Abstract
Mouse αA-crystallin gene encodes the most abundant protein of the mammalian lens. Expression of αA-crystallin is regulated temporally and spatially during lens development with initial expression in the lens vesicle followed by strong upregulation in the differentiating primary lens fibers. Lens-specific expression of αA-crystallin is mediated by DNA-binding transcription factors Pax6, c-Maf and CREB bound to its promoter region. Its 5′-distal enhancer, DCR1, mediates regulation of αA-crystallin via FGF signaling, while its 3′-distal enhancer, DCR3, functions only in elongated primary lens fibers via other lens differentiation pathways. DCR1 and DCR3 establish outside borders of a lens-specific chromatin region marked by histone H3 K9 acetylation. Here, we identified CREB-binding protein (CBP) and p300 as major histone acetyltransferases (HATs) associated in vivo with the mouse αA-crystallin locus. Both HATs are expressed in embryonic lens. Expression of CBP in primary lens fiber cells coincides with αA-crystallin. In the chromatin of lens epithelial cells, chromatin immunoprecipitations (ChIPs) show that the αA-crystallin promoter is notably devoid of any significant presence of CBP and p300, though DCR1 and a few other regions show the presence of these HATs. In the chromatin obtained from newborn lens, CBP was localized specifically at the promoter region with about ten times higher abundance compared to the entire αA-crystallin locus. In contrast, p300 is distributed more evenly across the entire locus. Analysis of total histone H3 and H3 K9 acetylation revealed potential lower density of nucleosomes 2 kb upstream from the promoter region. Collectively, our data suggest that moderate level of αA-crystallin gene expression in lens epithelial cells does not require the presence of CBP and p300 in the promoter. However, the lens-specific chromatin domain contains both promoter localized CBP on the “background” of locus-spread presence of CBP and p300.
Keywords: lens, crystallin, CBP/p300, chromatin, histone acetylation
Introduction
The eukaryotic genome is packed into a highly organized nucleoprotein structure, the chromatin fiber.1 The regulation of gene expression through chromatin requires coordinated actions of multiple enzymes and proteins to modify chromatin structure to either promote or repress transcription. These “chromatin remodeling” activities include local and global modifications of core histone proteins, change of positions of individual nucleosomes and removal of nucleosomes from promoter regions.2,3 It is hypothesized that a specific combination of these local/global activities are required for transcriptional regulation of any particular gene.4
Post-translationally modified core histones, with acetylated and methylated lysine and methylated arginine residues, serve as distinct marks to recruitproteins to the chromatin, and are thought to operate via the “histone code” of epigenetic gene regulation.5,6 Acetylation of lysine residues in histone tails is catalyzed by a family of histone acetyltransferases (HATs), including ATF2, CREB-binding protein (CBP), MOZ, p300, P/CAF, pCIP, SRC-1, TAF250 and Tip60.7 These HATs exhibit different substrate specificities, affinities for interacting proteins and can acetylate non-histone substrates.7 Acetylated histones are recognized by bromodomain-containing proteins such as ATP-dependent chromatin remodeling enzymes Brg1 and Snf2h, which are catalytic subunits of a variety of multiprotein chromatin remodeling complexes SWI/SNF and ISWI, respectively.8,9 These chromatin remodelers can move nucleosomes along the DNA or remove them completely to generate nucleosome-free regions in chromatin.10
During development, transcription of batteries of genes is controlled precisely in space and time. A large number of studies have shown that specific DNA-binding transcription factors bound to DNA in chromatin serve as platforms to recruit chromatin remodeling enzymes.4,11 In parallel, modifications of local chromatin structure facilitate binding of additional DNA-binding transcription factors and this process culminates with active transcription.4,10,12
