Abstract
During the last 30 years, several alterations to the standard genetic code have been discovered in various bacterial and eukaryotic species. Sense and nonsense codons have been reassigned or reprogrammed to expand the genetic code to selenocysteine and pyrrolysine. These discoveries highlight unexpected flexibility in the genetic code, but do not elucidate how the organisms survived the proteome chaos generated by codon identity redefinition. In order to shed new light on this question, we have reconstructed a Candida genetic code alteration in Saccharomyces cerevisiae and used a combination of DNA microarrays, proteomics and genetics approaches to evaluate its impact on gene expression, adaptation and sexual reproduction. This genetic manipulation blocked mating, locked yeast in a diploid state, remodelled gene expression and created stress cross-protection that generated adaptive advantages under environmental challenging conditions. This study highlights unanticipated roles for codon identity redefinition during the evolution of the genus Candida, and strongly suggests that genetic code alterations create genetic barriers that speed up speciation.
Keywords: Candida, gene expression, genetic code alterations, mRNA mistranslation, tRNA
Introduction
The discovery of alterations and expansions to the standard genetic code abolished the hypothesis of a frozen and universal genetic code (Crick, 1968) and raised important new questions, namely ‘(i) how do organisms survive codon identity redefinition? (ii) how and why do certain codons disappear while others alter their identity? and (iii) what is the evolutionary and physiological impact of codon identity redefinition?' Neutral (Codon Capture) evolutionary mechanisms postulate that highly biased G+C pressure can force codons to disappear and such unassigned codons can be captured by natural or mutant tRNAs from non-cognate codon families (Osawa and Jukes, 1989). This neutral mechanism explains some mitochondrial genetic code changes and unassignment of the CGG and AGA/AUA codons in Mycoplasma capricolum and Micrococcus luteus, which have 25% and 74% GC, respectively (Ohama et al, 1990; Oba et al, 1991).
Codon identity redefinition can also be driven by selection through codon misreading (Schultz and Yarus, 1994). This requires structural change of the translational machinery, in particular of tRNAs, aminoacyl-tRNA synthetases and release factors, which decrease codon decoding fidelity and generate codons with more than one identity (Schultz and Yarus, 1994). These selection-driven mechanisms support the expansion of the genetic code to selenocysteine (twenty-first amino acid) and pyrrolysine (twenty-second amino acid) (Chambers et al, 1986; Hao et al, 2002). Selenocysteine is used in both prokaryotic and eukaryotic selenoproteins and its insertion into the genetic code required reprogramming of UGA stop codons by novel translation elongation factors (SelB-prokaryotes; EF-sec and SBP2-eukaryotes), a new tRNA (tRNASec) and a selenocysteine mRNA insertion element (SECIS) (Namy et al, 2004). L-Pyrrolysine is used in the archeon Methanosarcina barkeri through reprogramming of UAG-stop codons in methylamine methyltransferase genes (Theobald-Dietrich et al, 2005). Codon ambiguity also explains the identity alterations of several sense and non-sense codons in bacteria, mitochondria and in the cytoplasm of eukaryotes. For example, the decoding of leucine CUN (N any nucleotide) codons as threonine in yeast mitochondria and the decoding of leucine CUG codons as serine in the cytoplasm of various Candida species (Schultz and Yarus, 1994; Massey et al, 2003; Miranda et al, 2006).
Genetic code alterations driven by codon ambiguity are most interesting because they should block lateral gene transfer and sexual reproduction. The former is highlighted in Candida albicans, where serine CUG decoding prevents the expression of functional wild-type green fluorescent protein (GFP), whose gene contains a single CUG codon, and of many other CUG-containing reporter genes, namely Escherichia coli β-galactosidase, Renilla reniformis luciferase, and Saccharomyces cerevisiae orotidine-5′-phosphate decarboxylase (Ura3p) (Cormack et al, 1997). Additionally, codon identity redefinition reshapes protein primary structure and may create novel protein functionalities, which on one hand may speed up the evolution of new phenotypes, but on the other may generate proteome and genetic incompatibilities to sexual reproduction. The latter is highlighted by the inability of using S. cerevisiae genes to complement C. albicans homologous gene disruptions (Sugiyama et al, 1995).
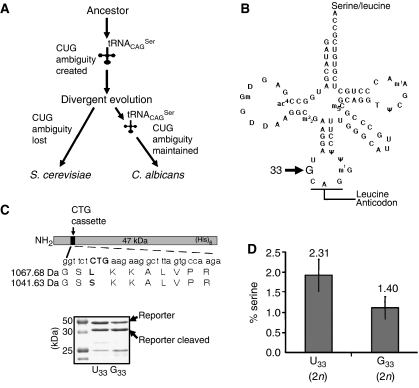
We are using C. albicans as a model system to elucidate the molecular mechanism of evolution of serine CUG decoding in various Candida species and to understand the cellular and evolutionary consequences of altering the genetic code (Santos et al, 1996, 1999; Massey et al, 2003; Silva et al, 2004; Miranda et al, 2006). In C. albicans and other Candida species, the leucine CUG codon is decoded as serine by a novel serine tRNA that appeared 272±25 million years ago in the ancestor of yeasts, before separation of Saccharomyces and Candida genera (Figure 1A; Massey et al, 2003). It originated through the insertion of an adenosine in the intron of a tRNACGASer gene (Massey et al, 2003; Miranda et al, 2006), a mutation that created a hybrid tRNA molecule containing the body of serine and anticodon (5′-CAG-3′) of leucine-tRNAs (tRNACAGSer) (Figure 1B). This unique tRNA could decode leucine CUG codons as serine (Santos and Tuite, 1995; Suzuki et al, 1997) and competed for approximately 100 million years with wild-type tRNACAGLeu for CUG decoding (Figure 1A). It was selected in Candida and eliminated in Saccharomyces lineages (Massey et al, 2003) and its atypical structure was gradually reshaped with functional consequences (Santos et al, 1997; Perreau et al, 1999). In particular, two novel mutations in the anticodon loop changed the conserved uridine at position 33 (U33) to guanosine (G33) and adenosine 37 (A37) to guanosine 37 (G37) (Figure 1B). These two mutations modulate the leucine mischarging and CUG decoding efficiency of tRNACAGSer (Santos et al, 1996; Suzuki et al, 1997; Miranda et al, 2006).
