Abstract
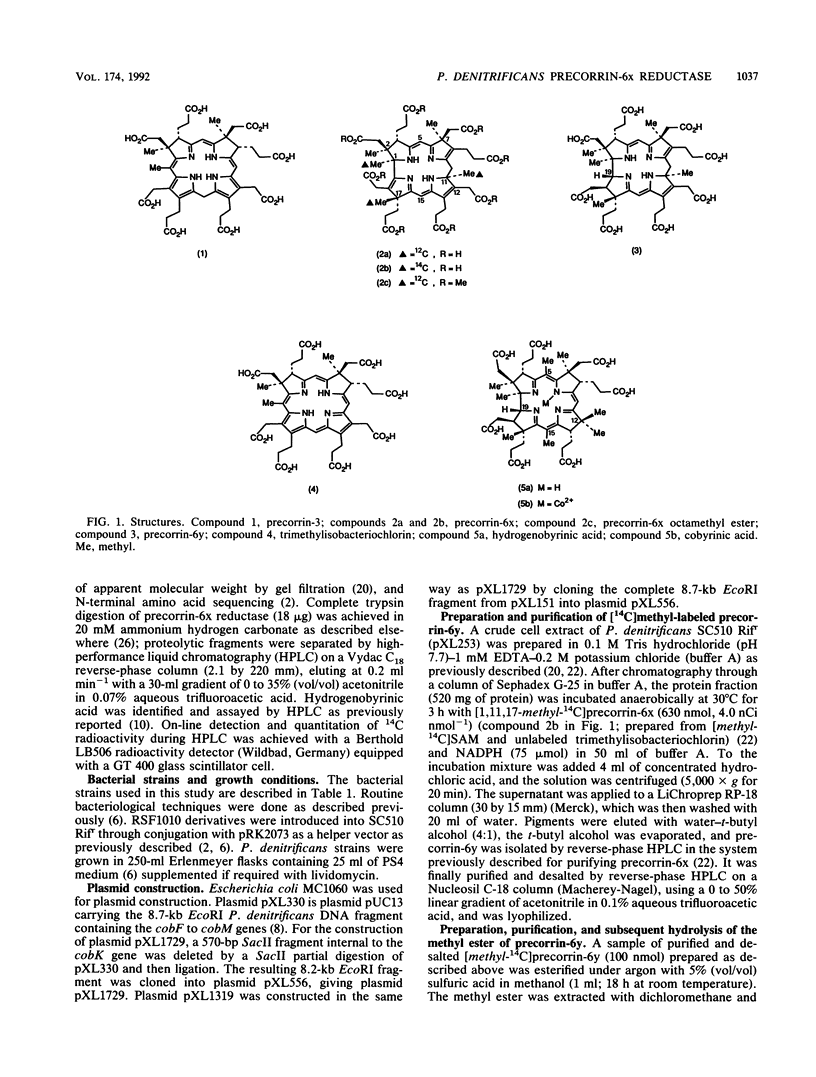
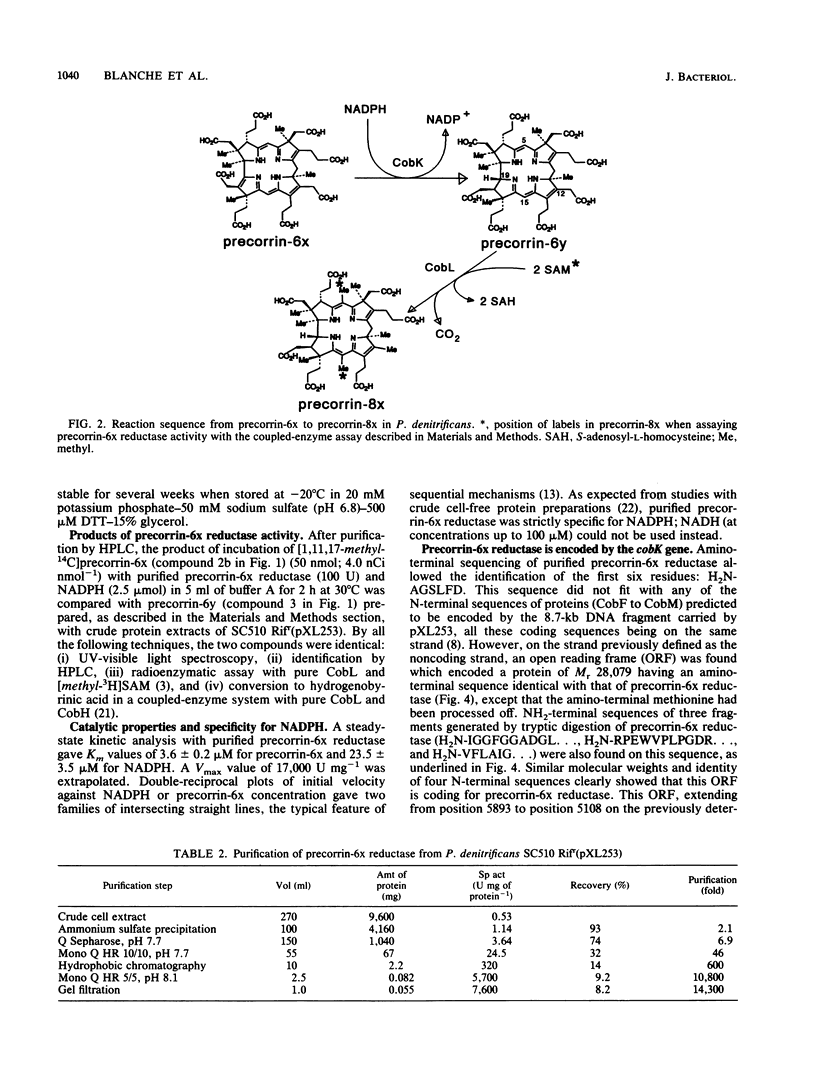
Precorrin-6x reductase, which catalyzes the NADPH-dependent reduction of precorrin-6x to a dihydro derivative named precorrin-6y, was purified 14,300-fold to homogeneity with an 8% yield from extracts of a recombinant strain of Pseudomonas denitrificans. Precorrin-6y was identified by fast atom bombardment-mass spectrometry. It was converted in high yield (90%) to hydrogenobyrinic acid by cell-free protein preparations from P. denitrificans. For the purification and characterization of precorrin-6x reductase, a coupled-enzyme radioenzymatic assay was developed in which precorrin-6y was methylated in situ by the cobL gene product (F. Blanche, A. Famechon, D. Thibaut, L. Debussche, B. Cameron, J. Crouzet, J. Bacteriol. 174:1050-1052, 1992) in the presence of [methyl-3H]S-adenosyl-L-methionine. Molecular weights of precorrin-6x reductase obtained by gel filtration (Mr congruent to 27,000) and by analytical sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Mr congruent to 31,000) were consistent with the enzyme being a monomer. Km values of 3.6 +/- 0.2 microM for precorrin-6x and 23.5 +/- 3.5 microM for NADPH and a Vmax value of 17,000 U mg-1 were obtained at pH 7.7. The N-terminal sequence (six amino acids) and three internal sequences obtained after tryptic digestion of the enzyme were determined by microsequencing and established that precorrin-6x reductase is encoded by the cobK gene, located on a previously described 8.7-kb EcoRI fragment (J. Crouzet, B. Cameron, L. Cauchois, S. Rigault, M.-C. Rouyez, F. Blanche, D. Thibaut, and L. Debussche, J. Bacteriol. 172:5980-5990, 1990). However, the coding sequence was shown to be on the strand complementary to the one previously proposed as the coding strand.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanche F., Debussche L., Thibaut D., Crouzet J., Cameron B. Purification and characterization of S-adenosyl-L-methionine: uroporphyrinogen III methyltransferase from Pseudomonas denitrificans. J Bacteriol. 