The key structural proteins of mammalian lens are 15 genes encoding the α- and β/γ-crystallins required for lens transparency and light refraction.13-15 The αA-crystallin is the most abundant crystallin in mammalian lens.13-15 Structurally and functionally, αA-crystallin belongs to a family of small heat shock proteins acting as molecular chaperones.13-15 Expression of αA-crystallin is regulated tightly both temporally and spatially at the level of transcription with the onset of expression in the lens vesicle around E10.5 of mouse embryonic development.16 αA-crystallin expression is boosted dramatically during lens fiber cell differentiation starting at E12.5.16 Using H3 K9 acetylated histone as a marker of transcriptionally active chromatin, we recently identified at least a 16 kb domain of lens-specific chromatin harboring the entire αA-crystallin (Cryaa) locus. Two enhancers, DCR1 and DCR3, are near its 5′/3′ borders.17 Responsive to FGF signaling in cultured lens explants, the DCR1 in combination with a 1.9 kb αA-crystallin promoter is able to recapitulate αA-crystallin endogenous expression pattern in the transgenic mouse model.17 In contrast, the 1.9 kb promoter/DCR3 transgene was activated only in more elongated primary lens fibers. Array of Pax6-binding, Maf-binding and CRE-binding sites have been identified within both DCR1 and the promoter.17-21 Local presence of chromatin remodeling enzymes Brg1 and Snf2h was linked to the presence of Pax6 and c-Maf in the αA-crystallin locus.17 Nevertheless, there remained the question of the possible roles of HATs, CBP and p300 suggested by earlier gene reporter and transgenic mouse overexpression studies.22,23
CBP and p300 are highly structurally homologous HATs involved in cell proliferation, differentiation and apoptosis.24,25 CBP/p300 have been shown to function as scaffolds or bridges between two DNA-binding transcription factors,26 and to acetylated histones via their internal bromodomains or other non-histone proteins.27-30 Mutations in human CBP cause Rubinstein-Taybi syndrome, characterized by a wide array of ocular defects, including cataracts and glaucoma together with mental retardation and malformed thumbs and toes.31,32
Although CBP and p300 are highly homologous and co-expressed in a variety of tissues, gene targeting studies suggest that their functions are not redundant.33-37 Here, we show that both CBP and p300 are expressed in lens with different expression patterns. Distinct physical recruitment patterns of CBP and p300 through the αA-crystallin locus in vivo by chromatin immunoprecipitation (ChIP) suggest that they play unique roles in regulating the αA-crystallin gene expression, in agreement with studies of other genes during neural and cardiac development.37
Results
CBP and p300 have distinct expression patterns during mouse lens development
Previously, CBP/p300 expression was reported in the epithelial cells of E15.5 lens.23 However, the antibody originally used recognized both CBP and p300 proteins. To identify precisely the expression patterns of CBP and p300 during mouse lens development, we analyzed CBP and p300 expression in earlier lens developmental stages using specific antibodies against CBP or p300. During the invagination stages of lens development from E10.5 to E11.5 (Figure 1(a) and (b), and (d) and (e)), both CBP and p300 are detected in the lens pit as well as lens vesicle. As expected, expression of both proteins is detected also in the non-lens surface ectoderm, developing optic cup and in the periocular mesenchyme. As primary lens fiber cells differentiate, they express high levels of αA-crystallin,16 and fill up the lens vesicle by E14.5. At this stage, stronger signals of CBP were detected predominantly in the primary lens fiber cells and transitional zone, while only a few cells were highlighted in the lens epithelium (Figure 1(c) and (g)). However, p300 appears to be expressed strongly in both the lens epithelium and the primary lens fiber cells (Figure 1(f) and (h)). Interestingly, cytoplasmic rather than nuclear localization of p300 was observed in E10.5 mouse lens pit (Figure 1(d)). This transient cytoplasm localization was observed only at this stage and was reproducible using a variety of modifications of the immunostaining procedures (data not shown).
Figure 1.
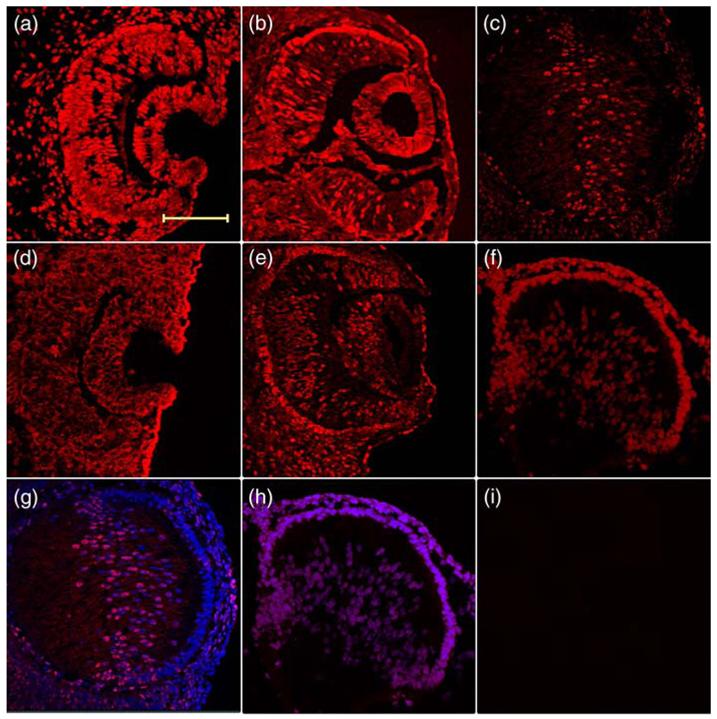
Different expression pattern of CBP and p300 during mouse lens development. Immunohistochemistry was used to test the expression of (a)-(c) CBP and (d)-(f) p300 on (a) and (d) E10.5, (b) and (e) E11.5 and (c), (f)-(i) E14.5 mouse lenses cryosections. Merged images with DAPI to show the (g) CBP and (h) p300 expression at E14.5. An experiment without the primary antibody is shown in (i). The scale bar represents 100 μm, and same magnification was used in all pictures.
Next, we analyzed the mRNA levels of CBP and p300 by quantitative RT-PCR using microdissected two days old rat lenses. We found approximately 2.4-fold higher expression of CBP in the epithelial compartment compared to the lens fiber mass (Figure 2(a)). However, p300 expression in both the epithelium and fibers showed no significant difference (Figure 2(b)). These data demonstrate that the main difference in temporal and spatial expression between CBP and p300 is much higher expression of CBP in differentiating lens fibers compared to the lens epithelium that coincides with the expression pattern of its presumptive target gene, the αA-crystallin.16
Figure 2.
