Abstract
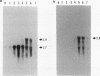
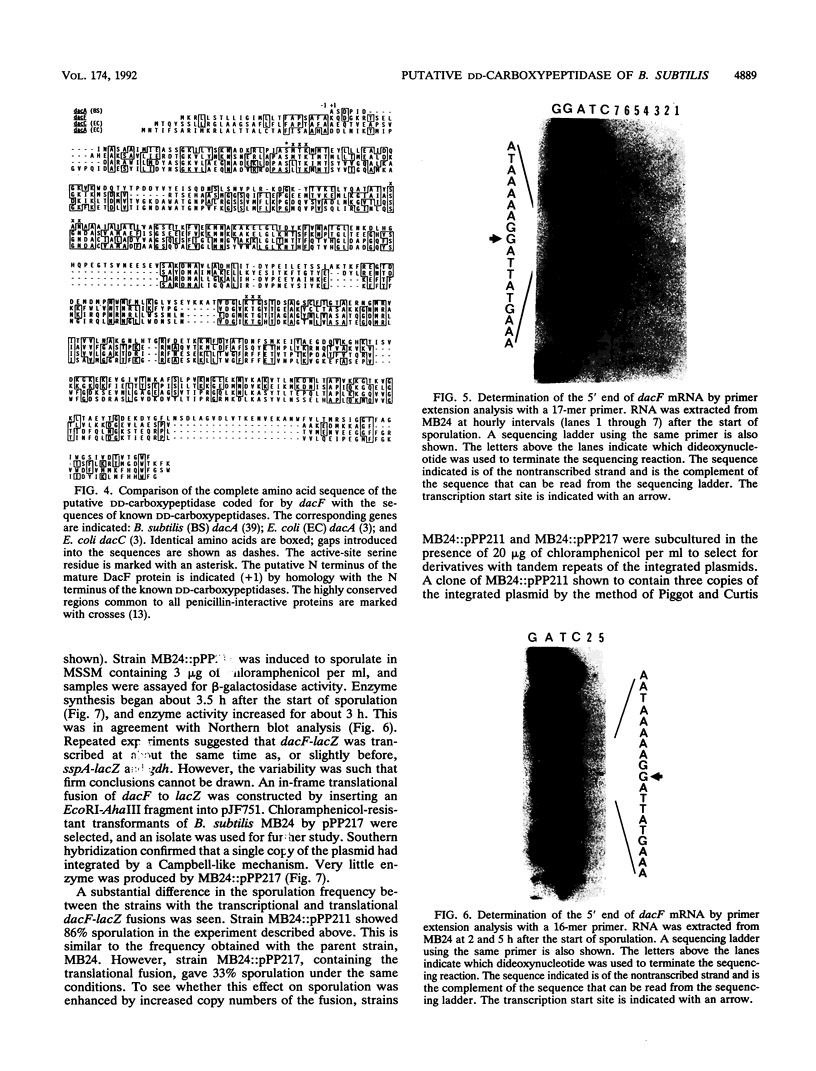
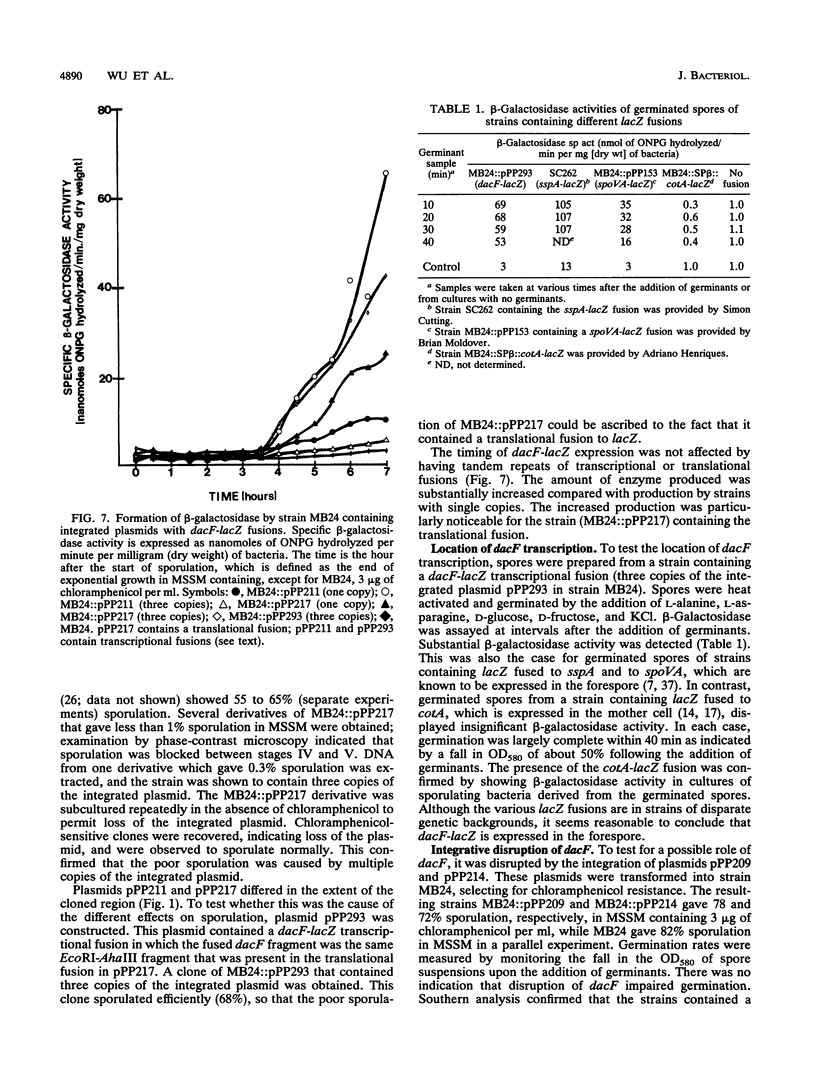
At early stages of sporulation, the spoIIA locus is transcribed as a tricistronic (1.7-kb) operon, coding for sigma F and for two proteins that modulate the activity of sigma F. The locus is transcribed as a longer (2.9-kb) transcript at the late stages of sporulation. We show here that the longer transcript contains an additional open reading frame whose product has extensive sequence homology with DD-carboxypeptidases; the corresponding gene is designated dacF. Cotranscription of a morphogene, such as dacF, with the gene for a sigma factor suggests a way to couple transcription regulation with morphogenesis. The predicted N-terminal sequence of the DacF protein and the inhibition of sporulation by a translational dacF-lacZ fusion both suggest that the protein has a signal peptide for transport into or across a membrane. Expression of a dacF-lacZ transcriptional fusion was in the forespore. The 5' end of the 2.9-kb transcript was determined by primer extension analysis. The region 5' to the end showed no homology to promoters recognized by known sigma factors but was homologous to the corresponding region of the forespore-specific 0.3-kb gene of Bacillus subtilis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beall B., Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991 Mar;5(3):447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Broome-Smith J. K. Construction of a mutant of Escherichia coli that has deletions of both the penicillin-binding protein 5 and 6 genes. J Gen Microbiol. 1985 Aug;131(8):2115–2118. doi: 10.1099/00221287-131-8-2115. [DOI] [PubMed] [Google Scholar]
- Broome-Smith J. K., Ioannidis I., Edelman A., Spratt B. G. Nucleotide sequences of the penicillin-binding protein 5 and 6 genes of Escherichia coli. Nucleic Acids Res. 1988 Feb 25;16(4):1617–1617. doi: 10.1093/nar/16.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan C. E., Ling M. L. Isolation and sequence analysis of dacB, which encodes a sporulation-specific penicillin-binding protein in Bacillus subtilis. J Bacteriol. 1992 Mar;174(6):1717–1725. doi: 10.1128/jb.174.6.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S., Oke V., Driks A., Losick R., Lu S., Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990 Jul 27;62(2):239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Perego M., Hoch J. A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988 Jan;170(1):289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Piggot P. J. Nucleotide sequence of sporulation locus spoIIA in Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2147–2153. doi: 10.1099/00221287-130-8-2147. [DOI] [PubMed] [Google Scholar]