Figure 1.
Reconstruction model of the Candida genetic code alteration. (A) Redefinition of the identity of the CUG codon from leucine to serine in Candida started with a novel serine tRNA (tRNACAGSer) and evolved gradually over the last 272±25 million years. tRNACAGSer disappeared and the cognate leucine CUG decoder (tRNACAGLeu) was maintained in the S. cerevisiae lineage (standard genetic code), while the converse occurred in the C. albicans lineage (altered genetic code). (B) The tRNACAGSer contains guanosine at position 33 (G33), which is a conserved position occupied by uridine (U33; U-turn) in other tRNAs. (C) The upper panel shows a diagram of the reporter system used to quantify serine misincorporation at CUG codons in vivo in S. cerevisiae. A CUG cassette inserted in the CaPGK gene was flanked by two thrombin cleavage sites to facilitate the purification of the short reporter peptide encoded by the cassette. The recombinant protein was expressed and purified from S. cerevisiae cultures using nickel affinity chromatography, and was then cleaved with thrombin for 16 h at 26°C, in solution. The resulting peptides were analysed by mass spectrometry. The lower panel shows a 12% SDS–PAGE of the reporter protein. (D) Serine and leucine incorporation at the CUG position (see panel C) was determined by quantitative MRM methodologies using a hybrid quadrupole/linear ion-trap mass spectrometer. Synthetic peptides with sequences identical to those of the serine and leucine peptides shown in panel C were used as external controls and to build the calibration curves used for quantification.
In the present study, we have reconstructed the early stages of CUG identity redefinition from leucine to serine in vivo in S. cerevisiae (Figure 1A). Such genetic manipulation was not lethal, but affected sporulation and mating severely and locked yeast in a diploid state. It altered the expression of molecular chaperones, cell wall and membrane proteins, increased proteasome activity and accumulation of glycogen and trehalose. These data support the hypothesis that this genetic code change altered physiology and created a diploid yeast lineage that gave rise to the genus Candida. It highlights unanticipated roles for genetic code alterations in speciation and as a hidden source of genetic and phenotypic diversity.
Results
Partial CUG identity redefinition affected sexual reproduction
We have already shown that the C. albicans tRNACAGSercan be expressed in S. cerevisiae from single-copy plasmids and that it is correctly processed and aminoacylated (Santos et al, 1996). Here, we used these plasmid-transformed strains to elucidate the impact of CUG identity redefinition on S. cerevisiae gene expression and physiology. The tRNACAGSer gene was also integrated into the genome of S. cerevisiae to evaluate the impact of this genetic code alteration on sexual reproduction. These S. cerevisiae clones expressed their own tRNAUAGLeu plus the C. albicans tRNACAGSerand incorporated leucine or serine randomly at CUG positions on a genome-wide scale. This mimicked the CUG ambiguity present in the Candida ancestor, where a cognate tRNALeu plus the novel mutant tRNACAGSer also competed for CUG codons (Figure 1A; Massey et al, 2003). Two versions of the C. albicans tRNACAGSer were used: the C. albicans wild-type tRNACAGSer containing G33, which is an inefficient decoder that appeared late in the evolutionary pathway of the genetic code alteration, plus a mutant tRNACAGSer containing the canonical U at position 33 (U33), which is an efficient decoder and represents the primordial tRNA (Santos et al, 1996; Perreau et al, 1999). The latter allowed us to confirm that the higher decoding efficiency of U33 tRNACAGSer was not an impediment to CUG identity redefinition. The levels of serine misincorporation in vivo at CUG positions were determined by mass spectrometry using a CUG-reporter system (Figure 1C; Supplementary Figures 1–3). In diploid cells, serine incorporation was 1.4% and 2.31% for the G33 and U33 tRNAGCAGSer, respectively (Figure 1D). Considering that background decoding error in vivo in yeast is in the order of 0.001% (Stansfield et al, 1998), those values represent 1400- and 2310-fold increase in decoding error and provide important insight into the level of ambiguity experienced by the Candida ancestor during the initial stages of the CUG identity change (Massey et al, 2003).
