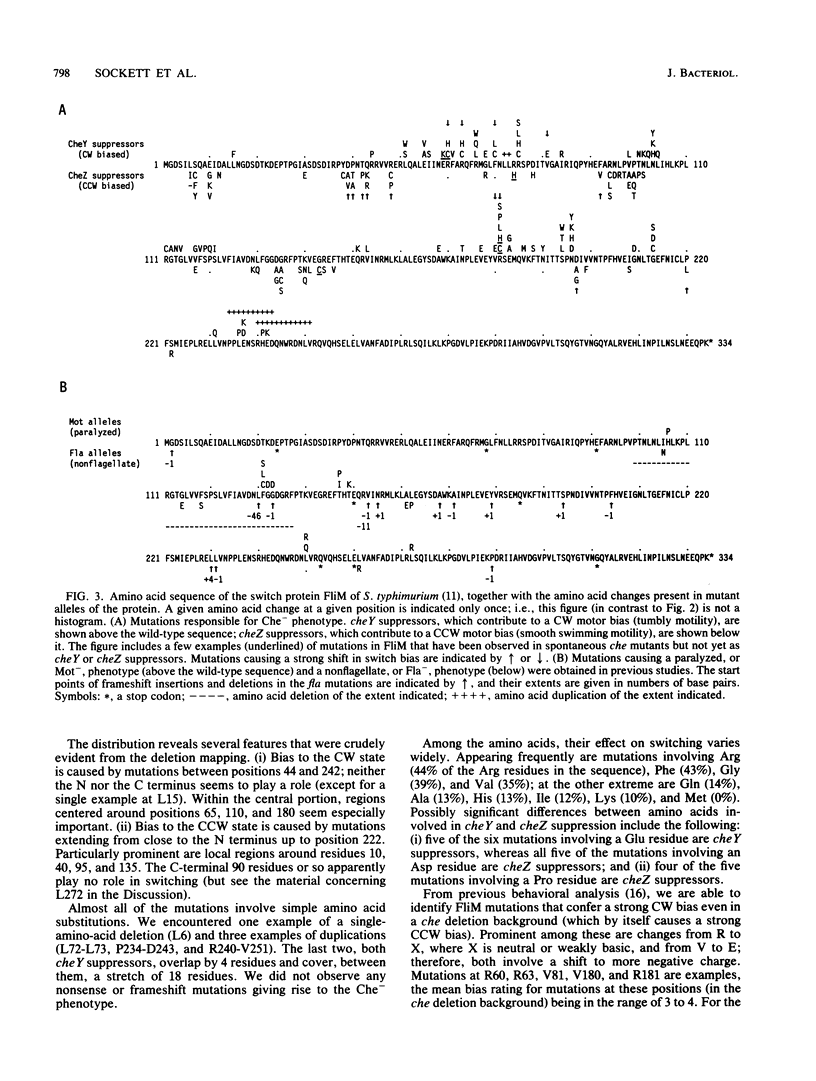
Defects in the chemotaxis proteins CheY and CheZ of Salmonella typhimurium can be suppressed by mutations in the flagellar switch, such that swarming of a pseudorevertant on semisolid plates is significantly better than that of its parent. cheY suppressors contribute to a clockwise switch bias, and cheZ suppressors contribute to a counterclockwise bias. Among the three known switch genes, fliM contributes most examples of such suppressor mutations. We have investigated the changes in FliM that are responsible for suppression, as well as the changes in CheY or CheZ that are being compensated for. Ten independently isolated parental cheY mutations represented nine distinct mutations, one an amino acid duplication and the rest missense mutations. Several of the altered amino acids lie on one face of the three-dimensional structure of CheY (A. M. Stock, J. M. Mottonen, J. B. Stock, and C. E. Schutt, Nature (London) 337:745-749, 1989; K. Volz and P. Matsumura, J. Biol. Chem. 266:15511-15519, 1991); this face may constitute the binding site for the switch. All 10 cheZ mutations were distinct, with several of them resulting in premature termination. cheY and cheZ suppressors in FliM occurred in clusters, which in general did not overlap. A few cheZ suppressors and one cheY suppressor involved changes near the N terminus of FliM, but neither cheY nor cheZ suppressors involved changes near the C terminus. Among the strongest cheY suppressors were changes from Arg to a neutral amino acid or from Val to Glu, suggesting that electrostatic interactions may play an important role in switching. A given cheY or cheZ mutation could be suppressed by many different fliM mutations; conversely, a given fliM mutation was often encountered as a suppressor of more than one cheY or cheZ mutation. The data suggest that an important factor in suppression is a balancing of the shift in switch bias introduced by alteration of CheY or CheZ with an appropriate opposing shift introduced by alteration of FliM. For strains with a severe parental mutation, such as the cheZ null mutations, adjustment of switch bias is