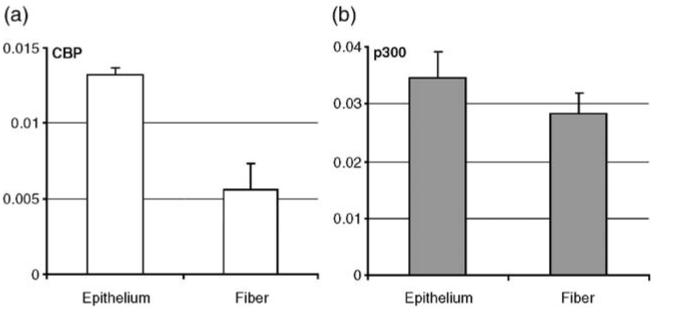
Quantitative RT-PCR of CBP and p300 in microdissected two day old rat lens epithelium and fiber cells. (a) Results for CBP and (b) for p300. Relative expressions were normalized against a pool of housekeeping genes, B2M, HPRT, G6PD and SDHA, as described in Materials and Methods.
Differential localization of CBP and p300 at 16 kb region of the mouse αA-crystallin locus
To understand the formation of the lens-specific chromatin domain marked by histone H3 K9ac shown earlier,17 we performed a series of ChIP studies in lens chromatins using antibodies specific to HATs: ATF2, CBP, p300, P/CAF, pCIP and SRC-1. The results showed the presence of CBP and p300 but not the other HATs (data not shown) at the mouse αA-crystallin locus (Figures 3 and 4). In cultured lens epithelial cells, we found generally lower levels of CBP and p300 compared to the chromatin isolated from newborn lenses. CBP was identified at several places, DCR1/-8 kb, -6 kb, exon1 and exon2/+2 kb (see Figure 3(b)). Similarly, p300 was found in these regions as well as at DCR2/-2 kb (see Figure 3(c)). Notably, the promoter region showed only a marginal presence of CBP and no presence of p300 (see Figure 3(b) and (c)).
Figure 3.
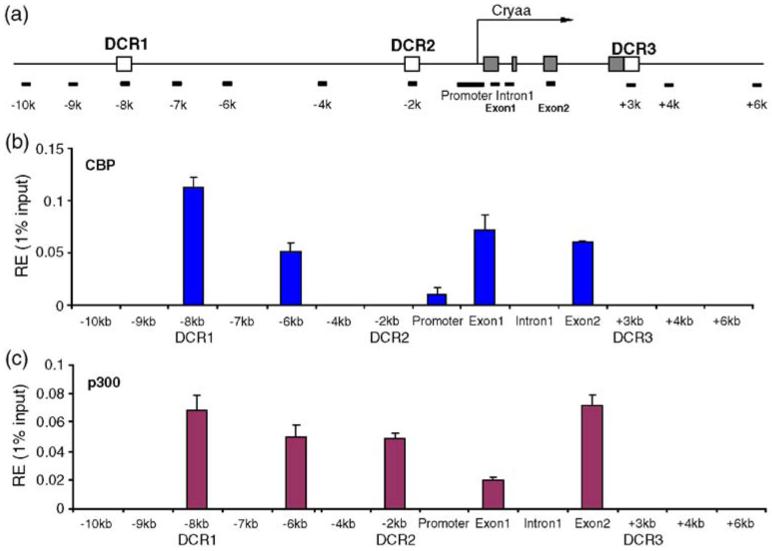
Interaction and distribution of CBP and p300 at the mouse αA-crystallin locus in chromatin of cultured lens epithelial cells. (a) A diagram of the αA-crystallin locus showing positions of PCR amplicons. Five partially overlapping promoter-specific primers were used and an average value of the relative enrichments was calculated as described.17 (b) Distribution of CBP, (c) distribution of p300. The relative enrichment unit represents 1% of the input. Background for each region was subtracted as described.17
Figure 4.
