Abstract
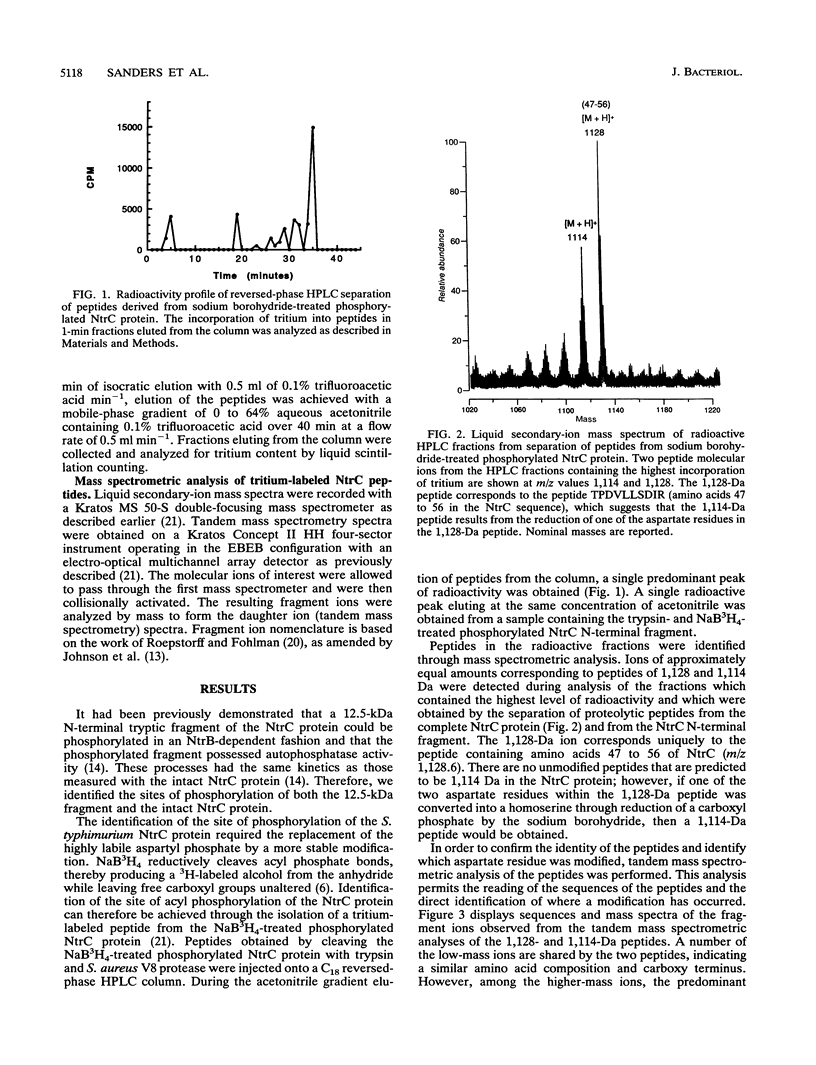
The NtrC transcription factor is a member of a family of homologous prokaryotic regulatory proteins that participate in the transduction of extracellular and nutritional signals. It has been demonstrated that the phosphate group from a histidine residue of the phosphorylated NtrB protein autokinase is transferred to the NtrC protein. Phosphorylation of the NtrC protein is transient and activates its transcriptional enhancement activity. We have investigated the site of phosphorylation of the Salmonella typhimurium NtrC protein and find that it is an aspartate residue (Asp-54) that is found within a sequence conserved in all of the members of the family of regulatory proteins. We propose that this phosphorylation is an NtrC protein histidine phosphatase catalytic intermediate. This conclusion suggests that the NtrC family should be viewed not as kinase substrates but as enzymes that can catalyze the hydrolysis of their activated forms in a concentration-independent fashion. They are similar in this sense to eukaryotic signal-transducing GTPases.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borkovich K. A., Simon M. I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990 Dec 21;63(6):1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulys D., Trach K. A., Hoch J. A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991 Feb 8;64(3):545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Degani C., Boyer P. D. A borohydride reduction method for characterization of the acyl phosphate linkage in proteins and its application to sarcoplasmic reticulum adenosine triphosphatase. J Biol Chem. 1973 Dec 10;248(23):8222–8226. [PubMed] [Google Scholar]
- Feuerstein J., Goody R. S., Webb M. R. The mechanism of guanosine nucleotide hydrolysis by p21 c-Ha-ras. The stereochemical course of the GTPase reaction. J Biol Chem. 1989 Apr 15;264(11):6188–6190. [PubMed] [Google Scholar]
- Gibbs J. B., Sigal I. S., Poe M., Scolnick E. M. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J. F., Bourret R. B., Simon M. I. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988 Nov 10;336(6195):139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Kaplan N., Simon M. I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Igo M. M., Ninfa A. J., Stock J. B., Silhavy T. J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989 Nov;3(11):1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- Jin S. G., Prusti R. K., Roitsch T., Ankenbauer R. G., Nester E. W. Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J Bacteriol. 1990 Sep;172(9):4945–4950. doi: 10.1128/jb.172.9.4945-4950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat G. S., Stock A. M., Stock J. B. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry. 1990 Jun 12;29(23):5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- Ninfa A. J., Bennett R. L. Identification of the site of autophosphorylation of the bacterial protein kinase/phosphatase NRII. J Biol Chem. 1991 Apr 15;266(11):6888–6893. [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Ninfa E. G., Lupas A. N., Stock A., Magasanik B., Stock J. Crosstalk between bacterial chemotaxis signal transduction proteins and regulators of transcription of the Ntr regulon: evidence that nitrogen assimilation and chemotaxis are controlled by a common phosphotransfer mechanism. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5492–5496. doi: 10.1073/pnas.85.15.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Roepstorff P., Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984 Nov;11(11):601–601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989 Dec 25;264(36):21770–21778. [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Weiss D. S., Batut J., Klose K. E., Keener J., Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991 Oct 4;67(1):155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- Weiss V., Magasanik B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]