1989 Aug;171(8):4222–4231. doi: 10.1128/jb.171.8.4222-4231.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F., Famechon A., Thibaut D., Debussche L., Cameron B., Crouzet J. Biosynthesis of vitamin B12 in Pseudomonas denitrificans: the biosynthetic sequence from precorrin-6y to precorrin-8x is catalyzed by the cobL gene product. J Bacteriol. 1992 Feb;174(3):1050–1052. doi: 10.1128/jb.174.3.1050-1052.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F., Thibaut D., Couder M., Muller J. C. Identification and quantitation of corrinoid precursors of cobalamin from Pseudomonas denitrificans by high-performance liquid chromatography. Anal Biochem. 1990 Aug 15;189(1):24–29. doi: 10.1016/0003-2697(90)90038-b. [DOI] [PubMed] [Google Scholar]
- Cameron B., Briggs K., Pridmore S., Brefort G., Crouzet J. Cloning and analysis of genes involved in coenzyme B12 biosynthesis in Pseudomonas denitrificans. J Bacteriol. 1989 Jan;171(1):547–557. doi: 10.1128/jb.171.1.547-557.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Crouzet J., Cameron B., Cauchois L., Rigault S., Rouyez M. C., Blanche F., Thibaut D., Debussche L. Genetic and sequence analysis of an 8.7-kilobase Pseudomonas denitrificans fragment carrying eight genes involved in transformation of precorrin-2 to cobyrinic acid. J Bacteriol. 1990 Oct;172(10):5980–5990. doi: 10.1128/jb.172.10.5980-5990.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J., Cauchois L., Blanche F., Debussche L., Thibaut D., Rouyez M. C., Rigault S., Mayaux J. F., Cameron B. Nucleotide sequence of a Pseudomonas denitrificans 5.4-kilobase DNA fragment containing five cob genes and identification of structural genes encoding S-adenosyl-L-methionine: uroporphyrinogen III methyltransferase and cobyrinic acid a,c-diamide synthase. J Bacteriol. 1990 Oct;172(10):5968–5979. doi: 10.1128/jb.172.10.5968-5979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debussche L., Thibaut D., Cameron B., Crouzet J., Blanche F. Purification and characterization of cobyrinic acid a,c-diamide synthase from Pseudomonas denitrificans. J Bacteriol. 