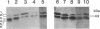Abstract
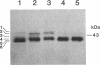
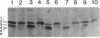
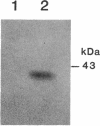
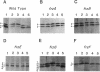
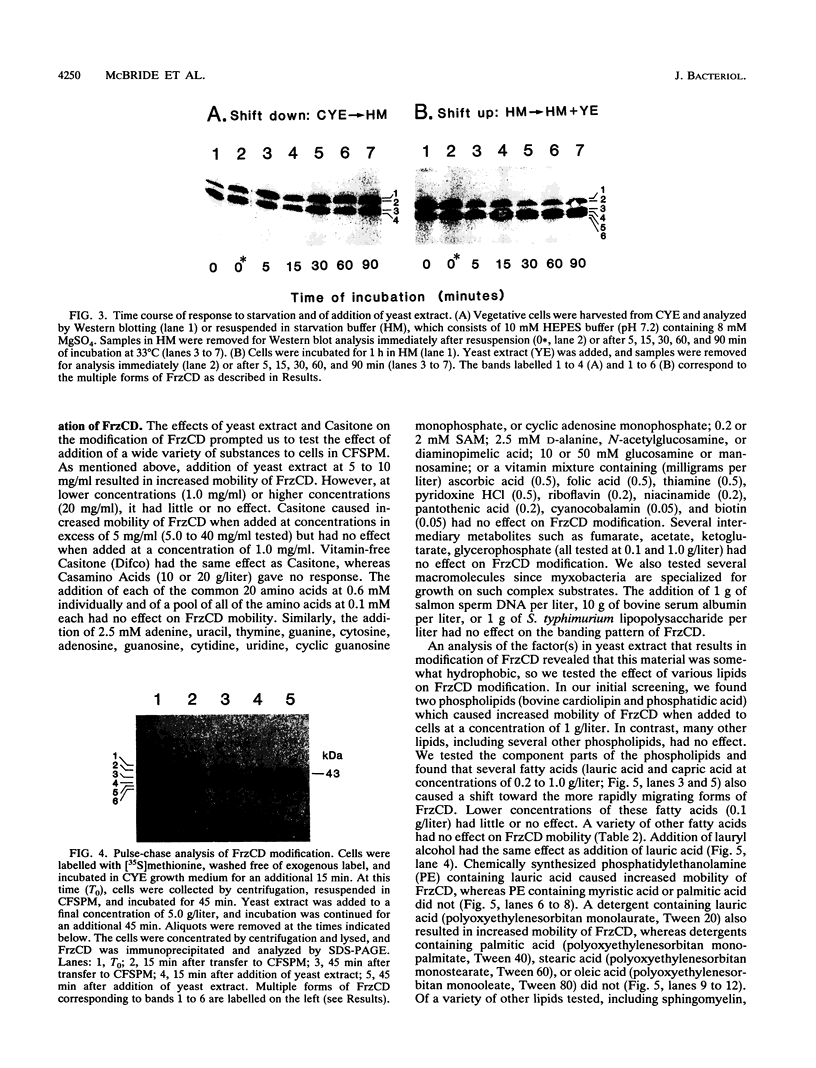
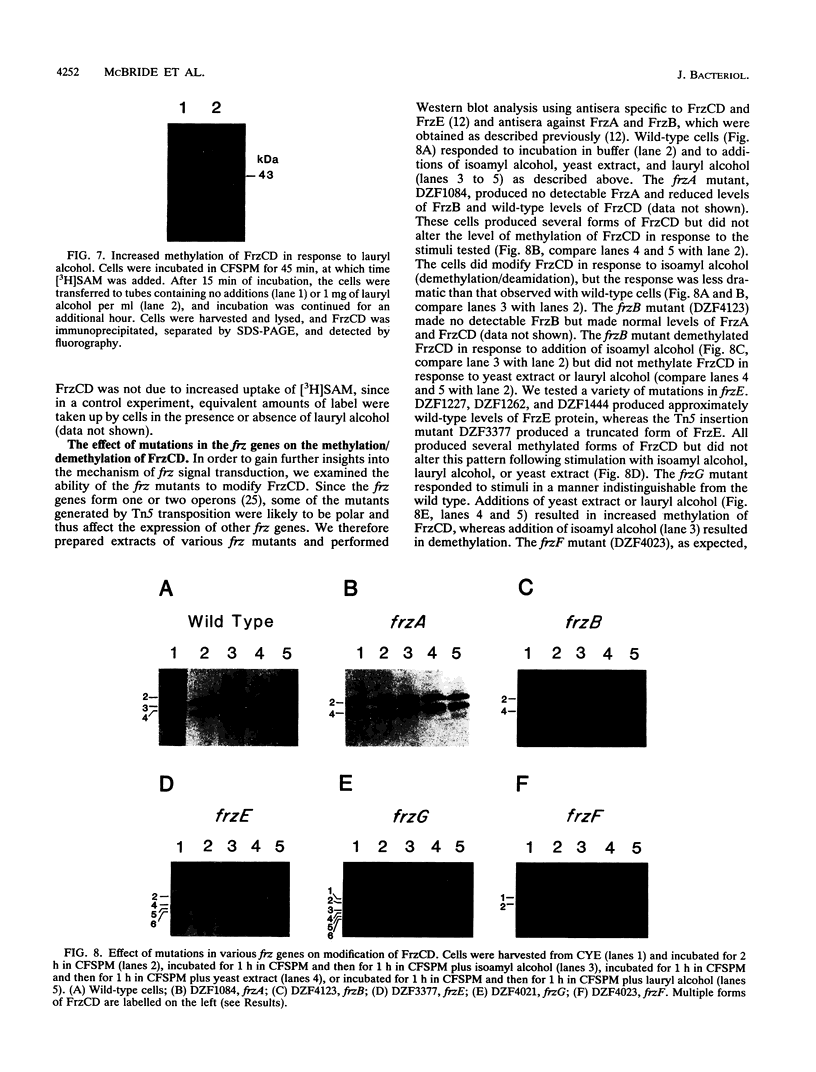
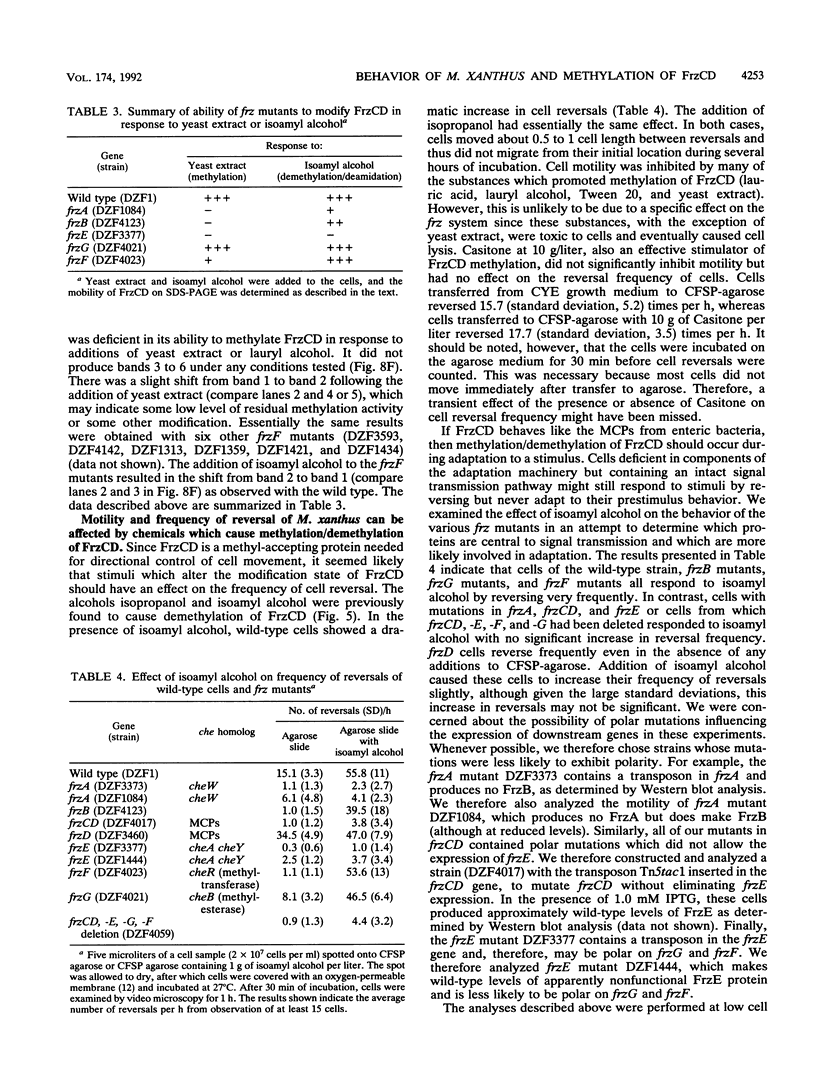
Myxococcus xanthus, a nonflagellated gliding bacterium, exhibits multicellular behavior during vegetative growth and fruiting body formation. The frizzy (frz) genes are required to control directed motility for these interactions. The frz genes encode proteins that are homologous to all of the major enteric chemotaxis proteins, with the exception of CheZ. In this study, we characterized FrzCD, a protein which is homologous to the methyl-accepting chemotaxis proteins from the enteric bacteria. FrzCD, unlike the other methyl-accepting chemotaxis proteins, was found to be localized primarily in the cytoplasmic fraction of cells. FrzCD migrates as a ladder of bands on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, reflecting heterogeneity due to methylation or demethylation and to deamidation. FrzCD was shown to be methylated in vivo when cells were exposed to yeast extract or Casitone and demethylated when starved in buffer. We used the methylation state of FrzCD as revealed by Western blot (immunoblot) analyses to search for stimuli that are recognized by the frz signal transduction system. Common amino acids, nucleotides, vitamins, and sugars were not recognized, but certain lipids and alcohols were recognized. For example, the saturated fatty acids capric acid and lauric acid stimulated FrzCD methylation, whereas a variety of other saturated fatty acids did not. Lauryl alcohol and lipoic acid also stimulated methylation, as did phospholipids containing lauric acid. In contrast, several short-chain alcohols, such as isoamyl alcohol, and some other solvents caused demethylation. The relatively high concentrations of the chemicals required for a response may indicate that these chemicals