Abstract
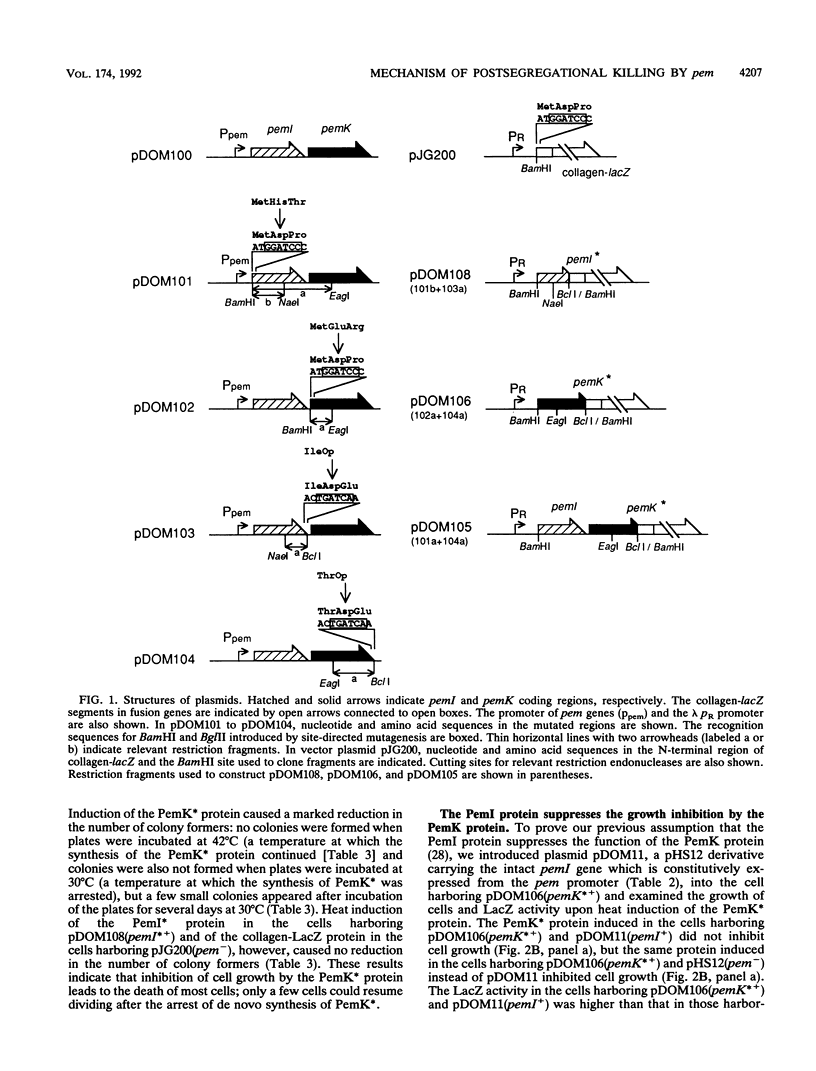
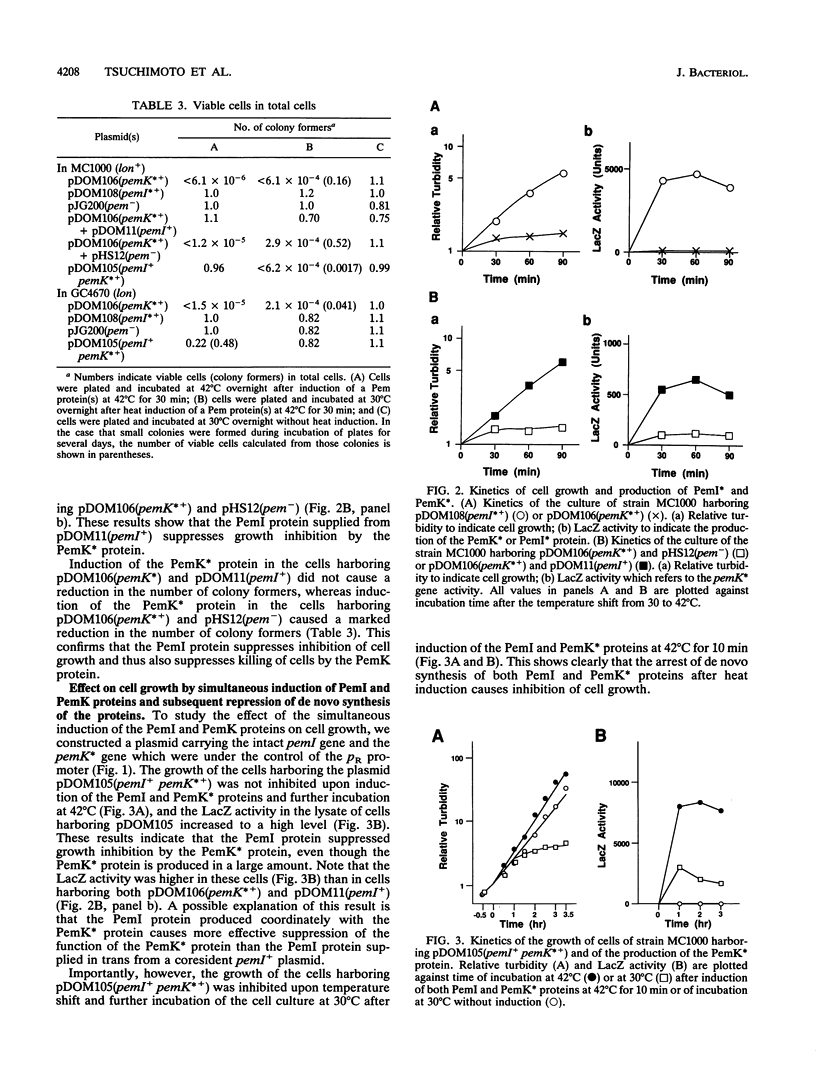
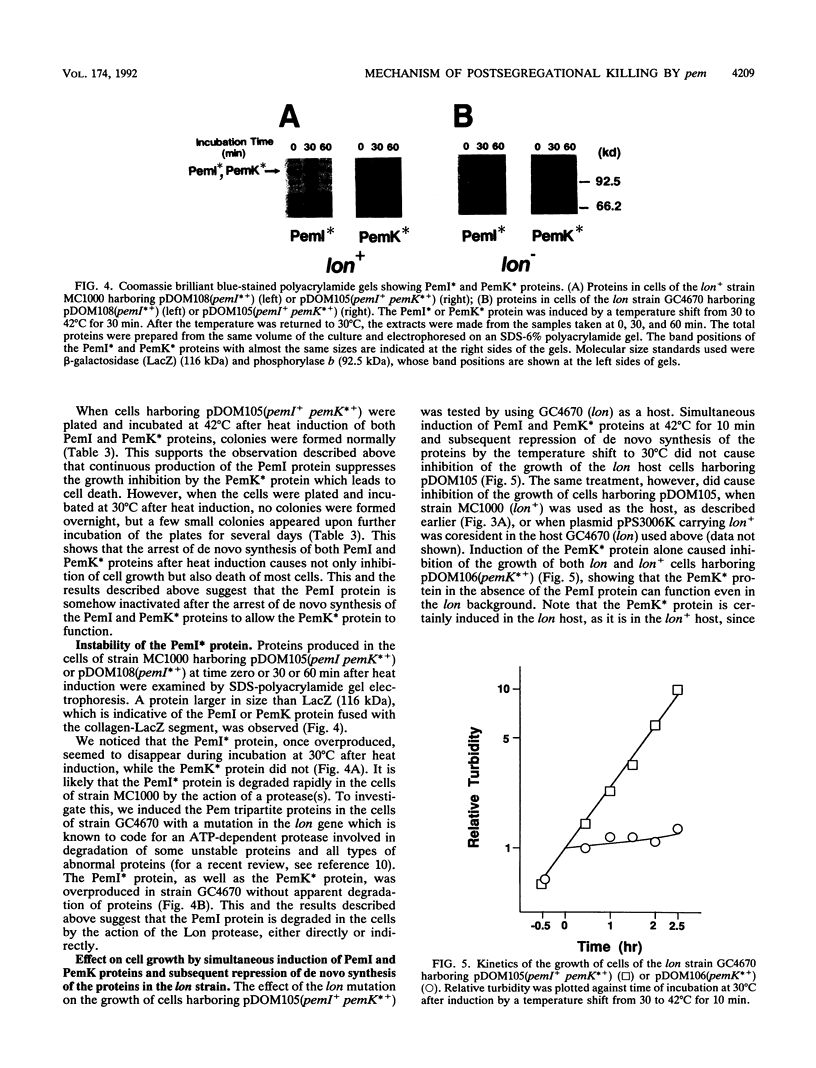
We constructed plasmids carrying heat-inducible pemI and pemK genes, which were fused with the collagen-lacZ sequence in frame. The PemK-collagen-LacZ (PemK*) protein produced from the fusion gene upon heat induction inhibited the growth of cells and killed most of the cells in the absence of the PemI protein but did not do so in the presence of the PemI protein. This supports our previous assumption that the PemK protein inhibits cell division, leading to cell death, whereas the PemI protein suppresses the function of the PemK protein. We also constructed the plasmid carrying the heat-inducible pem operon which consists of the intact pemI gene and the pemK gene fused with collagen-lacZ. The simultaneously induced PemI and PemK* proteins did not inhibit the growth of cells. However, the temperature shift to 30 degrees C after induction of both proteins at 42 degrees C caused inhibition of cell growth and death of most cells. This suggests that the PemI protein is somehow inactivated upon the arrest of de novo synthesis of the PemI and PemK* proteins, allowing the PemK* protein to function. We observed that the PemI-collagen-LacZ (PemI*) protein was degraded faster than the PemK* protein, perhaps by the action of a protease(s). In fact, the lon mutation, which caused no apparent degradation of the PemI* protein, did not allow the PemK* protein to function, supporting the suggestion described above. Instability of the PemI protein would explain why the cells which have