Abstract
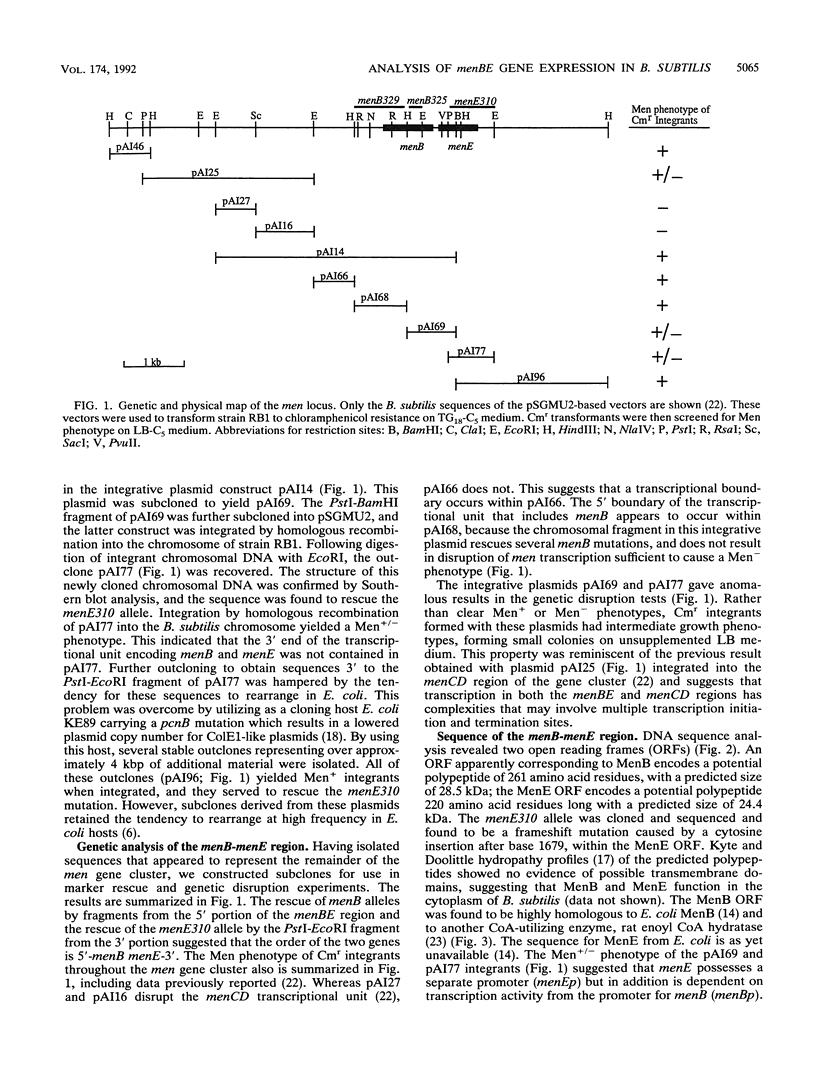
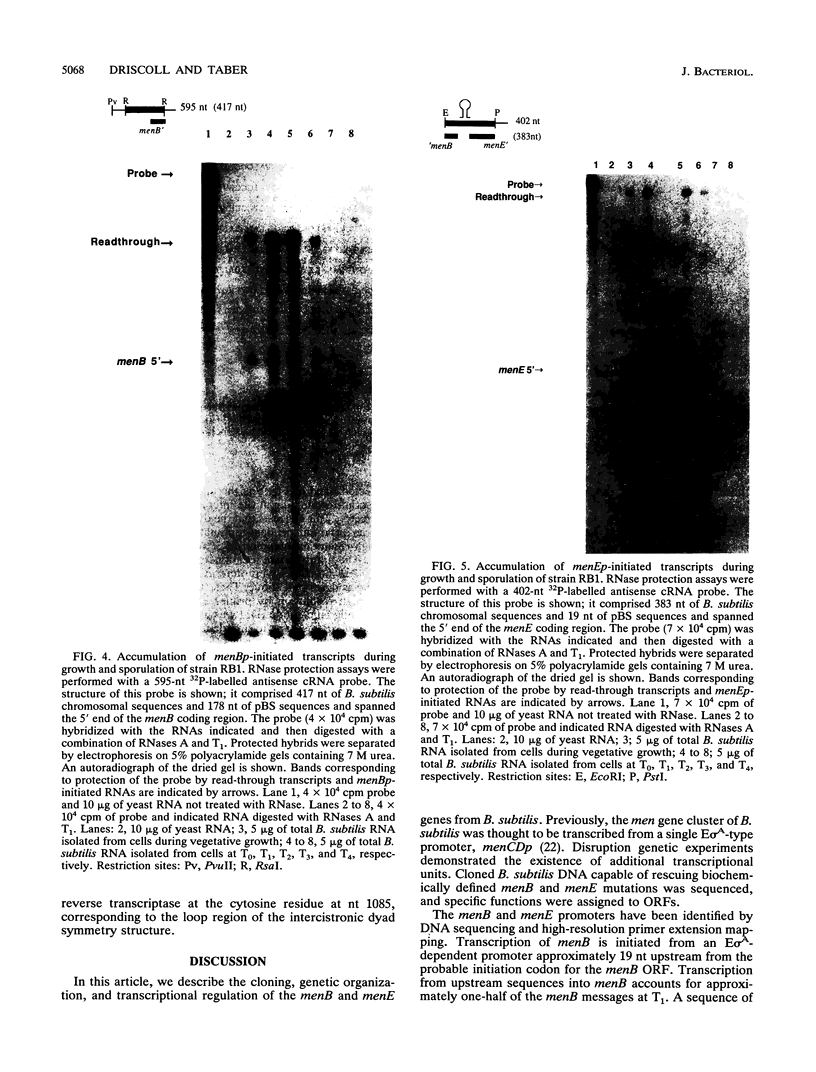
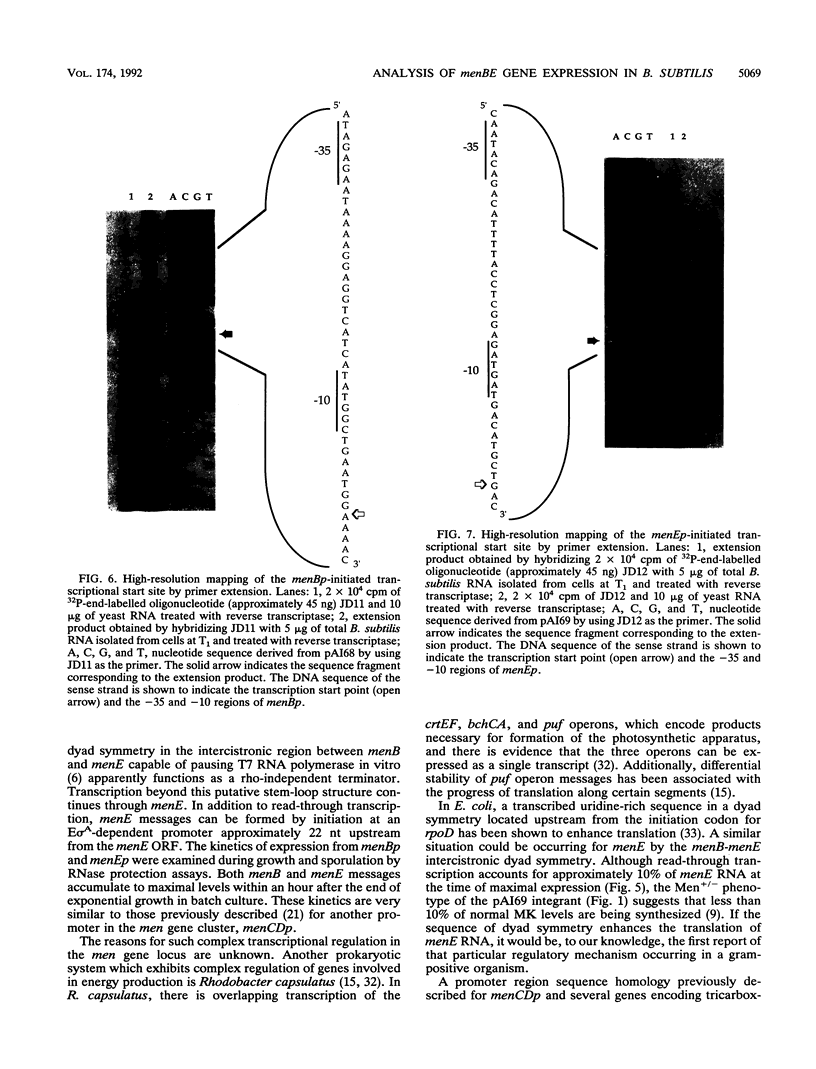
Menaquinone (MK) plays a central role in the respiratory chain of Bacillus subtilis. The biosynthesis of MK requires the formation of a naphthoquinone ring via a series of specific reactions branching from the shikimate pathway. "Early" MK-specific reactions catalyze the formation of o-succinylbenzoate (OSB) from isochorismate, and "late" reactions convert OSB to dihydroxynaphthoate, by utilizing an OSB-coenzyme A intermediate. We have cloned and sequenced the B. subtilis menE and menB genes encoding, respectively, OSB-coenzyme A synthase and dihydroxynaphthoate synthase. The MenB open reading frame encodes a potential polypeptide of 261 amino acid residues with a predicted size of 28.5 kDa, while the MenE open reading frame could encode a 24.4-kDa polypeptide of 220 amino acid residues. Probable promoter sequences were identified by high-resolution primer extension assays. Organization of these genes and regulatory regions was found to be menBp menB menEp menE. Expression of menE was dependent on both menEp and menBp, indicating an operonlike organization. A region of dyad symmetry capable of forming a stable RNA secondary structure was found between menB and menE. Culture cycle-dependent expression of menB and menE was measured by steady-state transcript accumulation. For both genes, maximal accumulation was found to occur within an hour after the end of exponential growth. The menBp and menEp promoters have sequences compatible with recognition by the major vegetative form of B. subtilis RNA polymerase, E sigma A. Both promoter regions also were found to contain homologies to a sequence motif previously identified in the menCDp region and in promoters for several B. subtilis tricarboxylic acid cycle genes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bentley R., Meganathan R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol Rev. 1982 Sep;46(3):241–280. doi: 10.1128/mr.46.3.241-280.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D. J., Zalkin H. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987 Jun 15;262(17):8274–8287. [PubMed] [Google Scholar]