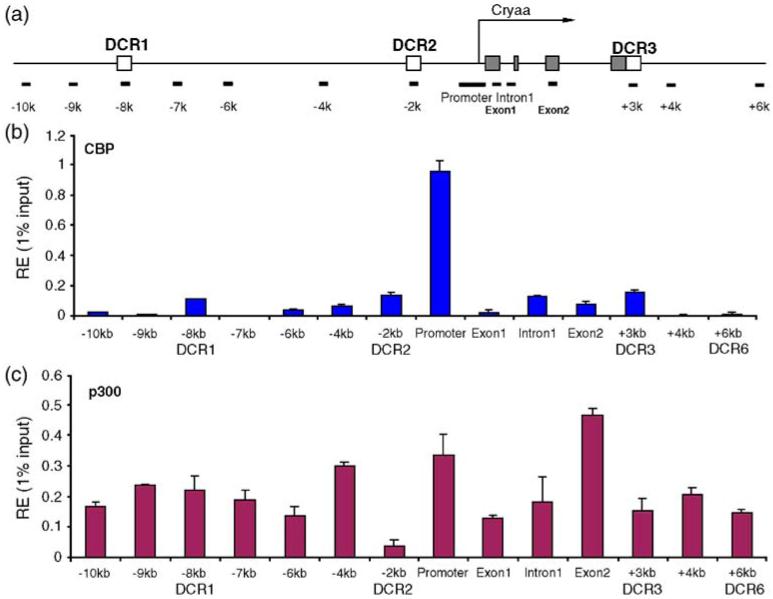
Interaction and distribution of CBP and p300 at the mouse αA-crystallin locus in chromatin of newborn lens. (a) A diagram of the αA-crystallin locus showing positions of PCR amplicons (see the legend to Figure 3(a)). (b) Distribution of CBP, (c) distribution of p300. The relative enrichment unit represents 1% of the input. The background for each region was subtracted as described.17
In contrast, in lens chromatin, we found high abundance of CBP at a single region, the promoter (see Figure 4(b); note the different y-axis scales in Figures 4(b) and (c), and 5(b) and (c)). Low abundance of CBP, comparable with its presence in DCR1 in lens epithelial chromatin, was found in other regions of the αA-crystallin locus. The distribution of p300 showed a more uniform pattern across the αA-crystallin locus. The exon2 region was an exception, with approximately twofold higher presence of p300 compared to the majority of adjacent regions of the locus. We conclude that the high expression of αA-crystallin in mouse newborn lens is marked by the localized presence of CBP in its promoter and more evenly spread p300 HAT in the locus. In contrast, in chromatin of cultured lens epithelial cells, both CBP and p300 are notably absent from the αA-crystallin promoter.
Figure 5.
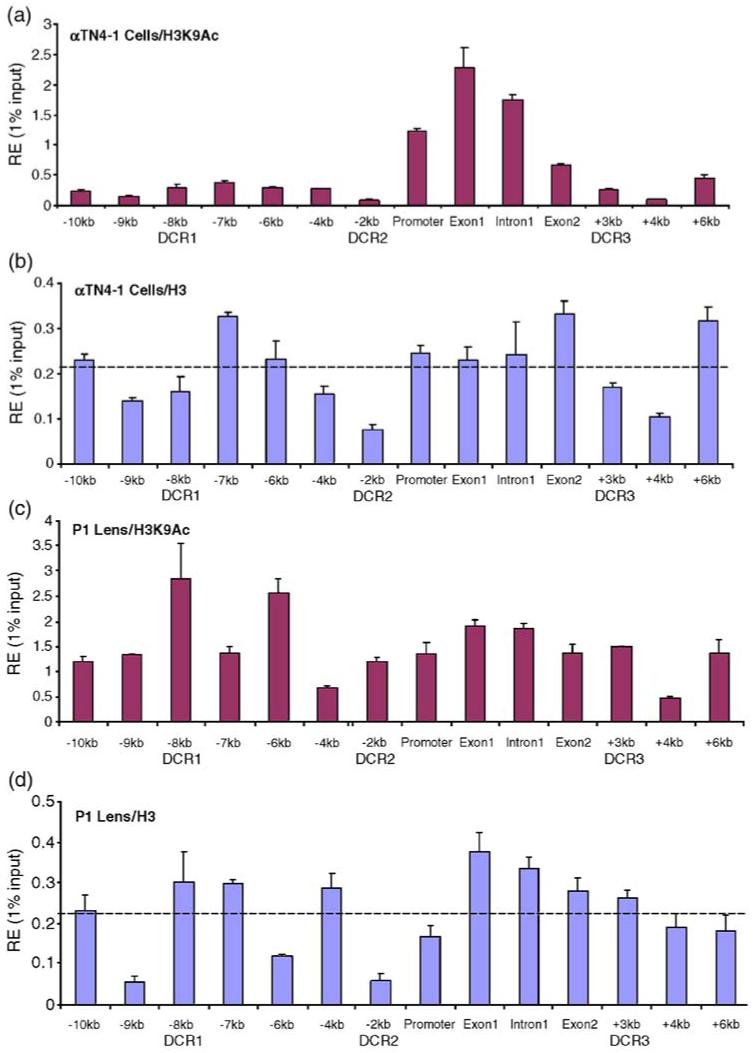
ChIP analyses of the distribution of the H3K9ac and total H3 at the mouse αA-crystallin locus. (a) Distribution of H3 K9ac and (b) total H3 signals in chromatin obtained from cultured lens epithelial cells. (c) Distribution of H3 K9ac and (d) total H3 signals in chromatin prepared from newborn mouse lenses. Dotted lines in (b) and (d) represent average signals of total H3 calculated from 14 positions across of the αA-crystallin locus.