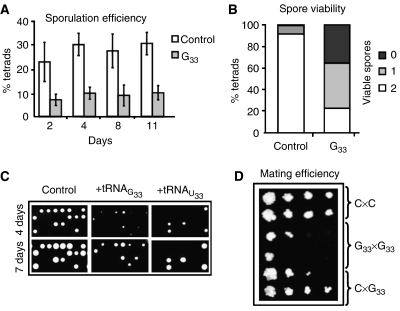
We have investigated the impact of partial CUG identity redefinition in sexual reproduction using S. cerevisiae clones expressing U33 tRNACAGSer and G33 tRNACAGSer. The inefficient G33 tRNACAGSer decoder decreased sporulation efficiency by 30% (Figure 2A), while clones expressing the efficient U33 tRNACAGSer decoder showed very high sporulation variability; 55% of the clones sporulated normally, 25% did so very poorly and 20% did not sporulate at all (data not shown). The impact of CUG ambiguity on fertility was also tested by germinating spores of dissected asci. In selective media (YEPD–geneticin), 95% and 44% of control and G33 tRNACAGSerspores were viable, respectively, but 42% of G33 tRNACAGSerasci had one viable spore and 23% had two viable spores (Figure 2B and C). In 80% of U33 tRNACAGSer clones that sporulated, 76% of the asci produced spores that did not germinate, 9% had one viable spore and only 15% had two viable spores (data not shown). Since CUG ambiguity is toxic and creates genetic instability (see below), the integrity of G33 and U33 tRNACAGSer genes was verified by PCR amplification and resequencing of the respective DNA fragments (8 G33 and 17 U33 in total), isolated from the colonies of germinated spores. Spores expressing G33 tRNACAGSer had the correct tRNA gene sequence, while all the spores expressing U33 tRNACAGSercontained mutations in positions that mapped to the extra-loop and anticodon stem of the mature tRNA (data not shown). That is, U33 tRNACAGSer is lethal in haploid backgrounds, since only cells containing mutations in U33 tRNACAGSer gene could sporulate. These mutations also explained the fast growth of the spores (Figure 2C) and the very high sporulation variability observed in diploid cells expressing U33 tRNACAGSer (see above). It also confirmed previous studies showing that haploid S. cerevisiae cannot be transformed with plasmids carrying U33 tRNACAGSer (Santos et al, 1996).
Figure 2.
Genetic code alterations act as a barrier for sexual reproduction. The ability of S. cerevisiae cells expressing C. albicans G33 tRNACAGSerto reproduce sexually was determined by their sporulation and mating efficiencies. (A) The number of tetrads produced by diploid ambiguous cells decreased by 30% when compared with control cells, indicating that sporulation efficiency was lower in CUG ambiguous cells. (B) Spore viability was reduced in ambiguous cells, since most tetrads yielded only one (42%) or no viable spores (35%), while tetrads from control cells produced mainly two viable spores (92%). (C) Ambiguous cells produced fewer viable spores and spores grew slower, as shown by growth on solid selective medium. The U33 tRNA spores shown had mutations in the tRNACAGSer gene. (D) Haploid control and ambiguous cells with opposite mating types were mixed and serial dilutions of the mixtures were plated onto selective media. C × C, G × G and C × G indicate the crosses between the control cells, or ambiguous G33 tRNA cells or control and ambiguous G33 tRNA cells, respectively. The reduced amount of diploids produced by crossing ambiguous (G33 tRNA) cell lines showed that mating efficiency was also negatively affected by CUG ambiguity. In all cases, the U33 tRNACAG was lethal in haploid backgrounds. Alternatively, its gene acquired mutations that inactivated the U33 tRNA.
Since U33 tRNACAGSer was lethal in haploid backgrounds, the impact of ambiguous CUG decoding on mating was evaluated using clones expressing the G33 tRNACAGSer gene. Crosses of G33 (MATa) × G33 (MATα) displayed low mating efficiency, which increased slightly for G33 (MATa) × control (MATα) crosses (Figure 2D). These mating differences are explained by gene dosage effects because diploids of G33 (MATα) × G33 (MATa) crosses had two copies of the G33 tRNACAGSer gene, while diploids of control (MATα) × G33 (MATa) crosses had a single copy of this tRNA gene and tRNA copy number determines tRNA abundance and CUG ambiguity levels.
Interestingly, flow cytometry analysis showed that expression of U33 and G33 tRNACAGSer in S. cerevisiae increased ploidy (Figure 3A and B). G33 tRNACAGSer induced ploidy increase from N to 2N in 56% of the haploid clones, while U33 tRNACAGSerincreased it from 2N to 4N in 50% of diploid clones (Figure 3C). Expression of G33 tRNACAGSer in diploid cells also shifted DNA content peaks slightly, suggesting that the cell population contained a significant number of aneuploid cells. Similar DNA peak shifts were observed in clones expressing U33 tRNACAGSer (Figure 3B). Expression of G33 and U33 tRNACAGSeralso generated highly heterogeneous colony and cell morphologies and increased cell size significantly, which is consistent with ploidy increase (Figure 3D). Nuclear DAPI staining showed the presence of micronuclei and two or more large nuclei in cells expressing G33 and U33 tRNACAGSer. Some daughter cells did not have nuclei, suggesting disruption of chromosome segregation or aberrant nuclear division during mitosis (Figure 3D).
Figure 3.
Genetic code alterations induce ploidy variation. Flow cytometry analysis of haploid (A) and diploid (B) S. cerevisiae cell lines expressing the C. albicans G33 or U33 tRNACAGSer had a general increase of the nuclear DNA content, providing evidence of polyploidy and aneuploidy events in CUG ambiguous cells. (C) Ploidy shift was observed in 56% of the haploid G33 tRNACAGSer clones and 50% of diploid U33 tRNACAGSer clones tested by flow cytometry analysis. (D) Heterogeneity of the ambiguous cell population is shown by the variability in colony, cell and bud size and shape. The increase in cell volume is consistent with polyploidization of the ambiguous clones. DAPI staining highlights ambiguous cells with two nuclei or without nucleus, suggesting the presence of polyploid and aneuploid cells.
CUG ambiguity altered gene expression and physiology
To shed further light on the impact of codon identity redefinition on physiology and evolution, gene expression was monitored in cells expressing G33 and U33 tRNACAGSer using DNA microarrays. Similar results were obtained for both tRNAs, although differences in the magnitude of changes were observed in some cases (Supplementary Table 1). Overall, DNA microarray profiling uncovered alterations in the expression of genes belonging to the stress response, carbohydrate and amino-acid metabolism, cell wall structure, protein synthesis and degradation (Figure 4A).
