Abstract
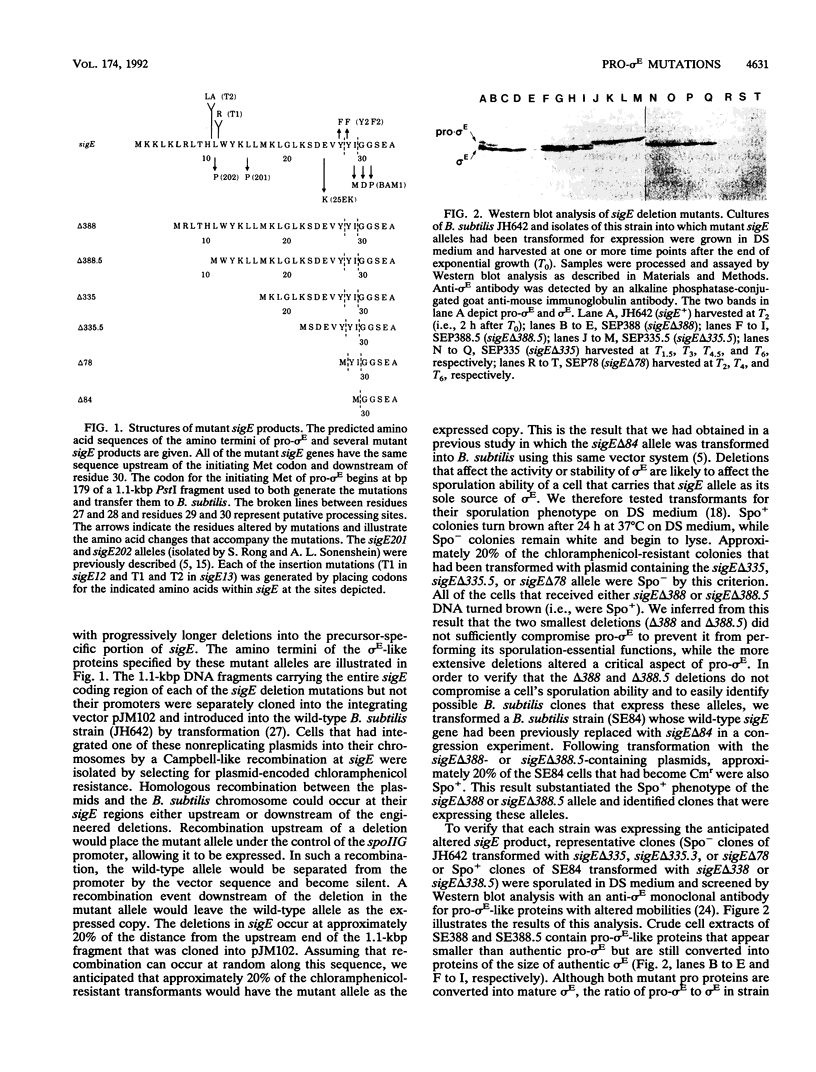
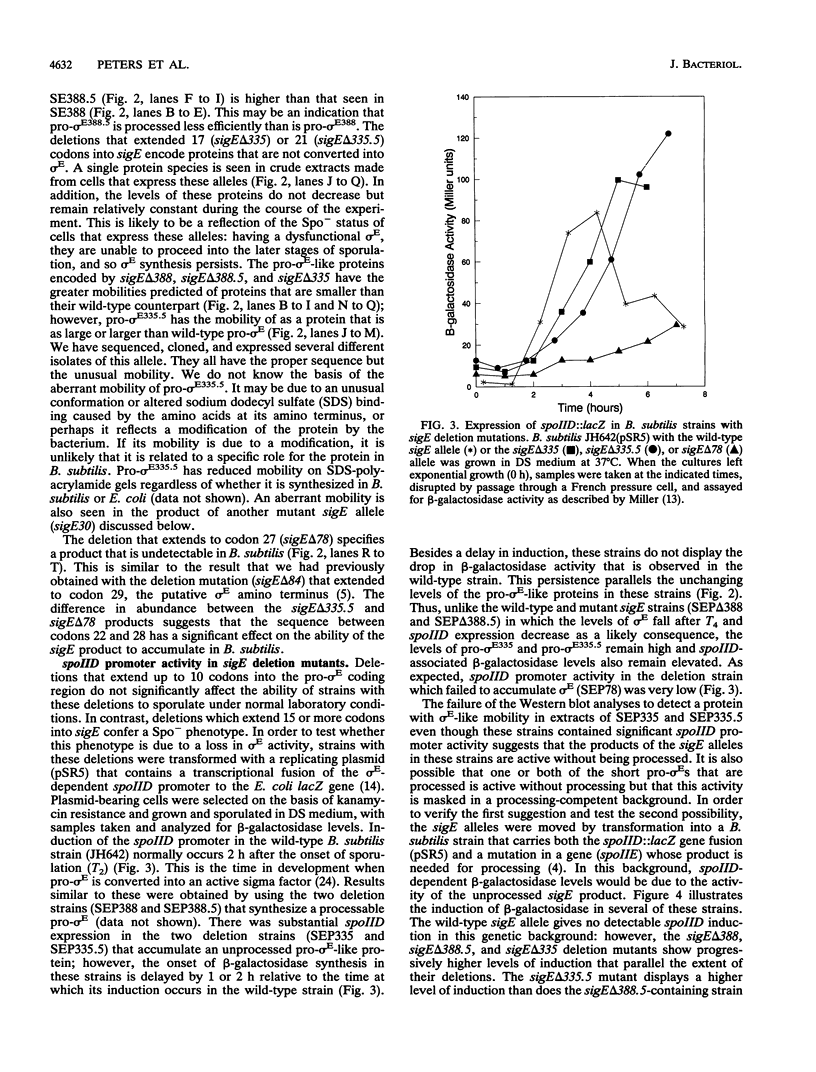
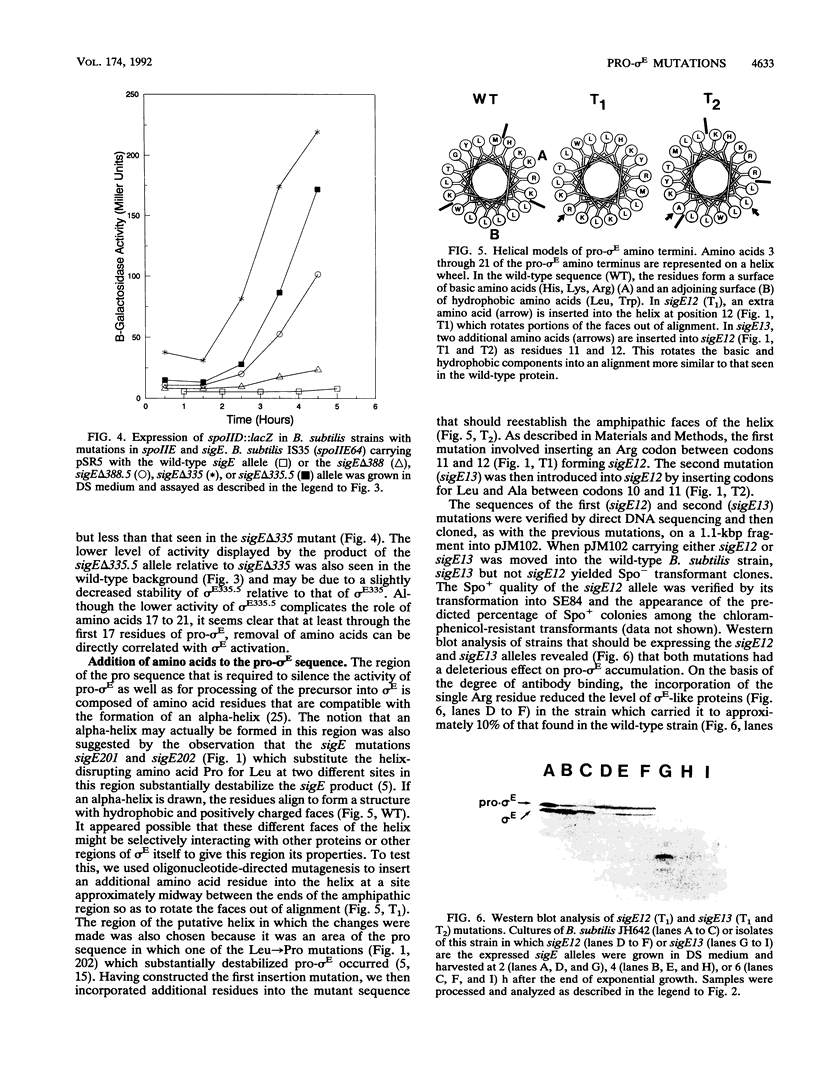
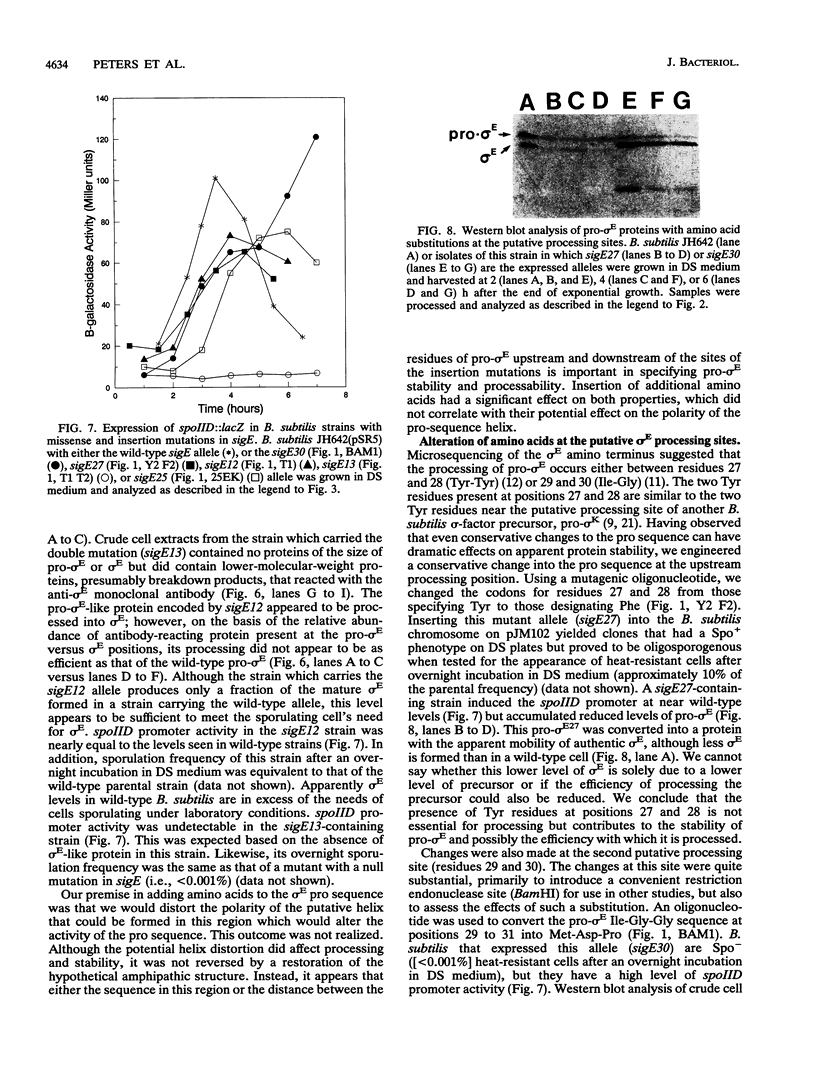
sigma E is a sporulation-specific sigma factor of Bacillus subtilis that is formed from an inactive precursor protein (pro-sigma E) by the removal of 27 to 29 amino acids from the pro-sigma E amino terminus. By using oligonucleotide-directed mutagenesis, sequential deletions were constructed in the precursor-specific region of sigE and analyzed for their effect on the gene product's activity, ability to accumulate, and susceptibility to conversion into mature sigma E. The results demonstrated that the first 17 residues of the pro sequence contribute to silencing the sigma-like activity of pro-sigma E and that the amino acids between positions 12 and 17 are also important for its conversion into sigma E. Deletions that remove 21 or more codons from sigE reduce sigma E activity in cells which carry it, presumably by affecting pro-sigma E stability. A 26-codon deletion results in a gene whose product is not detectable in B. subtilis by either reporter gene activity or Western blot (immunoblot) assay. The primary structure as well as the size of the pro region of sigma E contributes to the protein's stability. The placement of additional amino acids into the pro region reduces the cell's ability to accumulate pro-sigma E. Additional sigE mutations revealed