Analysis of distribution of H3 K9ac and total H3
Studies of H3 K9 acetylation and distribution of total H3 around promoter regions have shown that many active promoters are depleted of nucleosomes.38-40 To test this possibility at the αA-crystallin promoter, we used antibodies specific to H3 K9ac and total histones H3 in ChIPs. In chromatin of cultured lens epithelial cells, a significant build-up of H3 K9ac signal was found from the promoter to DCR3 (see Figure 5(a)). In contrast, the signal of total H3 between the promoter and DCR3 was similar to the average signal of relative enrichment through the entire region tested (see Figure 5(b)). Three areas of reduced total H3 signal included -9/-8, -4/-2 and +3/+4 kb regions. In lens chromatin, we found H3 K9ac signal spread all over the 16 kb of the αA-crystallin locus with a distinct peak at DCR1 (see Figure 5(c)). The 5 kb promoter-containing region from -2 to +3 kb (DCR3) showed expanded H3 K9ac signals in lens chromatin compared to the corresponding region examined in chromatin of cultured lens epithelial cells (compare Figure 5(a) and (c)). In addition, the pattern of total H3 distribution in lens chromatin changed between lens epithelial cells and lens. A notable difference in “nucleosome density” was found in the -2 kb/promoter region (compare dips/valleys in Figure 5(b) and (d)), suggesting possible loss of a few nucleosomes in approximately 2 kb of DNA. In contrast, the transcribed region (exon 1 to +3 kb/DCR3) did not show any sign of reduced nucleosomal density compared to the average signal. Note that the average values of relative enrichments of total H3 were identical between the chromatin models used.
Discussion
The high level of mouse αA-crystallin expression in lens is associated with the formation of at least a 16 kb long lens-specific chromatin domain marked by the presence of acetylated H3 K9 core histone.17 The main goal of the present study was to determine which HATs interact with the mouse αA-crystallin locus to generate this H3 K9 acetylated chromatin domain, and whether distribution and/ or abundance of these HATs would indicate any specific role of these enzymes in the regulation of αA-crystallin gene expression. Here, we show that CBP and p300 are associated with this locus. We show also that the developmental expression pattern of αA-crystallin correlates with the expression pattern of CBP, as both these genes are highly upregulated in differentiating primary lens fibers. It is indeed CBP identified here that is the most abundant HAT localized specifically at the promoter region of αA-crystallin in lens chromatin (see Figure 4(b)). Thus, our data suggest that CBP might have some specific roles in lens that cannot be executed by p300. To answer these questions directly, conditional inactivation of CBP and p300 using available mice models at different stages of lens development are necessary.36
Earlier studies showed that loss of expression of αA-crystallin leads to cataracts in mouse models.41 In addition, mutations in human CRYAA cause congenital cataract formation.42,43 Mutations in human and mouse PAX6 and c-MAF, two genes that regulate αA-crystallin expression, also promote cataractogenesis.44,45 A study of Rubinstein-Taybi syndrome caused by mutations in CBP found 15 congenital cataracts in 81 case reports.31 In contrast, human p300 mutations do not show any ocular phenotype.46,47 Although we do not know the expression levels of αA-crystallin in these human cataractous lenses, we do know that expression of this gene is reduced dramatically in mouse c-Maf null lenses.48 Expression of αA-crystallin was not detected in the lens primordium with conditionally deleted Pax6.49 Thus, these studies support genetic links between transcription factors CBP, c-Maf and Pax6 and expression of αA-crystallin gene in mammalian lens.
The function of CBP and p300 HATs in crystallin gene regulation was examined using lens-specific expression of T-antigen mutant (E107KΔ) in transgenic mouse lens.23 This mutant has a compromised ability to bind pRb proteins yet can associate with CBP/p300 to abrogate their functions in α- and β/γ-crystallin gene expression.23,50 Mechanistically, direct interaction of CBP/p300 with c-Maf was established in co-immunoprecipitation assays in lens nuclear extracts.22 Co-transfection of c-Maf and CBP/p300 activated two- to threefold αA-crystallin promoter in cell culture reporter assays.22 Interestingly, replacement of c-Maf transactivation domain with the p300 HAT catalytic subdomain generated a chimeric transcription factor with activity similar to that found with the wild type c-Maf in reporter assays.22 The present data show the presence in vivo of both CBP and p300 at the mouse αA-crystallin locus and support the earlier studies of CBP/p300 in lens suggesting that these HATs control expression of this gene directly.22,23,50 We found CBP highly abundant at the promoter region, suggesting a selective local mechanism of recruitment. DNA-binding factors CREB, c-Maf and Pax6 were shown to interact with CBP/p300 in cellular extracts.22,51 It is only the promoter region where high abundance of both CREB and c-Maf co-localized in vivo in lens chromatin.17 We propose that efficient recruitment of CBP to the promoter requires simultaneous presence of abundant amounts of CREB and c-Maf, a condition found only in lens chromatin and not in chromatin from cultured lens epithelial cells.17
On the basis of ChIP studies of Brg1 and Snf2h in chromatin prepared from lens epithelial cells and newborn lenses, we proposed that promoter-specific localization of Snf2h, mediated via promoter-bound c-Maf, is required for moderate levels of expression of αA-crystallin gene.17 The present data showed insignificant presence of both CBP and p300 at the promoter region, although some other regions, such as DCR1, were occupied by both these HATs. As mouse lens epithelial cell line αTN4-1 used in these studies is an SV40 T-antigen-transformed cell line, some function(s) of CBP and p300 are inhibited.22,23,52 Nevertheless, expression of αA-crystallin in these cells is about 18-fold lower compared to the newborn lens, and this ratio corresponds to the expression levels of this gene found in lens epithelium compared to lens fibers.17,19 Thus, our data suggest that a moderate level of αA-crystallin expression in cultured lens epithelial cells does not require promoter-localized CBP and p300 HATs. In contrast, ChIP results of the distribution of CBP and p300 in chromatin prepared from newborn mouse lenses showed promoter-localized CBP. The signal in the promoter region was approximately ten times higher compared to the average signals detected at remaining 13 regions examined. This signal was comparable/slightly higher with the highest signal of CBP found at DCR1 in the chromatin of lens epithelial cells. Studies of p300 in lens chromatin identified rather even distribution of this enzyme at the αA-crystallin locus, with exon2 showing about twofold increased abundance over the majority of the locus. These patterns of CBP and p300 suggest that promoter-enriched c-Maf and CREB selectively recruit CBP to the promoter. This possibility can be tested experimentally by analysis of chromatin from lenses with conditionally inactivated CREB and c-Maf genes.