Figure 4.
Genetic code alterations reprogramme gene expression. (A) Transcriptome analysis indicates the percentage of genes with altered expression levels in ambiguous strains. Genes whose expression was both up- and downregulated by CUG ambiguity were grouped according to their functions. The genes that are included in the stress group from the pie-chart were further divided into the functional categories displayed on the adjacent column. (B) Proteome data show the percentage of proteins whose expression was altered in S. cerevisiae cells expressing C. albicans U33 tRNACAGSer, distributed by functional categories. The proteins that are included in the stress group from the pie-chart were further divided into the functional categories displayed on the adjacent column. Both analyses indicate that genetic code ambiguity extensively remodelled gene expression, altered the expression of genes and proteins belonging to the stress response, protein synthesis, folding and degradation pathways, and general metabolism.
Expression of 58 genes was upregulated, whereas that of 21 genes was downregulated (Supplementary Table 1). Most upregulated genes were stress-response genes (34%), namely molecular chaperones HSP12 (12.4-fold), HSP26 (5.8-fold), HSP70 (SSA4) (3.9-fold) and HSP104 (2.4-fold). CUG ambiguity also resulted in the upregulation of drug-resistance genes, namely copper-binding metallothionein genes CRS5 (3 fold) and CUP1 (2.9-fold), as well as the membrane ABC (ATP-binding cassette) transporter gene PDR5 (2.2-fold). Other changes involved general stress-response genes, namely the multistress response protein genes DDR2 (9 fold), HSP42 (3.9-fold), HSP30 (3.7-fold), the stress-induced methylglyoxal reductase gene GRE2 (2.1-fold), the GPI-anchored cell-wall glycoprotein genes SED1 and SPI1 (2.8-fold) and the cell-wall genes TIP1 (6.2-fold), and CWP2 (4 fold), which encode major structural mannoproteins (Table I). These gene expression alterations are in line with our previous results showing that CUG ambiguity increases tolerance to arsenite, cadmium, cycloheximide, ethanol, oxidants and salt (Santos et al, 1999). Overexpression of the high-affinity inorganic phosphate transporter (PHO84; 17.6-fold); the acid phosphatases PHO12 (3.4-fold), PHO11 (3.1-fold) and PHO5 (2.9-fold); the hexokinase I (HXK1; 6.2-fold); glucokinase (GLK1; 2.3-fold); glycogen phosphorylase (GPH1; 4.7-fold) and the components of the trehalose-6-phosphate synthetase/phosphatase complex (TSL1 and TPS1; 3.2- and 2.1-fold) indicated that CUG ambiguity affected phosphate, glucose, glycogen and trehalose metabolism (see below).
Table 1.
Selected genes whose expression was altered by CUG ambiguity
| Function | Gene | Fold | Function | Gene | Fold |
|---|---|---|---|---|---|
| Chaperones | HSP12 | 12.4 | Carbohydrate metabolism | HXK1 | 6.2 |
| HSP26 | 5.8 | TSL1 | 3.2 | ||
| SSA4 | 3.9 | GLK1 | 2.3 | ||
| HSP104 | 2.4 | TPS1 | 2.1 | ||
| Stress response | CRS5 | 3.0 | Protein synthesis | RPL22B | −2.1 |
| CUP1 | 2.9 | YDR341C | −2.1 | ||
| PNC1 | 2.1 | RPL9B | −2.0 | ||
| Cell wall and transporters | PHO84 | 17.6 | Amino acid metabolism | PUT1 | 3.7 |
| PIR3 | 7.5 | LYS9 | −2.4 | ||
| TIP1 | 6.2 | SAM4 | −2.0 | ||
| CWP2 | 4.0 | ||||
| PDR5 | 2.2 | ||||
| Genes induced in ambiguous cells display positive fold variation and genes repressed are indicated by negative fold variation (FDR=0.001). | |||||
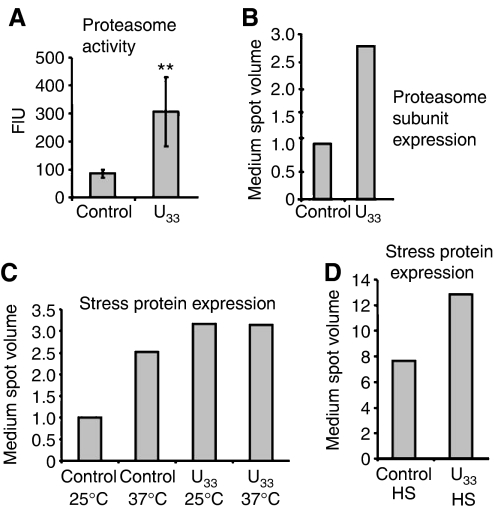
Since expression of G33 and U33 tRNACAGSer generated highly heterogeneous cell populations (Figure 3) and induced the stress response, which altered gene expression at the post-transcriptional level (DiDomenico et al, 1982), we wondered whether DNA microarray profiling was providing a complete view of the gene expression alterations induced by ambiguous CUG decoding. To clarify this question, quantification of protein expression was carried out using phosphorimaging of [35S]methionine-labelled proteins fractionated onto 2D-PAGE (Figure 4B; Supplementary Figure 4 and Table 2). Up-regulation of 43 proteins belonging to the stress response and protein degradation, and down-regulation of 34 proteins involved in amino-acid metabolism and protein synthesis was observed (Figure 4B). These data confirmed the DNA microarray data, but fold-induction differences were observed for several genes (Table II). For example, Hsp104p and Pnc1p were up-regulated 13.1- and 29.5-fold (Table II), while their mRNAs increased 2.4- and 2.1-fold only, respectively (Tables I and II). Two additional differences were found between the microarray and proteomics data. The latter, but not the former, showed up-regulation of several proteasome subunits, namely Rpn10p, Rpn12p, Pup2p and Scl1p (Table II), which was confirmed by increased proteasome activity (3.6-fold) in ambiguous cells (Figure 5A and B).
