Abstract
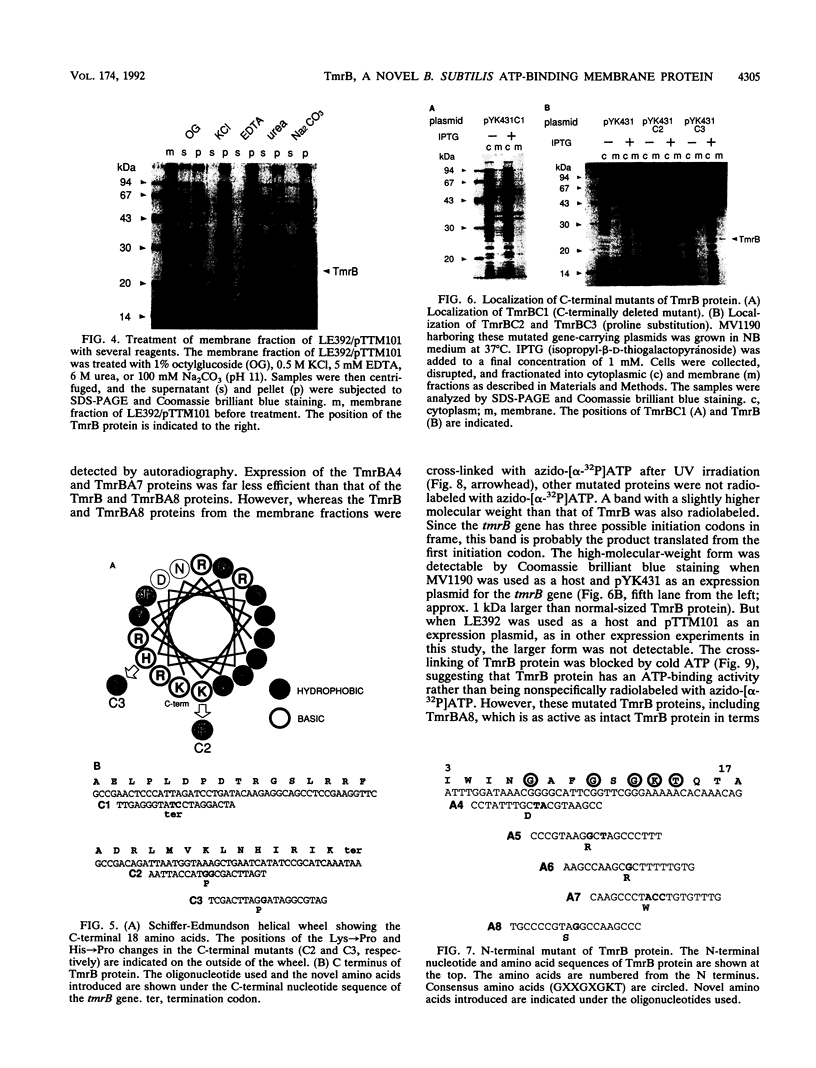
tmrB is the gene responsible for tunicamycin resistance in Bacillus subtilis. It is predicted that an increase in tmrB gene expression makes B. subtilis tunicamycin resistant. To examine the tmrB gene product, we produced the tmrB gene product in Escherichia coli by using the tac promoter. TmrB protein was found not only in the cytoplasm fraction but also in the membrane fraction. Although TmrB protein is entirely hydrophilic and has no hydrophobic stretch of amino acids sufficient to span the membrane, its C-terminal 18 amino acids could form an amphiphilic alpha-helix. Breaking this potential alpha-helix by introducing proline residues or a stop codon into this region caused the release of this membrane-bound protein into the cytoplasmic fraction, indicating that the C-terminal 18 residues were essential for membrane binding. On the other hand, TmrB protein has an ATP-binding consensus sequence in the N-terminal region. We have tested whether this sequence actually has the ability to bind ATP by photoaffinity cross-linking with azido-[alpha-32P]ATP. Wild-type protein bound azido-ATP well, but mutants with substitutions in the consensus amino acids were unable to bind azido-ATP. These C-terminal or N-terminal mutant genes were unable to confer tunicamycin resistance on B. subtilis in a multicopy state. It is concluded that TmrB protein is a novel ATP-binding protein which is anchored to the membrane with its C-terminal amphiphilic alpha-helix.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benveniste R., Davies J. Enzymatic acetylation of aminoglycoside antibiotics by Escherichia coli carrying an R factor. Biochemistry. 1971 May 11;10(10):1787–1796. doi: 10.1021/bi00786a009. [DOI] [PubMed] [Google Scholar]
- Bracha R., Glaser L. An intermediate in telchoic acid biosynthesis. Biochem Biophys Res Commun. 1976 Oct 4;72(3):1091–1098. doi: 10.1016/s0006-291x(76)80244-6. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Hancock I. C., Wiseman G., Baddiley J. Biosynthesis of the unit that links teichoic acid to the bacterial wall: inhibition by tunicamycin. FEBS Lett. 1976 Oct 15;69(1):75–80. doi: 10.1016/0014-5793(76)80657-6. [DOI] [PubMed] [Google Scholar]
- Hashiguchi K., Tanimoto A., Nomura S., Yamane K., Yoda K., Harada S., Mori M., Furusato T., Takatsuki A., Yamasaki M. Amplification of the amyE-tmrB region on the chromosome in tunicamycin-resistant cells of Bacillus subtilis. Mol Gen Genet. 1986 Jul;204(1):36–43. doi: 10.1007/BF00330184. [DOI] [PubMed] [Google Scholar]
- Hyde S. C., Emsley P., Hartshorn M. J., Mimmack M. M., Gileadi U., Pearce S. R., Gallagher M. P., Gill D. R., Hubbard R. E., Higgins C. F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990 Jul 26;346(6282):362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- Jackson M. E., Pratt J. M. An 18 amino acid amphiphilic helix forms the membrane-anchoring domain of the Escherichia coli penicillin-binding protein 5. Mol Microbiol. 1987 Jul;1(1):23–28. doi: 10.1111/j.1365-2958.1987.tb00522.x. [DOI] [PubMed] [Google Scholar]