are not the relevant signals recognized by M. xanthus in nature. Isoamyl alcohol and isopropanol also had profound effects on the behavior of wild-type cells, causing them to reverse continuously. Cells of frzB, frzF, and frzG mutants also reversed continuously in the presence of isoamyl alcohol, whereas cells of frzA, frzCD, or frzE mutants did not. On the basis of the data presented, we propose a model for the frz signal transduction pathway in M. xanthus.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bischoff D. S., Ordal G. W. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol Microbiol. 1992 Jan;6(1):23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. "Frizzy" genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. Cloning and complementation analysis of the "Frizzy" genes of Myxococcus xanthus. Mol Gen Genet. 1985;198(2):243–254. doi: 10.1007/BF00383002. [DOI] [PubMed] [Google Scholar]
- Burchard R. P. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- Chow W. Y., Berg D. E. Tn5tac1, a derivative of transposon Tn5 that generates conditional mutations. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6468–6472. doi: 10.1073/pnas.85.17.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M., Eide D. Myxococcus xanthus does not respond chemotactically to moderate concentration gradients. J Bacteriol. 1983 Apr;154(1):437–442. doi: 10.1128/jb.154.1.437-442.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Control of multicellular development: Dictyostelium and Myxococcus. Annu Rev Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- McBride M. J., Weinberg R. A., Zusman D. R. "Frizzy" aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci U S A. 1989 Jan;86(2):424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., McBride M. J., Zusman D. R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990 Sep;172(9):4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., Zusman D. R. FrzE of Myxococcus xanthus is homologous to both CheA and CheY of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5898–5902. doi: 10.1073/pnas.87.