Table 2.
Selected proteins whose expression was altered by CUG ambiguity
| Function | Protein | Fold | P-value | Function | Protein | Fold | P-value |
|---|---|---|---|---|---|---|---|
| Chaperones | Hsp104 | 13.1 | — | Amino-acid metabolism | Met17 | −5.3 | 0.0004 |
| Ssa1 | 4.7 | 0.0007 | Aro8 | −5.1 | 0.000007 | ||
| Ssa2 | 2.5 | 0.0091 | Arg1 | −3.4 | 0.005 | ||
| Ssa4 | N | — | Lys9 | −2.8 | 0.006 | ||
| Stress | Pnc1 | 29.5 | 0.0154 | Leu2 | −2.3 | 0.0001 | |
| Response | Ahp1 | 3.4 | 0.0021 | ||||
| Carbohydrate metabolism | Glk1 | 5.0 | 0.0091 | Protein synthesis | Krs1 | −2.7 | 0.0011 |
| Hxk1 | N | — | Ssb1 | −2.1 | 0.0167 | ||
| Hor2 | N | — | Ssb2 | −2.1 | 0.0052 | ||
| Protein degradation | Rpn12 | 5.2 | 0.0230 | ||||
| Rpn10 | 3.6 | 0.0065 | |||||
| Pup2 | 3.6 | 0.0010 | |||||
| Scl1 | 3.6 | 0.0228 | |||||
| Proteins whose expression was altered in ambiguous cells, indicating the respective fold variation and statistical significance. Proteins induced display positive fold variation and proteins repressed are represented by negative fold variation; N stands for new spots (proteins that were not expressed in the control condition and, therefore, their fold variation could not be accurately determined). An average fold represents proteins that are present in the 2D gel by more than one spot. | |||||||
Figure 5.
Genetic code alterations reprogramme the stress response. (A) Proteasome activity increased 3.6-fold in S. cerevisiae cells expressing C. albicans tRNACAGSer (U33), as shown by enhanced proteolysis of the chymotrypsin-like substrate SucLLVY-AMC. The results are expressed as mean±s.d. of 4–6 independent experiments (**P<0.01 by Student's t-test). Fluorescence intensity (FIU) is shown in arbitrary units. (B) Expression of proteasome subunits (Supplementary Table 3) was threefold induced by CUG ambiguity, as measured by proteome analysis. Control (C) and ambiguous (U33) cells were grown at 25°C (25), 37°C (37) or heat shocked (HS). Proteins were labelled in vivo with L-[35S]methionine and separated by 2D-PAGE as described in Materials and methods. The medium expression level of the selected proteins was calculated and normalized to the control to deduce general folds. (C) Ambiguity pre-adapted cells to tolerate adverse growth conditions. Expression of stress proteins (Supplementary Tables 3 and 4) increased twofold in control cells at 37°C, but not in ambiguous cells that already had increased amounts (3 fold) of these stress-protective proteins. (D) Ambiguous cells retained the capacity to respond to additional stress. Expression of stress proteins (Supplementary Tables 3 and 4) was induced in both strains under heat-shock (8- and 13-fold for the control and ambiguous cells, respectively).
Strong up-regulation (29.5-fold) of the enzyme nicotinamidase (PNC1), which converts nicotinamide into nicotinic acid in the NAD+ salvage pathway (Anderson et al, 2003), was unanticipated, as NAD+ is a cofactor of the histone deacetylase Sir2p. In yeast, Sir2p is involved in silencing chromatin at telomeres, ribosomal DNA (rDNA) and mating-type loci, and deletion of the SIR2 gene promotes aging by increasing recombination at the rDNA locus (Sinclair and Guarente, 1997). Since PNC1 integrates multiple stimuli and modulates environmental adaptation through Sir2p (Anderson et al, 2003), its strong upregulation by codon misreading (Table II) suggests that aberrant proteins activate conserved mechanisms that stabilize the genome. Indeed, PNC1 may counteract genome-destabilizing effects caused by CUG ambiguity by stabilizing the rDNA locus.
Constitutive CUG ambiguity does not impair long-term adaptation
Redefinition of codon identity evolved gradually over millions of years (272±25 million years in Candida) and results in constitutive synthesis of aberrant proteins, generating a unique form of intracellular stress. An important question came to mind, namely ‘would ambiguous cells loose their capacity to respond to additional environmental challenges?' In order to shed light on this question, various stress proteins, including proteasome subunits, were labelled with [35S]Met and quantified by phosphorimaging of 2D protein maps, as before (Supplementary Tables 3 and 4). Ambiguous cells had higher proteasome activity and displayed increased expression of proteasome subunits (Figure 5A and B) and increased levels of stress proteins at 25°C, in particular of molecular chaperones (Figure 5C). However, the latter were not upregulated at 37°C (Figure 5C) and their expression only increased when cells were exposed to heat shock (Figure 5D). This capacity of ambiguous cells (permanently stressed) to respond to heat stress by further upregulating critical components of the stress response supports the hypothesis that codon identity redefinition did not compromise adaptation to environmental change.