essentially the only factor in suppression, since the attractant L-aspartate caused at most a slight further enhancement of the swarming rate over that occurring in the absence of a chemotactic stimulus. We discuss a model for switching in which there are distinct interactions for the counterclockwise and clockwise states, with suppression occurring by impairment of one of the states and hence by relative enhancement of the other state. FliM can also undergo amino acid changes that result in a paralyzed (Mot-) phenotype; these changes were confined to a very few residues in the protein.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. L., Stocker B. A. Salmonella typhimurium mutants generally defective in chemotaxis. J Bacteriol. 1976 Dec;128(3):754–765. doi: 10.1128/jb.128.3.754-765.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G. E., Aizawa S. I., Macnab R. M. flaAII (motC, cheV) of Salmonella typhimurium is a structural gene involved in energization and switching of the flagellar motor. J Bacteriol. 1983 Apr;154(1):84–91. doi: 10.1128/jb.154.1.84-91.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto M. Genetic studies of paralyzed mutant in Salmonella. I. Genetic fine structure of the mot loci in Salmonella typhimurium. Genetics. 1966 Sep;54(3):715–726. doi: 10.1093/genetics/54.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Failure of sensory adaptation in bacterial mutants that are defective in a protein methylation reaction. Cell. 1978 Dec;15(4):1231–1240. doi: 10.1016/0092-8674(78)90049-1. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Kaplan N., Simon M. I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Homma M., Iino T., Macnab R. M. Identification and characterization of the products of six region III flagellar genes (flaAII.3 through flaQII) of Salmonella typhimurium. J Bacteriol. 1988 May;170(5):2221–2228. doi: 10.1128/jb.170.5.2221-2228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Neumann R., Keyte J. Amplification of human minisatellites by the polymerase chain reaction: towards DNA fingerprinting of single cells. Nucleic Acids Res. 1988 Dec 9;16(23):10953–10971. doi: 10.1093/nar/16.23.10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Macnab R. M., DeFranco A. L., Koshland D. E., Jr Inversion of a behavioral response in bacterial chemotaxis: explanation at the molecular level. