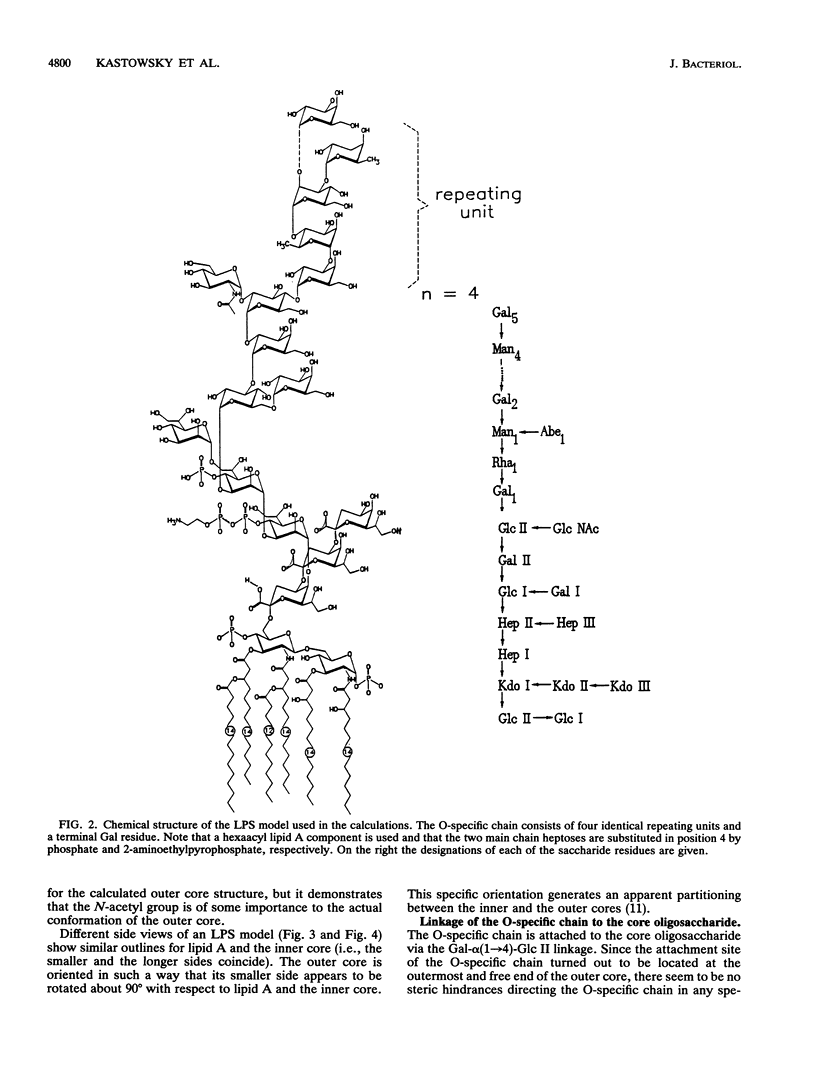
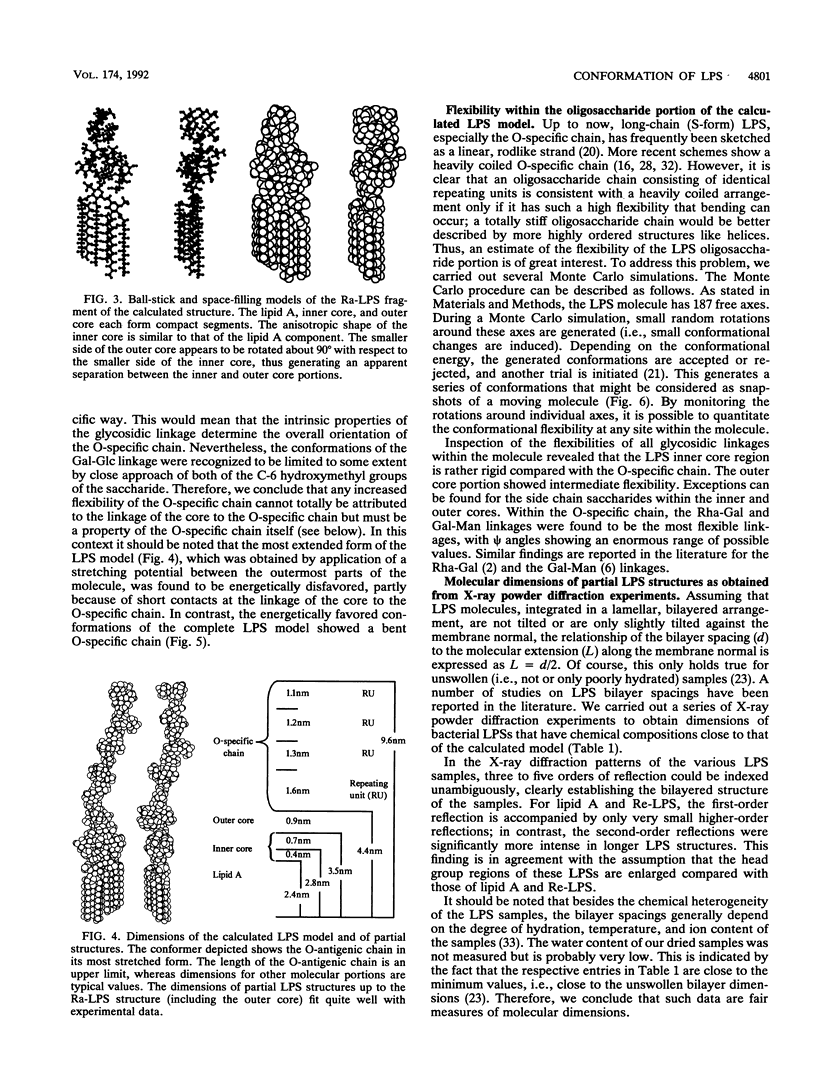
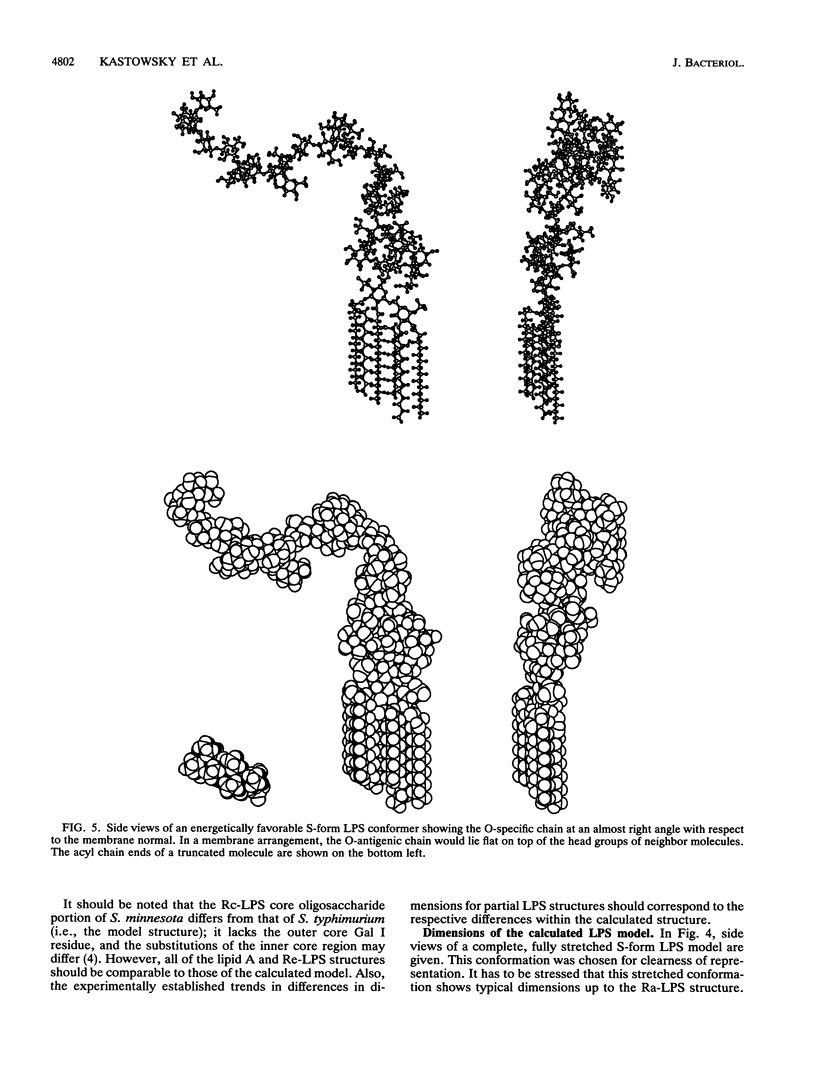
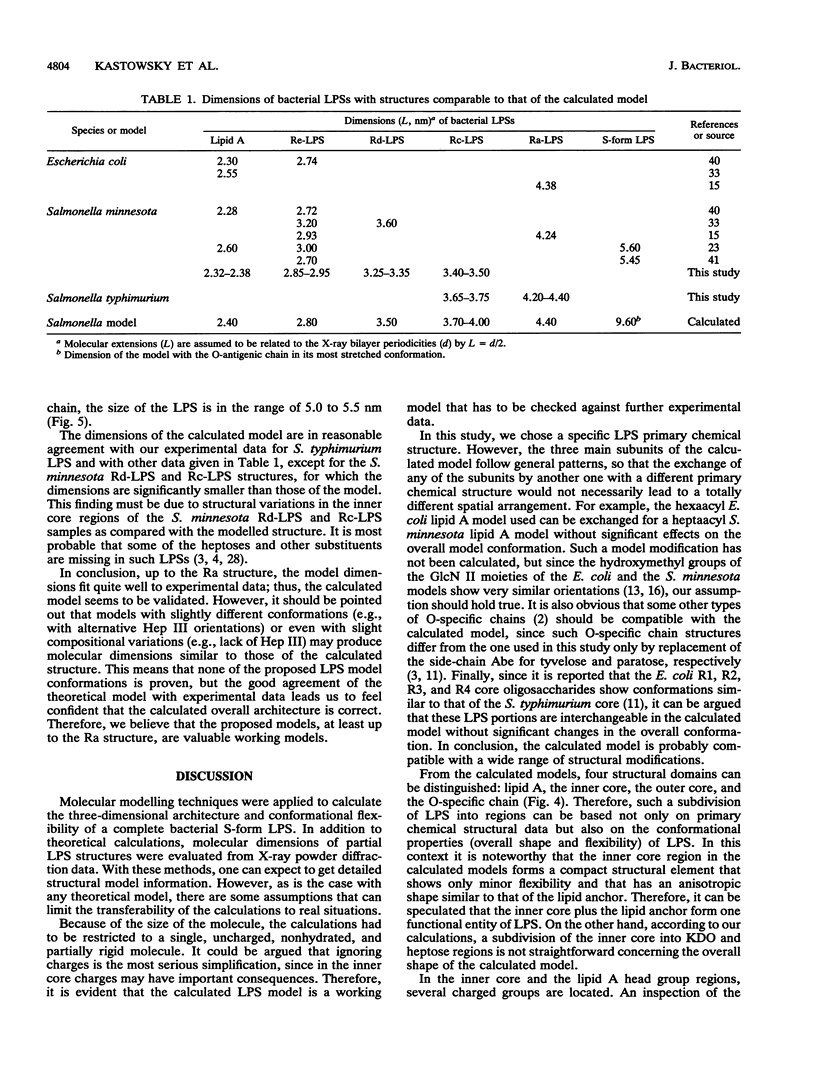
Abstract
Molecular modelling techniques have been applied to calculate the three-dimensional architecture and the conformational flexibility of a complete bacterial S-form lipopolysaccharide (LPS) consisting of a hexaacyl lipid A identical to Escherichia coli lipid A, a complete Salmonella typhimurium core oligosaccharide portion, and four repeating units of the Salmonella serogroup B O-specific chain. X-ray powder diffraction experiments on dried samples of LPS were carried out to obtain information on the dimensions of the various LPS partial structures. Up to the Ra-LPS structure, the calculated model dimensions were in good agreement with experimental data and were 2.4 nm for lipid A, 2.8 nm for Re-LPS, 3.5 nm for Rd-LPS, and 4.4 nm for Ra-LPS. The maximum length of a stretched S-form LPS model bearing four repeating units was evaluated to be 9.6 nm; however, energetically favored LPS conformations showed the O-specific chain bent with respect to the Ra-LPS portion and significantly smaller dimensions (about 5.0 to 5.5 nm). According to the calculations, the Ra-LPS moiety has an approximately cylindrical shape and is conformationally well defined, in contrast to the O-specific chain, which was found to be the most flexible portion within the molecule.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bock K., Meldal M., Bundle D. R., Iversen T., Garegg P. J., Norberg T., Lindberg A. A., Svenson S. B. The conformation of Salmonella O-antigenic polysaccharide chains of serogroups A, B, and D1 predicted by semi-empirical, Hard-Sphere (HSEA) calculations. Carbohydr Res. 1984 Jul 15;130:23–34. doi: 10.1016/0008-6215(84)85267-2. [DOI] [PubMed] [Google Scholar]