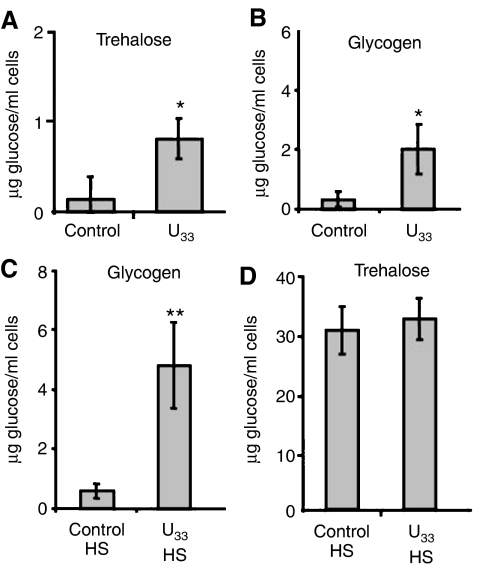
Glycogen and trehalose are reserve carbohydrates that accumulate under stress as energy reserves. Trehalose also stabilizes protein structure at high temperatures and decreases the aggregation of unfolded or heat-denatured proteins (Singer and Lindquist, 1998; Ueda et al, 2001). Accordingly, ambiguous cells accumulated them at higher levels than control cells (Figure 6A and B), due to upregulation of the components of the trehalose synthase complex, namely TPS1 (2 fold) and TSL1 (3 fold) (Table I). Under heat shock, ambiguous cells had higher levels of both glycogen and trehalose (Figure 6C and D), and proteome analysis under heat shock confirmed that both control and U33 tRNACAGSer cells upregulated proteins belonging to the trehalose biosynthesis pathway (Supplementary Tables 4 and 5). Previous studies showed that glycogen concentration plateaus after 60 min under heat shock and its level is maintained up to 120 min (Parrou et al, 1997). In our experimental conditions (30 min of heat shock) control cells accumulated small amounts while ambiguous cells already had significant amounts of glycogen (Figure 6B and C). Genes encoding enzymes of the glycogen biosynthesis pathways, namely GSY2 and GLC3, were not significantly upregulated in cells expressing U33 tRNACAGSer (Supplementary Table 5); however GPH1 involved in glycogen degradation was upregulated 4 fold, pointing towards increased simultaneous recycling of this reserve carbohydrate, as previously described (Parrou et al, 1997).
Figure 6.
Genetic code alterations increase trehalose and glycogen accumulation. C and U33 represent S. cerevisiae control cells and cells expressing C. albicans U33 tRNACAGSer, respectively. The results are expressed as mean±s.d. of four independent experiments (*P<0.05 and **P<0.01 by Student's t-test). Trehalose (A) and glycogen (B) content were 6 fold higher in exponentially growing CUG ambiguous cells (OD=0.5), when compared with control cells. (C, D) Glycogen and trehalose accumulation were induced in ambiguous cells after a 30-min heat shock; glycogen content increased 8 fold under heat shock when compared with control cells, while trehalose accumulation increased in both ambiguous and control strains under heat stress.
Since codon-decoding ambiguity causes major proteome disruption and reduces fitness, genetic code alterations should have been eliminated by natural selection. The gene and protein expression profiling data described herein provide an explanation for this apparent paradox. Up-regulation of proteasome activity, induction of stress proteins, cell-wall remodelling and accumulation of trehalose and glycogen contributed to elimination or recovery of aberrant proteins. Furthermore, gene expression and physiological remodelling created stress cross-protection and pre-adapted cells to severe environmental change, allowing them to explore new ecological niches. Indeed, previous studies have shown that ambiguous S. cerevisiae were able to outcompete wild-type cells under stress (Santos et al, 1999). In this study, spores of S. cerevisiae expressing G33 tRNACAGSer, plated in rich glucose media (YEPD) grew much slower than non-ambiguous spores (Figure 7), but stress severity reduced colony size differences between control and ambiguous clones, namely in synthetic media (SD) and in minimal media (MM) containing 100 μM CdCl2 or 1.5 mM H2O2 (Figure 7). This supports the hypothesis that, to a certain extent, stress tolerance and stress cross-protection overcome the negative impact of proteome disruption caused by mistranslation.
Figure 7.
Ambiguous cells have selective advantage under stress. Spores of S. cerevisiae control cells and cells expressing the G33 tRNACAGSer were grown for 7 days in agar plates of rich medium (YEPD), synthetic medium (SD) and minimal medium (MM) containing 100 μM CdCl2 or 1.5 mM H2O2. Spores from ambiguous cells grew much slower than non-ambiguous spores in rich medium, but recovered competitive capacity when growing under stress conditions, as shown by a similar colony size of control and ambiguous cells.