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C. E., Zusman D. R. Myxococcus xanthus mutants with temperature-sensitive, stage-specific defects: evidence for independent pathways in development. J Bacteriol. 1979 Dec;140(3):1036–1042. doi: 10.1128/jb.140.3.1036-1042.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlin D. M., Bollinger J., Hazelbauer G. L. Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins. 1988;3(2):102–112. doi: 10.1002/prot.340030205. [DOI] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Behavior of peripheral rods and their role in the life cycle of Myxococcus xanthus. J Bacteriol. 1991 Jun;173(11):3342–3355. doi: 10.1128/jb.173.11.3342-3355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983 Jul;155(1):317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J Bacteriol. 1991 Jun;173(11):3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Genetic analysis of Myxococcus xanthus and isolation of gene replacements after transduction under conditions of limited homology. J Bacteriol. 1986 Aug;167(2):744–748. doi: 10.1128/jb.167.2.744-748.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Protein phosphorylation in bacterial chemotaxis. Cell. 1988 Apr 8;53(1):1–2. doi: 10.1016/0092-8674(88)90478-3. [DOI] [PubMed] [Google Scholar]
- Rampersaud A., Inouye M. Procaine, a local anesthetic, signals through the EnvZ receptor to change the DNA binding affinity of the transcriptional activator protein OmpR. J Bacteriol. 1991 Nov;173(21):6882–6888. doi: 10.1128/jb.173.21.6882-6888.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990 Dec;54(4):473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman D. R. "Frizzy" mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982 Jun;150(3):1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman D. R., McBride M. J. Sensory transduction in the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1991 Oct;5(10):2323–2329. doi: 10.1111/j.1365-2958.1991.tb02077.x. [DOI] [PubMed] [Google Scholar]