- Gholamhoseinian A., Piggot P. J. Timing of spoII gene expression relative to septum formation during sporulation of Bacillus subtilis. J Bacteriol. 1989 Oct;171(10):5747–5749. doi: 10.1128/jb.171.10.5747-5749.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBell T. L., Trempy J. E., Haldenwang W. G. Sporulation-specific sigma factor sigma 29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. M., Chak K. F., Piggot P. J. Isolation and characterization of a recombinant plasmid carrying a functional part of the Bacillus subtilis spoIIA locus. J Gen Microbiol. 1982 Nov;128(11):2805–2812. doi: 10.1099/00221287-128-11-2805. [DOI] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Lu S., Halberg R., Kroos L. Processing of the mother-cell sigma factor, sigma K, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S. C., Sokolov N. V., He C. M., Setlow P. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):77–81. doi: 10.1073/pnas.88.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer S., Losick R., Sun D., Setlow P. Evidence for an additional temporal class of gene expression in the forespore compartment of sporulating Bacillus subtilis. J Bacteriol. 1989 Jan;171(1):561–564. doi: 10.1128/jb.171.1.561-564.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Curtis C. A. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J Bacteriol. 1987 Mar;169(3):1260–1266. doi: 10.1128/jb.169.3.1260-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Curtis C. A., de Lencastre H. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2123–2136. doi: 10.1099/00221287-130-8-2123. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva D., Mandelstam J. Synthesis of spoIIA and spoVA mRNA in Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):3005–3011. doi: 10.1099/00221287-132-11-3005. [DOI] [PubMed] [Google Scholar]
- Schmidt R., Margolis P., Duncan L., Coppolecchia R., Moran C. P., Jr, Losick R. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9221–9225. doi: 10.1073/pnas.87.23.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Beckwith J. R. Uses of lac fusions for the study of biological problems. Microbiol Rev. 1985 Dec;49(4):398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. A., Lang N., Sandman K., Losick R. A promoter whose utilization is temporally regulated during sporulation in Bacillus subtilis. J Mol Biol. 1984 Jul 5;176(3):333–348. doi: 10.1016/0022-2836(84)90493-5. [DOI] [PubMed] [Google Scholar]
- Stragier P., Bonamy C., Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988 Mar 11;52(5):697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Sun D. X., Stragier P., Setlow P. Identification of a new sigma-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 1989 Feb;3(2):141–149. doi: 10.1101/gad.3.2.141. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Todd J. A., Roberts A. N., Johnstone K., Piggot P. J., Winter G., Ellar D. J. Reduced heat resistance of mutant spores after cloning and mutagenesis of the Bacillus subtilis gene encoding penicillin-binding protein 5. J Bacteriol. 1986 Jul;167(1):257–264. doi: 10.1128/jb.167.1.257-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. J., Howard M. G., Piggot P. J. Regulation of transcription of the Bacillus subtilis spoIIA locus. J Bacteriol. 1989 Feb;171(2):692–698. doi: 10.1128/jb.171.2.692-698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre H., Piggot P. J. Identification of different sites of expression for spo loci by transformation of Bacillus subtilis. J Gen Microbiol. 1979 Oct;114(2):377–389. doi: 10.1099/00221287-114-2-377. [DOI] [PubMed] [Google Scholar]