- Brade H., Brade L., Rietschel E. T. Structure-activity relationships of bacterial lipopolysaccharides (endotoxins). Current and future aspects. Zentralbl Bakteriol Mikrobiol Hyg A. 1988 Apr;268(2):151–179. doi: 10.1016/s0176-6724(88)80001-4. [DOI] [PubMed] [Google Scholar]
- Cygler M., Rose D. R., Bundle D. R. Recognition of a cell-surface oligosaccharide of pathogenic Salmonella by an antibody Fab fragment. Science. 1991 Jul 26;253(5018):442–445. doi: 10.1126/science.1713710. [DOI] [PubMed] [Google Scholar]
- Haishima Y., Holst O., Brade H. Structural investigation on the lipopolysaccharide of Escherichia coli rough mutant F653 representing the R3 core type. Eur J Biochem. 1992 Jan 15;203(1-2):127–134. doi: 10.1111/j.1432-1033.1992.tb19837.x. [DOI] [PubMed] [Google Scholar]
- Jansson P. E., Wollin R., Bruse G. W., Lindberg A. A. The conformation of core oligosaccharides from Escherichia coli and Salmonella typhimurium lipopolysaccharides as predicted by semi-empirical calculations. J Mol Recognit. 1989 Jul;2(1):25–36. doi: 10.1002/jmr.300020105. [DOI] [PubMed] [Google Scholar]
- Katowsky M., Sabisch A., Gutberlet T., Bradaczek H. Molecular modelling of bacterial deep rough mutant lipopolysaccharide of Escherichia coli. Eur J Biochem. 1991 May 8;197(3):707–716. doi: 10.1111/j.1432-1033.1991.tb15962.x. [DOI] [PubMed] [Google Scholar]