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4150–4154. doi: 10.1073/pnas.75.9.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
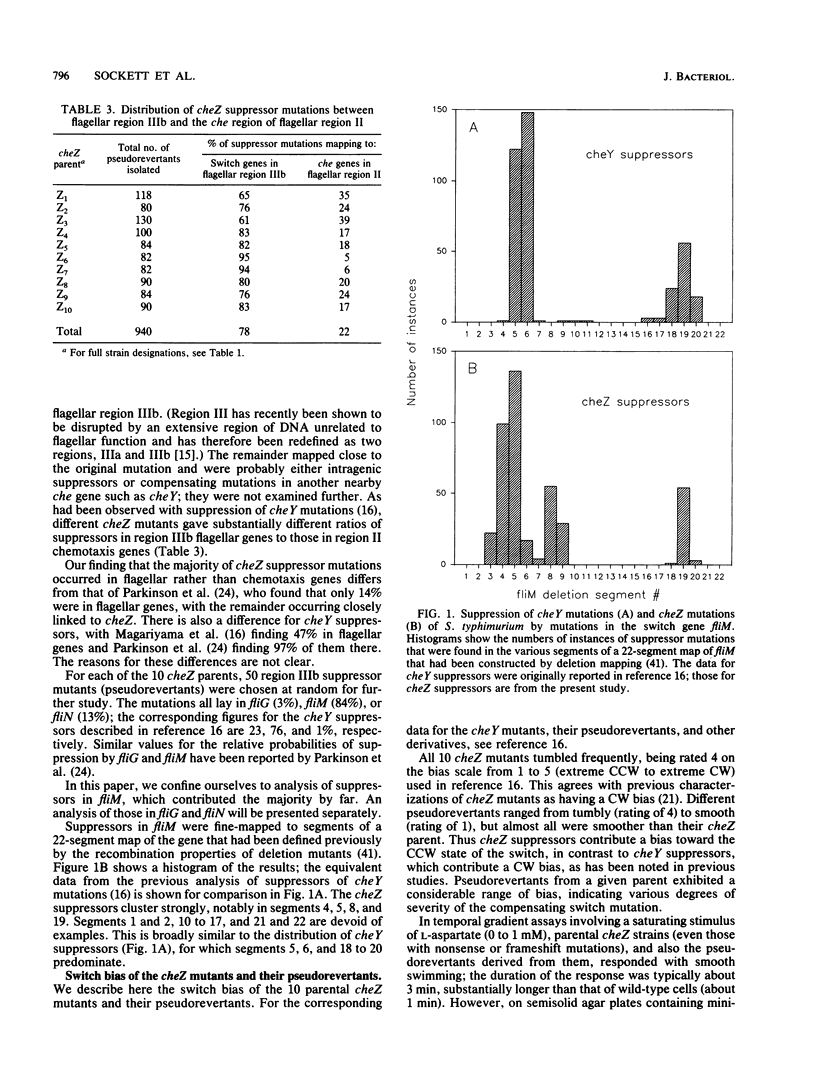
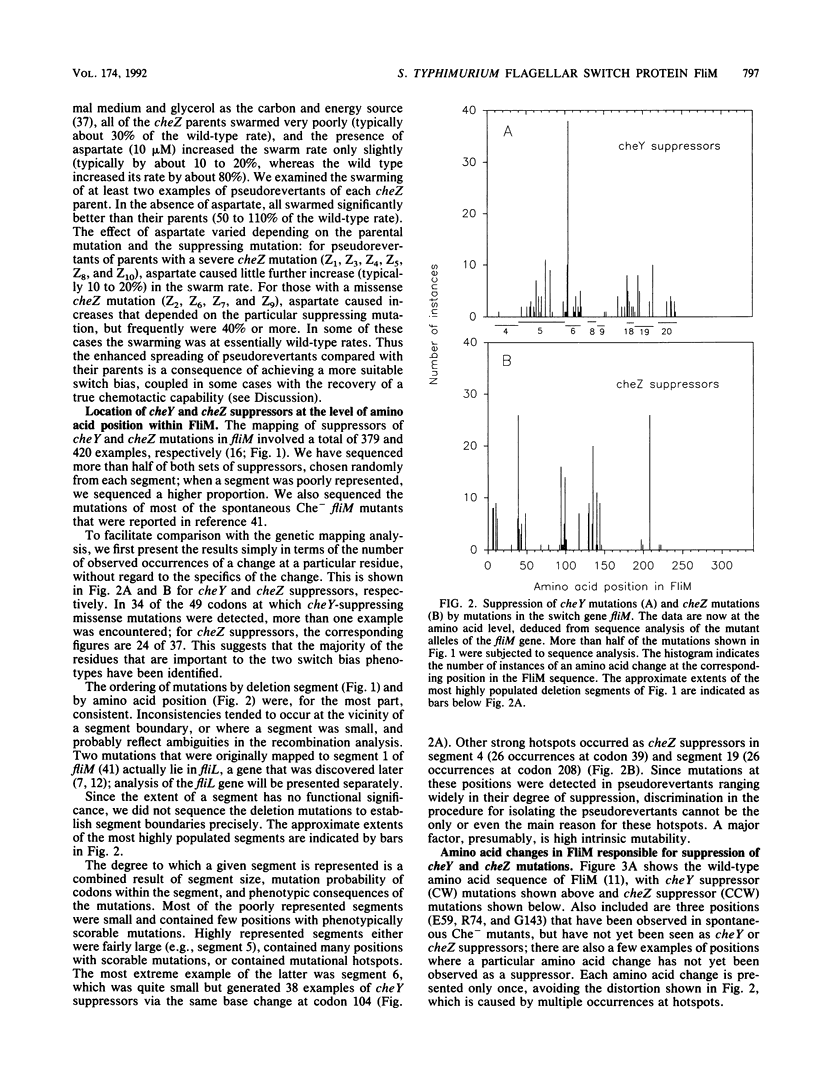
- Kihara M., Homma M., Kutsukake K., Macnab R. M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jun;171(6):3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J Bacteriol. 1987 Mar;169(3):1307–1314. doi: 10.1128/jb.169.3.1307-1314.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986 Jun;166(3):1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S. H., Reader R. W., Kort E. N., Tso W. W., Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974 May 3;249(452):74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- Magariyama Y., Yamaguchi S., Aizawa S. Genetic and behavioral analysis of flagellar switch mutants of Salmonella typhimurium. J Bacteriol. 1990 Aug;172(8):4359–4369. doi: 10.1128/jb.172.8.4359-4369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierendorf R. C., Pfeffer D. Direct sequencing of denatured plasmid DNA. Methods Enzymol. 1987;152:556–562. doi: 10.1016/0076-6879(87)52061-4. [DOI] [PubMed] [Google Scholar]
- Ninfa E. G., Stock A., Mowbray S., Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991 May 25;266(15):9764–9770. [PubMed] [Google Scholar]
- Parkinson J. S. Behavioral genetics in bacteria. Annu Rev Genet. 1977;11:397–414. doi: 10.1146/annurev.ge.11.120177.002145. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R. Interaction of the cheC and cheZ gene products is required for chemotactic behavior in Escherichia coli. Proc Natl Acad Sci U S A. 1979 May;76(5):2390–2394. doi: 10.1073/pnas.76.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R., Talbert P. B., Houts S. E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983 Jul;155(1):265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J Bacteriol. 1976 May;126(2):758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid S., Matsumura P., Eisenbach M. Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7157–7161. doi: 10.1073/pnas.83.19.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989 Dec 25;264(36):21770–21778. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A. M., Mottonen J. M., Stock J. B., Schutt C. E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989 Feb 23;337(6209):745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- Stock A. M., Stock J. B. Purification and characterization of the CheZ protein of bacterial chemotaxis. J Bacteriol. 1987 Jul;169(7):3301–3311. doi: 10.1128/jb.169.7.3301-3311.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A., Koshland D. E., Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Stock A. M., Mottonen J. M. Signal transduction in bacteria. Nature. 1990 Mar 29;344(6265):395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- Tindall K. R., Stankowski L. F., Jr Molecular analysis of spontaneous mutations at the gpt locus in Chinese hamster ovary (AS52) cells. Mutat Res. 1989 Mar-May;220(2-3):241–253. doi: 10.1016/0165-1110(89)90028-6. [DOI] [PubMed] [Google Scholar]
- Volz K., Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991 Aug 15;266(23):15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- Wolfe A. J., Berg H. C. Migration of bacteria in semisolid agar. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie D., Stock A., Wong C. Y., Stock J. Sensory transduction in bacterial chemotaxis involves phosphotransfer between Che proteins. Biochem Biophys Res Commun. 1988 Mar 15;151(2):891–896. doi: 10.1016/s0006-291x(88)80365-6. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Aizawa S., Kihara M., Isomura M., Jones C. J., Macnab R. M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986 Dec;168(3):1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Ishihara A., Aizawa S., Macnab R. M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986 Apr;166(1):187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Sugata K., Taira T., Iino T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol. 1984 Feb;130(2):255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]