that the amino acids normally found at the putative processing site(s) of pro-sigma E are not essential to the processing reaction; however, a Glu residue upstream of these sites (position 25) was found to be important for processing. These last results suggest that the pro-sigma E processing apparatus does not recognize the actual site within pro-sigma E at which cleavage occurs but rater sequence elements that are upstream of this site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beall B., Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991 Mar;5(3):447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Lang N., Losick R. A sporulation-induced sigma-like regulatory protein from B. subtilis. Cell. 1981 Feb;23(2):615–624. doi: 10.1016/0092-8674(81)90157-4. [DOI] [PubMed] [Google Scholar]
- Jonas R. M., Haldenwang W. G. Influence of spo mutations on sigma E synthesis in Bacillus subtilis. J Bacteriol. 1989 Sep;171(9):5226–5228. doi: 10.1128/jb.171.9.5226-5228.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas R. M., Peters H. K., 3rd, Haldenwang W. G. Phenotypes of Bacillus subtilis mutants altered in the precursor-specific region of sigma E. J Bacteriol. 1990 Aug;172(8):4178–4186. doi: 10.1128/jb.172.8.4178-4186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas R. M., Weaver E. A., Kenney T. J., Moran C. P., Jr, Haldenwang W. G. The Bacillus subtilis spoIIG operon encodes both sigma E and a gene necessary for sigma E activation. J Bacteriol. 