In conclusion, the Cryaa promoter occupancy in lens epithelial cells is marked by the local presence of ATP-dependent chromatin remodeling enzyme Snf2h,17 and the notable absence of CBP and p300 (see Figure 3). Analysis of chromatin prepared from newborn lenses showed the promoter-specific presence of CBP on the “background” of locus-spread CBP and p300 (see Figure 4) and at least a 16 kb long lens-specific H3 K9 acetylation domain (see Figure 5). The presence and distribution of other acetylated H3 species, such as K18ac and K23ac, H2A K5ac, H2B K12ac and K15ac, and H4 K5ac and K8ac, at the αA-crystallin locus remain to be determined. Acetylated lysine residues in histones can serve for recruitment of bromodomain-containing proteins such as Brg1.9,53 High abundance of Brg1 is indeed found through the entire 16 kb region of the mouse αA-crystallin locus.17 Thus, our model suggests that after the initial chromatin remodeling catalyzed by Snf2h, recruitment of CBP to the promoter via c-Maf22 and CREB25,54 occurs in differentiating lens fibers, followed by spreading the H3 K9ac domain that stimulates recruitment of Brg1. The availability of floxed alleles in mouse genes encoding Brg1,55 CBP,36 CREB,56 and p300,36 and a variety of cre-lines expressed in differentiating lens in combination with chromatin studies will be useful to validate the above model experimentally.
Materials and Methods
Quantitative reverse transcriptase-polymerase chain reaction
Two days old rat lenses were microdissected into lens epithelium and fiber cells. Rat lenses were used in lieu of mouse lenses as their microdissection generates more materials for RNA isolation. Total RNA of lens epithelial and fiber cells were isolated and cDNA was amplified as described.19 Expression levels of transcripts encoding B2M, HPRT, G6PD and SDHA were pooled together as the internal control for normalization.57,58 The quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) primers were synthesized by Invitrogen (Gaithersburg, MD):
-
CBP forward:
5′-AAACCATGAGTGAACGCCTC
and reverse:
5′-TGAACACTCGTTGCAAGTCC
-
p300 forward:
5′-AGTGCAAACACCATGTGGAA
and reverse:
5′-ATGGGTGTGGCTCTTTGTGT
-
B2M forward:
5′-CACTCTGAAGGAGCCCAAAA
and reverse:
5′-CTGGTCCAGATGATTCAGAGC
-
HPRT forward:
5′-TGTTGTTGGATATGCCCTTG
and reverse:
5′-CCGCTGTCTTTTAGGCTTTG
-
G6PD forward:
5′-CCCATCCCGTATGTCTATGG
and reverse:
5′-TGGAAGCCCACTCTCTTCAT
-
SDHA forward:
5′-AACACTGGAGGAAGCACACC
and reverse:
5′-GCACAGTCAGCCTCATTCAA
Immunofluorescence
Primary antibodies used for immunofluorescence are anti-CBP (A-22) (1:100 (v/v) dilution; Santa Cruz Biotechnology, Santa Cruz, CA), anti-p300 (C-20) (1: 500 (v/v) dilution; Santa Cruz Biotechnolgy) and Alexa Fluor 568 goat anti-rabbit IgG (1: 300 (v/v) dilution; Molecular Probes, OR) was used as the secondary antibody. Embryos were fixed in 4% (v/v) paraformaldehyde at 4 °C for 5-12 h, and then cryoprotected in 30% (w/v) sucrose. Cryostat sections 5 μm thick were used for CBP staining. For p300 staining, fresh embryos were embedded and 5 μm thick cryosections were fixed in acetone for 10 min at 4 °C. Immunoflouroscence was conducted as described.59 Briefly, sections were washed twice in PBS and blocked for 30 min with Image-iT™ FX signal enhancer (Molecular Probes, Eugene, OR). Sections were then washed twice in PBS, incubated with the primary antibody at the concentration described above, in 1% Bovine Albumin Fraction Solution (Invitrogen, Grand Island, NY) and 0.1% (v/v) Triton X in PBS overnight at 4 °C. The second day, sections were washed twice in PBS and incubated with the secondary antibody in a humidified chamber for 30 min. After washing in PBS, sections were mounted with fluorescence preserve mounting medium (Vector Laboratories, Burlingame, CA) and imaged using a Leica AOBS laser scanning confocal microscope.
