Abstract
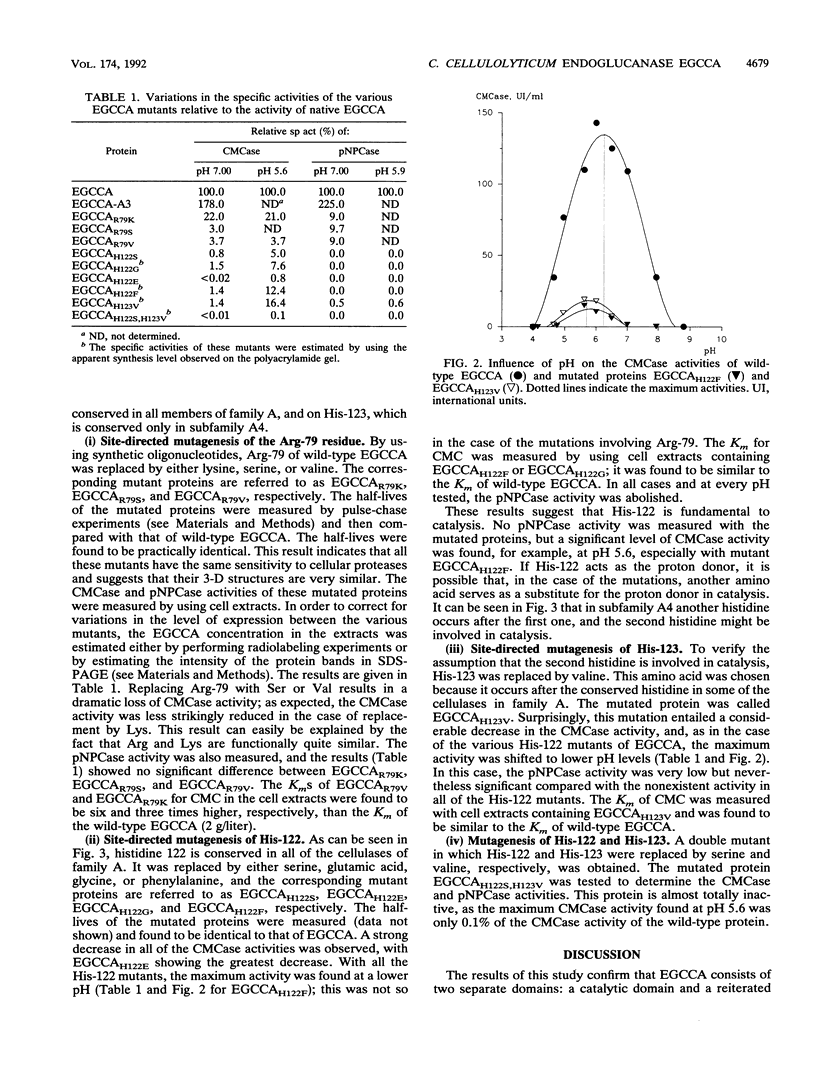
Sequence analysis of the endoglucanase EGCCA of Clostridium cellulolyticum indicates the existence of two domains: a catalytic domain extending from residue 1 to residue 376 and a reiterated domain running from residue 390 to 450. A small deletion in the C terminal end of the catalytic domain inactivated the protein. From the analysis of the sequences of 26 endoglucanases belonging to family A, we focused on seven amino acids which were totally conserved in all the catalytic domains compared. The roles of two of these, Arg-79 and His-122, were studied and defined on the basis of the mutants obtained by introducing various substitutions. Our findings suggest that Arg-79 is involved in the structural organization of the protein; the His-122 residue seems to be more essential for catalysis. The role of His-123, which is conserved only in subfamily A4, was also investigated.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Banner D. W., Bloomer A. C., Petsko G. A., Phillips D., Rivers P. S., Wilson I. A. On the three-dimensional structure and catalytic mechanism of triose phosphate isomerase. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):159–171. doi: 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- Busetta B. The use of folding patterns in the search of protein structural similarities; a three-dimensional model of phosphoribosyl transferases. Biochim Biophys Acta. 1988 Nov 2;957(1):21–33. doi: 10.1016/0167-4838(88)90153-7. [DOI] [PubMed] [Google Scholar]
- Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- Carrell H. L., Glusker J. P., Burger V., Manfre F., Tritsch D., Biellmann J. F. X-ray analysis of D-xylose isomerase at 1.9 A: native enzyme in complex with substrate and with a mechanism-designed inactivator. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4440–4444. doi: 10.1073/pnas.86.12.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E., Bagnara C., Belaich A., Belaich J. P. Cloning and expression of two cellulase genes of Clostridium cellulolyticum in Escherichia coli. Gene. 1988 May 15;65(1):51–58. doi: 10.1016/0378-1119(88)90416-7. [DOI] [PubMed] [Google Scholar]
- Faure E., Belaich A., Bagnara C., Gaudin C., Belaich J. P. Sequence analysis of the Clostridium cellulolyticum endoglucanase-A-encoding gene, celCCA. Gene. 1989 Dec 7;84(1):39–46. doi: 10.1016/0378-1119(89)90137-6. [DOI] [PubMed] [Google Scholar]
- Fierobe H. P., Gaudin C., Belaich A., Loutfi M., Faure E., Bagnara C., Baty D., Belaich J. P. Characterization of endoglucanase A from Clostridium cellulolyticum. J Bacteriol. 1991 Dec;173(24):7956–7962. doi: 10.1128/jb.173.24.7956-7962.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foong F., Hamamoto T., Shoseyov O., Doi R. H. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J Gen Microbiol. 1991 Jul;137(7):1729–1736. doi: 10.1099/00221287-137-7-1729. [DOI] [PubMed] [Google Scholar]