- Kalmar G. B., Kay R. J., Lachance A., Aebersold R., Cornell R. B. Cloning and expression of rat liver CTP: phosphocholine cytidylyltransferase: an amphipathic protein that controls phosphatidylcholine synthesis. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6029–6033. doi: 10.1073/pnas.87.16.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S., Yamada H. Isolation and characterization of two outer membrane preparations from Escherichia coli. Biochim Biophys Acta. 1975 Jan 14;375(1):44–53. doi: 10.1016/0005-2736(75)90071-1. [DOI] [PubMed] [Google Scholar]
- Neyfakh A. A., Bidnenko V. E., Chen L. B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okanishi K., Kondo S., Utahara R., Umezawa H. Phosphorylation and inactivation of aminoglycosidic antibiotics by E. coli carrying R factor. J Antibiot (Tokyo) 1968 Jan;21(1):13–21. doi: 10.7164/antibiotics.21.13. [DOI] [PubMed] [Google Scholar]
- Ozaki M., Mizushima S., Nomura M. Identification and functional characterization of the protein controlled by the streptomycin-resistant locus in E. coli. Nature. 1969 Apr 26;222(5191):333–339. doi: 10.1038/222333a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasawa S., Utahara R., Okanishi M., Maeda K., Umezawa H. Studies on adenylylstreptomycin, a product of streptomycin inactivation by E. coli carrying the R-factor. J Antibiot (Tokyo) 1968 Aug;21(8):477–484. doi: 10.7164/antibiotics.21.477. [DOI] [PubMed] [Google Scholar]
- Takatsuki A., Arima K., Tamura G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J Antibiot (Tokyo) 1971 Apr;24(4):215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Primary structure of the COOH-terminal membranous segment of a penicillin-sensitive enzyme purified from two Bacilli. J Biol Chem. 1981 Feb 25;256(4):2067–2077. [PubMed] [Google Scholar]
- Yamada H., Tokuda H., Mizushima S. Proton motive force-dependent and -independent protein translocation revealed by an efficient in vitro assay system of Escherichia coli. J Biol Chem. 1989 Jan 25;264(3):1723–1728. [PubMed] [Google Scholar]
- Yue V. T., Schimmel P. R. Direct and specific photochemical cross-linking of adenosine 5'-triphosphate to an aminoacyl-tRNA synthetase. Biochemistry. 1977 Oct 18;16(21):4678–4684. doi: 10.1021/bi00640a023. [DOI] [PubMed] [Google Scholar]
- Zoumbos N. C., Gascon P., Djeu J. Y., Young N. S. Interferon is a mediator of hematopoietic suppression in aplastic anemia in vitro and possibly in vivo. Proc Natl Acad Sci U S A. 1985 Jan;82(1):188–192. doi: 10.1073/pnas.82.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]