Chromatin immunoprecipitations (ChIPs)
Antibodies used for ChIPs are: anti-CBP (A-22) and anti-p300 (N-15) from Santa Cruz Biotechnology (St. Cruz, CA); anti-Histone H3 from Abcam (Cambridge, UK); anti-acetyl-histone H3 (Lys9) ChIP grade from Upstate (Lake Placid, NY); normal rabbit IgG from Oncogene Research Products (San Diego, CA). ChIPs were performed in mouse lens epithelial αTN4-1 cells and in microdissected mouse P1 lens as described.17,19 Briefly, formaldehyde-crosslinked chromatin was sonicated to generate chromatin comprised mostly of mono-, di- and trinucleosomes. Antibodies (2 μg) were incubated with protein A and G beads (Sigma, St. Louis, MI) for 5 h and then incubated with the precleared chromatin overnight at 4 °C. After three washes and treatment with proteinase K overnight, chromatin fragments were eluted, purified with the Qiagen QIAquick PCR purification kit (Qiagen, Germantown, MD) and analyzed by quantitative PCR as described.17,19 A serial dilution of input (e.g. 0.05%, 0.2%, 1% and 5%) was used to generate a standard curve for each probe in every ChIP experiment. The Ct value of quantitative PCR was normalized to the percentage of input according to the standard curve. The relative enrichments and standard deviation (SD) were calculated from at least two independent ChIPs, and every ChIP was tested by real time PCR in triplicate. Background from independent non-specific antibody (IgG) ChIP was subtracted from each probe and the final signals were normalized and presented versus 1% input.17,60
In addition to the published primers,17 we included primers (exon1 and intron 1) that improve coverage of the αA-crystallin locus. Within the -500 bp to +100 bp promoter region, five primers were used to fine map the chromatin distribution changes. The averages of these primers were calculated and are shown in Figures 3 to 5. There are five additional primer sequences within the promoter region:
-
forward
5′-AGTCATGTCGGGAAGACCTG
and reverse
5′-TGTGTGCTGGATGTGGTTCT
-
forward
5′-GGAGGGTGCCTCAGTACAAT
and reverse
5′-AGCAGCAGAGGCTCTCAGAC
-
forward
5′-CCCGAGCTGAGCATAGACAT
and reverse
5′-AGTCAGACAGGAGCCTCTGG
-
forward
5′-CTACCTCTCCCCACCTGTGA
and reverse
5′-GCCAAGGGACATCACTGTTT
-
forward
5′-TGAGCATTCCAGCTGCTGAC
and reverse
5′-CCTGCACAGAATGGAGGAAT
as well as exon 1 forward
5′-CCTTCCTGTCTTCCACCATC
and reverse
5′-GCAGCTAGGAGGAACCAGTG
and intron 1 forward
5′-GACCCTGTGCTCTCCTCAAG
and reverse
5′-TGGCTTCAGCATAGGCATGT
Acknowledgements
We thank Dr Tomas Stopka for critical reading of the manuscript. This work was supported by NIH grants EY12200 and 14237. A.C. is a recipient of the Irma T. Hirschl Career Scientist Award.
Glossary
Abbreviations used
- CBP
CREB-binding protein
- HATs
histone acetyltransferases
- ChIP
chromatin immunoprecipitation
- qRT-PCR
quantitative reverse transcriptase-polymerase chain reaction.
References
- 1.Wu C. Chromatin remodeling and the control of gene expression. J. Biol. Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 2.Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding-wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 3.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 4.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nature Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 5.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 6.Nightingale KP, O’Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr. Opin. Genet. Dev. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Marmorstein R. Structure of histone acetyltransferases. J. Mol. Biol. 2001;311:433–444. doi: 10.1006/jmbi.2001.4859. [DOI] [PubMed] [Google Scholar]
- 8.Muchardt C, Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol. 1999;293:187–198. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 9.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? BioEssays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 10.Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 11.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 12.Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 13.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog. Biophys. Mol. Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Bhat SP. Crystallins, genes and cataract. Prog. Drug Res. 2003;60:205–262. doi: 10.1007/978-3-0348-8012-1_7. [DOI] [PubMed] [Google Scholar]
- 15.Wistow GJ, Piatigorsky J. Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu. Rev. Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 16.Robinson ML, Overbeek PA. Differential expression of alpha A- and alpha B-crystallin during murine ocular development. Invest. Ophthalmol. Vis. Sci. 1996;37:2276–2284. [PubMed] [Google Scholar]
- 17.Yang Y, Stopka T, Golestaneh N, Wang Y, Wu K, Li A, et al. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cvekl A, Kashanchi F, Sax CM, Brady JN, Piatigorsky J. Transcriptional regulation of the mouse alpha A-crystallin gene: activation dependent on a cyclic AMP-responsive element (DE1/CRE) and a Pax-6-binding site. Mol. Cell Biol. 1995;15:653–660. doi: 10.1128/mcb.15.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Cvekl A. Tissue-specific regulation of the mouse alphaA-crystallin gene in lens via recruitment of Pax6 and c-Maf to its promoter. J. Mol. Biol. 2005;351:453–469. doi: 10.1016/j.jmb.2005.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kantorow M, Cvekl A, Sax CM, Piatigorsky J. Protein-DNA interactions of the mouse alpha A-crystallin control regions. Differences between expressing and non-expressing cells. J. Mol. Biol. 1993;230:425–435. doi: 10.1006/jmbi.1993.1160. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Cvekl A. Large Maf transcription factors: cousins of AP-1 proteins and important regulators of cellular differentiation. Einstein J. Biol. Med. 2007;23:2–11. doi: 10.23861/ejbm20072347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q, Dowhan DH, Liang D, Moore DD, Overbeek PA. CREB-binding protein/p300 co-activation of crystallin gene expression. J. Biol. Chem. 2002;277:24081–24089. doi: 10.1074/jbc.M201821200. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q, Liang D, Fromm LD, Overbeek PA. Inhibition of lens fiber cell morphogenesis by expression of a mutant SV40 large T antigen that binds CREB-binding protein/p300 but not pRb. J. Biol. Chem. 2004;279:17667–17673. doi: 10.1074/jbc.M311678200. [DOI] [PubMed] [Google Scholar]