Discussion
A role for CUG identity redefinition in Candida evolution
New species may arise from ecological differentiation, hybridization, chromosomal and ploidy alterations and gradual evolution of genetic incompatibilities (Otto and Whitton, 2000; Storchova and Pellman, 2004). Our data show that, beyond blocking lateral gene transfer, genetic code alterations remodel gene expression and physiology and are able to create immediate genetic barriers to sexual reproduction. Indeed, partial redefinition of CUG identity in S. cerevisiae had a strong negative impact on sporulation efficiency, fertility and mating. Only half of the spores (44%) expressing G33 tRNACAGSer were viable and U33 tRNACAGSer was lethal in haploid backgrounds. Mating was severely affected in clones expressing G33 tRNACAGSer and completely disrupted in clones expressing U33 tRNACAGSer (Figure 2). More importantly, tRNACAGSer generated high cell heterogeneity and created diploid and tetraploid lineages with nuclear aberrations (micronulei) (Figure 3). Since diploid S. cerevisiae cells tolerated U33 tRNACAGSer, which mimics the ancestral tRNACAGSer (Massey et al, 2003), and haploid cells did not, it is likely that CUG identity redefinition locked the Candida ancestor in a diploid or polyploid state and blocked sexual reproduction altogether. This is supported by diploidy and lack of sexual reproduction in extant Candida species, and by the observation that polyploid plants and animals are normally parthenogenic (asexual) (Otto and Whitton, 2000). Ploidy increase and population heterogeneity induced by codon identity redefinition is of further evolutionary consequence, as ploidy differences alter gene expression, developmental, physiological, cell size and other unique morphological characteristics in yeast and polyploid tumour cells (Galitski et al, 1999). Intriguingly, in E. coli, codon-decoding ambiguity generated translational stress-induced mutagenesis (TSM) and created hypermutagenic cells (Al Mamun et al, 2002). If TSM occurred in the Candida ancestor, it is likely that diploidy and polyploidy may have buffered the genome against rapid accumulation of degenerative mutations. Alternatively, ploidy increase may have allowed rapid evolution of new gene functions by freeing one copy of duplicated genes for evolutionary experimentation. This hypothesis is strongly supported by very high genome heterozygosity and rapid functional divergence of a number of gene families in most Candida species (Zhao et al, 2003; Jones et al, 2004).
Genetic code alterations remodel gene expression
Yeast cells respond to sudden exposure to environmental stress by transiently (60 min) changing the expression of an array of genes (approximately 600 downregulated and 300 upregulated). As cells adapt to stress, gene expression recovers to a new steady state and transcript abundance differences between normal and stress conditions become much lower (Gasch et al, 2000). This so-called environmental stress response (ESR) is mediated by the transcription factors Msn2p and Msn4p through the general stress-responsive element (STRE; CCCCT) (Gasch et al, 2000; Causton et al, 2001). Unicellular organisms also respond to specific stresses by activating unique cellular programs that are controlled by other transcription factors, namely Yap1p (oxidative), Hog1p (osmotic) and Hsf1p (Heat shock) (Brewster et al, 1993; Kuge et al, 1997; Boy-Marcotte et al, 1999). Exposure to mild stress activates ESR, creating stress cross-protection and resistance to severe stress conditions (Mizzen and Welch, 1988; Gasch et al, 2000). Redefinition of codon identity resulted in constitutive stress due to permanent synthesis of aberrant proteins (misfolded) over millions of years, but how this stress response was (de)regulated remains to be studied.
Partial CUG identity redefinition in S. cerevisiae induced a stress response similar to that induced by heat shock and the proline analogue azetidine-2-carboxylic acid (AZC), two stress agents that also generate protein misfolding (Trotter et al, 2002). The 81 genes altered in response to CUG mistranslation were a subset of the 217 and 293 genes up- and downregulated by AZC, respectively (Trotter et al, 2002). However, AZC induced genes whose expression is controlled by the heat-shock factor (HSF) through the heat-shock element (HSE), while CUG ambiguity induced genes whose expression is controlled by Fkh1p, Fkh2p, Hsf1p, Msn2p and Msn4p (Supplementary Table 6). The latter (but not AZC) also induced the accumulation of trehalose and glycogen, which are under the control of Msn2p and Msn4p (Boy-Marcotte et al, 1999; Trotter et al, 2002). The transcription factors Fkh1p and Fkh2p regulate the yeast cell cycle, pseudohyphal growth and are involved in chromatin silencing at HML and HMR (Hollenhorst et al, 2000; Zhu et al, 2000; Sun et al, 2002). Interestingly, mistranslation of threonine ACA codons as alanine in Schizosaccharomyces pombe resulted in late mitotic defects, abnormal chromosome segregation, aneuploidy and decreased cell viability (Kimata and Yanagida, 2004). Therefore, it is now important to elucidate how CUG mistranslation triggered cell morphology and ploidy alterations and clarify how it created cell-cycle defects and mating impairment in S. cerevisiae.
The stress responses induced by CUG ambiguity and heat shock were rather similar, since the latter is also mediated by HSF, Msn2p and Msn4p. HSF activation by heat shock is due to titration by misfolded proteins of its chaperone HSP70, while Msn2p and Msn4p are activated by phosphorylation by protein kinase A (PKA) (Thevelein and de Winde, 1999). Also, like heat shock, CUG ambiguity induced thermotolerance (Santos et al, 1996), which is dependent on the activation of the protein kinase C–mitogen-activated protein kinase pathway (PKC1–MAP) (Kamada et al, 1995). Therefore, CUG identity redefinition may have interfered with important signalling pathways that modulated the general stress response in the Candida ancestor. If so, it is likely that such pathways have been remodelled in Candida over the last 272±25 million years of CUG identity redefinition. This hypothesis is strongly supported by major differences in the S. cerevisiae and C. albicans stress responses. For example, the former has a general stress response while the latter does not. C. albicans does respond to stress, but each stress response is unique and there is no cross-protection (Enjalbert et al, 2003). Furthermore, CaMsn4p and CaMnl1p (homologues of S. cerevisiae Msn2p) do not play an active role in the C. albicans stress response and their functions and those of their S. cerevisiae homologues diverged significantly (Nicholls et al, 2004). Also, C. albicans does not accumulate trehalose and glycogen in response to stress (Enjalbert et al, 2003).