- Escamilla J. E., Barquera B., Ramírez R., García-Horsman A., del Arenal P. Role of menaquinone in inactivation and activation of the Bacillus cereus forespore respiratory system. J Bacteriol. 1988 Dec;170(12):5908–5912. doi: 10.1128/jb.170.12.5908-5912.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand S. K., Taber H. W. Changes in menaquinone concentration during growth and early sporulation in Bacillus subtilis. J Bacteriol. 1974 Jan;117(1):324–326. doi: 10.1128/jb.117.1.324-326.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand S. K., Taber H. W. Physiological effects of menaquinone deficiency in Bacillus subtilis. J Bacteriol. 1973 Sep;115(3):1035–1044. doi: 10.1128/jb.115.3.1035-1044.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. F., Mueller J. P., Taber H. W. The Bacillus subtilis menCD promoter is responsive to extracellular pH. Arch Microbiol. 1990;153(4):355–359. doi: 10.1007/BF00249005. [DOI] [PubMed] [Google Scholar]
- Klug G., Cohen S. N. Effects of translation on degradation of mRNA segments transcribed from the polycistronic puf operon of Rhodobacter capsulatus. J Bacteriol. 1991 Feb;173(4):1478–1484. doi: 10.1128/jb.173.4.1478-1484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug G., Gad'on N., Jock S., Narro M. L. Light and oxygen effects share a common regulatory DNA sequence in Rhodobacter capsulatus. Mol Microbiol. 1991 May;5(5):1235–1239. doi: 10.1111/j.1365-2958.1991.tb01897.x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989 Mar;171(3):1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan R., Bentley R., Taber H. Identification of Bacillus subtilis men mutants which lack O-succinylbenzoyl-coenzyme A synthetase and dihydroxynaphthoate synthase. J Bacteriol. 1981 Jan;145(1):328–332. doi: 10.1128/jb.145.1.328-332.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P., Mueller J., Hill K., Taber H. Transcriptional regulation of a promoter in the men gene cluster of Bacillus subtilis. J Bacteriol. 1988 Jun;170(6):2742–2748. doi: 10.1128/jb.170.6.2742-2748.