- Giallo J., Gaudin C., Belaich J. P. Metabolism and Solubilization of Cellulose by Clostridium cellulolyticum H10. Appl Environ Microbiol. 1985 May;49(5):1216–1221. doi: 10.1128/aem.49.5.1216-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R., Warren R. A., Miller R. C., Jr, Kilburn D. G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988 Jul 25;263(21):10401–10407. [PubMed] [Google Scholar]
- Gräbnitz F., Seiss M., Rücknagel K. P., Staudenbauer W. L. Structure of the beta-glucosidase gene bglA of Clostridium thermocellum. Sequence analysis reveals a superfamily of cellulases and beta-glycosidases including human lactase/phlorizin hydrolase. Eur J Biochem. 1991 Sep 1;200(2):301–309. doi: 10.1111/j.1432-1033.1991.tb16186.x. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Joliff G., Béguin P., Millet J., Aubert J. P., Alzari P., Juy M., Poljak R. J. Crystallization and preliminary X-ray diffraction study of an endoglucanase from Clostridium thermocellum. J Mol Biol. 1986 May 5;189(1):249–250. doi: 10.1016/0022-2836(86)90396-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lüthi E., Love D. R., McAnulty J., Wallace C., Caughey P. A., Saul D., Bergquist P. L. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Apr;56(4):1017–1024. doi: 10.1128/aem.56.4.1017-1024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B., Bortoli-German I., Haiech J., Chippaux M., Barras F. Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu133 residues for catalysis. Protein Eng. 1991 Feb;4(3):325–333. doi: 10.1093/protein/4.3.325. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A. Carbohydrate-binding proteins: tertiary structures and protein-sugar interactions. Annu Rev Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- Rouvinen J., Bergfors T., Teeri T., Knowles J. K., Jones T. A. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990 Jul 27;249(4967):380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokatlidis K., Salamitou S., Béguin P., Dhurjati P., Aubert J. P. Interaction of the duplicated segment carried by Clostridium thermocellum cellulases with cellulosome components. FEBS Lett. 1991 Oct 21;291(2):185–188. doi: 10.1016/0014-5793(91)81279-h. [DOI] [PubMed] [Google Scholar]
- Tomme P., Chauvaux S., Béguin P., Millet J., Aubert J. P., Claeyssens M. Identification of a histidyl residue in the active center of endoglucanase D from Clostridium thermocellum. J Biol Chem. 1991 Jun 5;266(16):10313–10318. [PubMed] [Google Scholar]
- Wain-Hobson S., Sonigo P., Danos O., Cole S., Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985 Jan;40(1):9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]