1988 Feb;170(2):507–511. doi: 10.1128/jb.170.2.507-511.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney T. J., Kirchman P. A., Moran C. P., Jr Gene encoding sigma E is transcribed from a sigma A-like promoter in Bacillus subtilis. J Bacteriol. 1988 Jul;170(7):3058–3064. doi: 10.1128/jb.170.7.3058-3064.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney T. J., Moran C. P., Jr Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kunkel B., Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989 Jan 27;243(4890):526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBell T. L., Trempy J. E., Haldenwang W. G. Sporulation-specific sigma factor sigma 29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992 Feb 13;355(6361):601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- Rong S., Rosenkrantz M. S., Sonenshein A. L. Transcriptional control of the Bacillus subtilis spoIID gene. J Bacteriol. 1986 Mar;165(3):771–779. doi: 10.1128/jb.165.3.771-779.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S., Sonenshein A. L. Mutations in the precursor region of a Bacillus subtilis sporulation sigma factor. J Bacteriol. 1992 Jun;174(11):3812–3817. doi: 10.1128/jb.174.11.3812-3817.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satola S. W., Baldus J. M., Moran C. P., Jr Binding of Spo0A stimulates spoIIG promoter activity in Bacillus subtilis. J Bacteriol. 1992 Mar;174(5):1448–1453. doi: 10.1128/jb.174.5.1448-1453.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini J. M., Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969 Jun;113(1):29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Bouvier J., Bonamy C., Szulmajster J. A developmental gene product of Bacillus subtilis homologous to the sigma factor of Escherichia coli. Nature. 1984 Nov 22;312(5992):376–378. doi: 10.1038/312376a0. [DOI] [PubMed] [Google Scholar]
- Stragier P., Kunkel B., Kroos L., Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989 Jan 27;243(4890):507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- Trempy J. E., Bonamy C., Szulmajster J., Haldenwang W. G. Bacillus subtilis sigma factor sigma 29 is the product of the sporulation-essential gene spoIIG. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4189–4192. doi: 10.1073/pnas.82.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempy J. E., Morrison-Plummer J., Haldenwang W. G. Synthesis of sigma 29, an RNA polymerase specificity determinant, is a developmentally regulated event in Bacillus subtilis. J Bacteriol. 1985 Jan;161(1):340–346. doi: 10.1128/jb.161.1.340-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Wilson G. A., Young F. E. Transformation and transfection in lysogenic strains of Bacillus subtilis 168. J Bacteriol. 1973 Feb;113(2):540–548. doi: 10.1128/jb.113.2.540-548.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]