- Labischinski H., Barnickel G., Bradaczek H., Naumann D., Rietschel E. T., Giesbrecht P. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer membrane. J Bacteriol. 1985 Apr;162(1):9–20. doi: 10.1128/jb.162.1.9-20.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labischinski H., Vorgel E., Uebach W., May R. P., Bradaczek H. Architecture of bacterial lipid A in solution. A neutron small-angle scattering study. Eur J Biochem. 1990 Jun 20;190(2):359–363. doi: 10.1111/j.1432-1033.1990.tb15583.x. [DOI] [PubMed] [Google Scholar]
- Leps B., Barnickel G., Bradaczek H. Structural studies on the bacterial cell wall peptidoglycan pseudomurein. I. Conformational energy calculations on the glycan strands in C1 conformation and comparison with murein. J Theor Biol. 1984 Mar 7;107(1):85–114. doi: 10.1016/s0022-5193(84)80123-x. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Van Alphen L. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim Biophys Acta. 1983 Mar 21;737(1):51–115. doi: 10.1016/0304-4157(83)90014-x. [DOI] [PubMed] [Google Scholar]
- Peterson A. A., McGroarty E. J. High-molecular-weight components in lipopolysaccharides of Salmonella typhimurium, Salmonella minnesota, and Escherichia coli. J Bacteriol. 1985 May;162(2):738–745. doi: 10.1128/jb.162.2.738-745.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus M. R., Burgess A. W., Scheraga H. A. Conformational energy calculations of enzyme-substrate complexes of lysozyme. I. Energy minimization of monosaccharide and oligosaccharide inhibitors and substrates of lysozyme. Biopolymers. 1976 Dec;15(12NA-NA-770103-770104):2485–2521. doi: 10.1002/bip.1976.360151212. [DOI] [PubMed] [Google Scholar]
- Qureshi N., Takayama K., Heller D., Fenselau C. Position of ester groups in the lipid A backbone of lipopolysaccharides obtained from Salmonella typhimurium. J Biol Chem. 1983 Nov 10;258(21):12947–12951. [PubMed] [Google Scholar]