lost the pem+ plasmid are preferentially killed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Ohtsubo H., Bauer W. R., Yoshioka Y., Miyazaki C., Maeda Y., Ohtsubo E. Characterization of the gene products produced in minicells by pSM1, a derivative of R100. Mol Gen Genet. 1986 Oct;205(1):56–65. doi: 10.1007/BF02428032. [DOI] [PubMed] [Google Scholar]
- Bravo A., Ortega S., de Torrontegui G., Díaz R. Killing of Escherichia coli cells modulated by components of the stability system ParD of plasmid R1. Mol Gen Genet. 1988 Dec;215(1):146–151. doi: 10.1007/BF00331316. [DOI] [PubMed] [Google Scholar]
- Bravo A., de Torrontegui G., Díaz R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet. 1987 Nov;210(1):101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Helin K., Christensen O. W., Løbner-Olesen A. Translational control and differential RNA decay are key elements regulating postsegregational expression of the killer protein encoded by the parB locus of plasmid R1. J Mol Biol. 1988 Sep 5;203(1):119–129. doi: 10.1016/0022-2836(88)90096-4. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino J., Bastia D. Rapid purification of a cloned gene product by genetic fusion and site-specific proteolysis. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4692–4696. doi: 10.1073/pnas.81.15.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Genetics of proteolysis in Escherichia coli*. Annu Rev Genet. 1989;23:163–198. doi: 10.1146/annurev.ge.23.120189.001115. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Jaffé A., Ogura T., Mori H., Takahashi H. F plasmid ccd mechanism in Escherichia coli. J Bacteriol. 