- 24.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 25.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 26.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 27.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 28.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 29.Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 30.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 31.van Genderen MM, Kinds GF, Riemslag FC, Hennekam RC. Ocular features in Rubin-stein-Taybi syndrome: investigation of 24 patients and review of the literature. Br. J. Ophthalmol. 2000;84:1177–1184. doi: 10.1136/bjo.84.10.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennekam RC, Van Den Boogaard MJ, Sibbles BJ, Van Spijker HG. Rubinstein-Taybi syndrome in The Netherlands. Am. J. Med. Genet. Suppl. 1990;6:17–29. doi: 10.1002/ajmg.1320370604. [DOI] [PubMed] [Google Scholar]
- 33.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 34.Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 35.Partanen A, Motoyama J, Hui CC. Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int. J. Dev. Biol. 1999;43:487–494. [PubMed] [Google Scholar]
- 36.Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol. Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 38.Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells-insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr. Opin. Cell. Biol. 2006;18:291–298. doi: 10.1016/j.ceb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 40.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 41.Brady JP, Garland D, Duglas-Tabor Y, Robison WG, Jr, Groome A, Wawrousek EF. Targeted disruption of the mouse alpha A-crystallin gene induces cataract and cytoplasmic inclusion bodies containing the small heat shock protein alpha B-crystallin. Proc. Natl Acad. Sci. USA. 1997;94:884–889. doi: 10.1073/pnas.94.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graw J. The genetic and molecular basis of congenital eye defects. Nature Rev. Genet. 2003;4:876–888. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- 43.Graw J, Klopp N, Illig T, Preising MN, Lorenz B. Congenital cataract and macular hypoplasia in humans associated with a de novo mutation in CRYAA and compound heterozygous mutations in P. Graefes Arch. Clin. Expt. Ophthalmol. 2006;244:912–919. doi: 10.1007/s00417-005-0234-x. [DOI] [PubMed] [Google Scholar]
- 44.Hever AM, Williamson KA, van Heyningen V. Developmental malformations of the eye: the role of PAX6, SOX2 and OTX2. Clin. Genet. 2006;69:459–470. doi: 10.1111/j.1399-0004.2006.00619.x. [DOI] [PubMed] [Google Scholar]
- 45.Graw J. Congenital hereditary cataracts. Int. J. Dev. Biol. 2004;48:1031–1044. doi: 10.1387/ijdb.041854jg. [DOI] [PubMed] [Google Scholar]
- 46.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong JM, et al. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 47.Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- 49.Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Q, Ash JD, Branton P, Fromm L, Overbeek PA. Inhibition of crystallin expression and induction of apoptosis by lens-specific E1A expression in transgenic mice. Oncogene. 2002;21:1028–1037. doi: 10.1038/sj.onc.1205050. [DOI] [PubMed] [Google Scholar]
- 51.Hussain MA, Habener JF. Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J. Biol. Chem. 1999;274:28950–28957. doi: 10.1074/jbc.274.41.28950. [DOI] [PubMed] [Google Scholar]
- 52.Cho S, Tian Y, Benjamin TL. Binding of p300/CBP co-activators by polyoma large T antigen. J. Biol. Chem. 2001;276:33533–33539. doi: 10.1074/jbc.M102906200. [DOI] [PubMed] [Google Scholar]
- 53.Hassan AH, Awad S, Al-Natour Z, Othman S, Mustafa F, Rizvi TA. Selective recognition of acetylated histones by bromodomains in transcriptional co-activators. Biochem. J. 2007;402:125–133. doi: 10.1042/BJ20060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 55.Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Expt. Med. 2003;198:1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, et al. Disruption of CREB function in brain leads to neurodegeneration. Nature Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 57.Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. (Lond) 2005;109:365–379. doi: 10.1042/CS20050086. [DOI] [PubMed] [Google Scholar]
- 58.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F.Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes Genome Biol 20023 RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed NA, Oh DJ, Czymmek KJ, Duncan MK. An immunohistochemical method for the detection of proteins in the vertebrate lens. J. Immunol. Methods. 2001;253:243–252. doi: 10.1016/s0022-1759(01)00374-x. [DOI] [PubMed] [Google Scholar]
- 60.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]