- Rozalski A., Brade L., Kuhn H. M., Brade H., Kosma P., Appelmelk B. J., Kusumoto S., Paulsen H. Determination of the epitope specificity of monoclonal antibodies against the inner core region of bacterial lipopolysaccharides by use of 3-deoxy-D-manno-octulosonate-containing synthetic antigens. Carbohydr Res. 1989 Oct 31;193:257–270. doi: 10.1016/0008-6215(89)85124-9. [DOI] [PubMed] [Google Scholar]
- Seydel U., Brandenburg K., Koch M. H., Rietschel E. T. Supramolecular structure of lipopolysaccharide and free lipid A under physiological conditions as determined by synchrotron small-angle X-ray diffraction. Eur J Biochem. 1989 Dec 8;186(1-2):325–332. doi: 10.1111/j.1432-1033.1989.tb15212.x. [DOI] [PubMed] [Google Scholar]
- Shands J. W., Jr, Graham J. A., Nath K. The morphologic structure of isolated bacterial lipopolysaccharide. J Mol Biol. 1967 Apr 14;25(1):15–21. doi: 10.1016/0022-2836(67)90275-6. [DOI] [PubMed] [Google Scholar]
- Stuike-Prill R., Meyer B. A new force-field program for the calculation of glycopeptides and its application to a heptacosapeptide-decasaccharide of immunoglobulin G1. Importance of 1-6-glycosidic linkages in carbohydrate.peptide interactions. Eur J Biochem. 1990 Dec 27;194(3):903–919. doi: 10.1111/j.1432-1033.1990.tb19485.x. [DOI] [PubMed] [Google Scholar]
- Svensson G., Albertsson J., Svensson C., Magnusson G., Dahmén J. X-ray crystal structure of galabiose, O-alpha-D-galactopyranosyl-(1---4)-D-galactopyranose. Carbohydr Res. 1986 Jan 15;146(1):29–38. doi: 10.1016/0008-6215(86)85021-2. [DOI] [PubMed] [Google Scholar]