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P., Rabinowitz A., Taber H. Molecular cloning and preliminary genetic analysis of the men gene cluster of Bacillus subtilis. J Bacteriol. 1988 Jun;170(6):2735–2741. doi: 10.1128/jb.170.6.2735-2741.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami-Ishii N., Taketani S., Osumi T., Hashimoto T. Molecular cloning and sequence analysis of the cDNA for rat mitochondrial enoyl-CoA hydratase. Structural and evolutionary relationships linked to the bifunctional enzyme of the peroxisomal beta-oxidation system. Eur J Biochem. 1989 Oct 20;185(1):73–78. doi: 10.1111/j.1432-1033.1989.tb15083.x. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Curtis C. A., de Lencastre H. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2123–2136. doi: 10.1099/00221287-130-8-2123. [DOI] [PubMed] [Google Scholar]
- Saunders C. W., Schmidt B. J., Mirot M. S., Thompson L. D., Guyer M. S. Use of chromosomal integration in the establishment and expression of blaZ, a Staphylococcus aureus beta-lactamase gene, in Bacillus subtilis. J Bacteriol. 1984 Mar;157(3):718–726. doi: 10.1128/jb.157.3.718-726.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H. W., Dellers E. A., Lombardo L. R. Menaquinone biosynthesis in Bacillus subtilis: isolation of men mutants and evidence for clustering of men genes. J Bacteriol. 1981 Jan;145(1):321–327. doi: 10.1128/jb.145.1.321-327.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington C. L., Beatty J. T. Overlapping mRNA transcripts of photosynthesis gene operons in Rhodobacter capsulatus. J Bacteriol. 1991 Feb;173(4):1432–1443. doi: 10.1128/jb.173.4.1432-1443.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Deutscher M. P. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2605–2609. doi: 10.1073/pnas.89.7.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Aubenton Carafa Y., Brody E., Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990 Dec 20;216(4):835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- de Vrij W., van den Burg B., Konings W. N. Spectral and potentiometric analysis of cytochromes from Bacillus subtilis. Eur J Biochem. 1987 Aug 3;166(3):589–595. doi: 10.1111/j.1432-1033.1987.tb13554.x. [DOI] [PubMed] [Google Scholar]