1990 Nov;172(11):6239–6244. doi: 10.1128/jb.172.11.6239-6244.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann H. C., Cagen L. M. Microbial biosynthesis of B12-like compounds. Annu Rev Microbiol. 1970;24:159–208. doi: 10.1146/annurev.mi.24.100170.001111. [DOI] [PubMed] [Google Scholar]
- Fromm H. J. Summary of kinetic reaction mechanisms. Methods Enzymol. 1979;63:42–53. doi: 10.1016/0076-6879(79)63005-7. [DOI] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Leong S. A., Ditta G. S., Helinski D. R. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem. 1982 Aug 10;257(15):8724–8730. [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Thibaut D., Blanche F., Debussche L., Leeper F. J., Battersby A. R. Biosynthesis of vitamin B12: structure of precorrin-6x octamethyl ester. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8800–8804. doi: 10.1073/pnas.87.22.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut D., Couder M., Crouzet J., Debussche L., Cameron B., Blanche F. Assay and purification of S-adenosyl-L-methionine:precorrin-2 methyltransferase from Pseudomonas denitrificans. J Bacteriol. 1990 Nov;172(11):6245–6251. doi: 10.1128/jb.172.11.6245-6251.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut D., Couder M., Famechon A., Debussche L., Cameron B., Crouzet J., Blanche F. The final step in the biosynthesis of hydrogenobyrinic acid is catalyzed by the cobH gene product with precorrin-8x as the substrate. J Bacteriol. 1992 Feb;174(3):1043–1049. doi: 10.1128/jb.174.3.1043-1049.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut D., Debussche L., Blanche F. Biosynthesis of vitamin B12: isolation of precorrin-6x, a metal-free precursor of the corrin macrocycle retaining five S-adenosylmethionine-derived peripheral methyl groups. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8795–8799. doi: 10.1073/pnas.87.22.8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