1986 Apr;166(1):100–104. doi: 10.1128/jb.166.1.100-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé A., Ogura T., Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985 Sep;163(3):841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miki T., Chang Z. T., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. II. Identification of genes for inhibitor protein and trigger protein on the 42.84-43.6 F segment. J Mol Biol. 1984 Apr 25;174(4):627–646. doi: 10.1016/0022-2836(84)90087-1. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Yoshioka K., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. I. The 42.84-43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. J Mol Biol. 1984 Apr 25;174(4):605–625. doi: 10.1016/0022-2836(84)90086-x. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ryder T. B., Maeda Y., Armstrong K., Ohtsubo E. DNA replication of the resistance plasmid R100 and its control. Adv Biophys. 1986;21:115–133. doi: 10.1016/0065-227x(86)90018-3. [DOI] [PubMed] [Google Scholar]
- Rasmussen P. B., Gerdes K., Molin S. Genetic analysis of the parB+ locus of plasmid R1. Mol Gen Genet. 1987 Aug;209(1):122–128. doi: 10.1007/BF00329846. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto S., Ohtsubo E. Effect of the pem system on stable maintenance of plasmid R100 in various Escherichia coli hosts. Mol Gen Genet. 1989 Feb;215(3):463–468. doi: 10.1007/BF00427044. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto S., Ohtsubo H., Ohtsubo E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988 Apr;170(4):1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]