The mobilization of molecular chaperones to correct misfolded proteins was prominent, but it did not compromise the capacity to respond to additional stress. Indeed, S. cerevisiae cells expressing C. albicans tRNACAGSer were able to further induce the expression of heat-shock proteins when exposed to a temperature up-shift from 25 to 37°C during 30 min (Figure 5D). In other words, ambiguous cells were not impaired in their capacity to respond to different unfavourable conditions that require gene expression reprogramming. However, S. cerevisiae cells expressing C. albicans tRNACAGSergrown under an additional permanent stress did not show further induction of the stress-protective proteins (Figure 5C), as observed for control cells, suggesting that these cells are pre-adapted to adverse environments.
Conclusions
Genetic code alterations create major proteome chaos and should be eliminated by natural selection. The robust cellular response to codon-decoding ambiguity and the selective advantages observed under stress in yeast (Santos et al, 1999), explain this apparent paradox. Despite this, genetic code changes have a major impact on protein primary structure on a proteome-wide scale, and one would expect that they create genetic barriers to sexual reproduction on an evolutionary time scale. This is of relevance to the early evolution of the genetic code, since incorporation of new amino acids into the primordial code would have also created codon ambiguity and generated genetic barriers to exchange of genetic information between ‘old code' and ‘new code' lineages, contributing to the elimination of less competitive ‘old code' lineages. Why code expansion stopped at the twenty-second amino acid remains to be elucidated, but it is likely that increased proteome complexity may have been the critical factor that prevented further expansion.
Materials and methods
Strains and growth conditions
S. cerevisiae strains used were based on the CEN.PK2 background (MATa/MATα; ura3-52/ura3-52; trp1-289/trp1-289; leu2-3_112/leu2-3_112; his3Δ1/his3Δ1). CUG ambiguous cells were obtained by expressing C. albicans tRNACAGSer either by transformation, performed according to the lithium acetate method (Gietz and Woods, 1994), or by integration in the genome, carried out by PCR-based gene disruption method (Lorenz et al, 1995). Unless otherwise stated, control cells are S. cerevisiae cells transformed with the single-copy vector pRS315 (vector alone), and ambiguous strains carry the plasmids pUKC715 and pUKC716, containing G33 and U33 tRNACAGSer, respectively (Santos et al, 1996). Cells were grown to OD600 0.5 at 25°C, in minimal medium containing 0.67% yeast nitrogen base without amino acids, buffered at pH 5.8 with 1% succinate and 0.6% NaOH, and 2% glucose, supplemented with 100 μg/ml of the required amino acids. For heat-shock experiments, cells were grown at 25°C, transferred to new flasks preheated at 37°C and grown for additional 30 min at 37°C. For sporulation, mating and stress plate assays, DNA fragments containing S. cerevisiae HIS3 and URA3 genes were integrated in the genome, and this strain was used to integrate the PCR products of the gene KanMX (control) or KanMX+tRNACAGSer (G33 or U33) at the LEU2 locus. All integrations were confirmed by PCR and sequencing.
CUG ambiguity reporter system
To quantify CUG ambiguity in S. cerevisiae cell lines expressing the C. albicans tRNACAGSer G33 and U33, a reporter protein was engineered. Briefly, this protein is based on the CaPGK1 gene, has one CUG codon flanked by thrombin cleavage sites and a 6-histidine tail for purification by affinity chromatography. The CaPGK1 promoter and ORF were cloned on pRS425 between the HindIII and PstI restriction sites. External XhoI and SacII restriction sites were eliminated using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and the CUG-reporter cassette was inserted between internal XhoI and SacII restriction sites, generating the plasmid pUA202. S. cerevisiae cells transformed with pUA202 were grown to OD600 between 1 and 1.5 and harvested by centrifugation. The pellet was washed with mQ water and frozen at −80°C. Cells were lysed in ice-cold buffer A (6 M urea, 15 mM Na–HEPES, 150 mM NaCl, 0.01% Triton X-100, 10% glycerol, pH 7.8) supplemented with protease inhibitors (Complete EDTA-free protease inhibitor cocktail tablets (Roche) and 2 mM PMSF). Cells were broken by agitation in the presence of glass beads and lysates were clarified by centrifugation. The supernatant was transferred to a new, clean tube and incubated with 1 ml of Ni–Sepharose (Amersham) for overnight at 4°C with agitation. The His-tagged reporter protein was purified by Ni-NTA agarose chromatography as described by the resin manufacturer. The purified protein was dialysed against thrombin cleavage buffer (20 mM Tris–Cl pH 8.4, 150 mM NaCl, 2.5 mM CaCl2, from Novagen) and digested with thrombin (Novagen) at 26°C for 16 h.
Mass spectrometry analysis
Mutiple Reaction Monitoring (MRM) experiments were carried out on a hybrid quadrupole/linear ion-trap mass spectrometer (4000 QTrap; Applied Biosystems/MDS Sciex) using electrospray source and sample flow injection (FIA, Ultimate 3000; Dionex). The mass spectrometer was operated in triple quadrupole mode, with Q1 set to the specific precursor m/z value and Q3 set to the specific m/z value corresponding to a specific fragment of that peptide. The collision energy was tuned to optimize the intensity of the fragment ions of interest. In the MRM mode, a series of single reactions (precursor/fragment ion transitions) were measured sequentially and the cycle (2 s) was looped throughout the entire time of the FIA. MRM transitions were determined from the MS/MS spectra of the existing peptides (Supplementary Figures 1–3). Other parameters were tuned for best performance (Stahl-Zeng et al, 2007).
Sporulation assays
To determine sporulation efficiency, integrated cells were grown on sporulation medium (1% potassium acetate, 0.1% yeast extract, 0.05% glucose) for up to 12 days. At each time point, appropriate culture dilutions were prepared and both cells and tetrads were counted using a Neubauer chamber. For spore viability studies, aliquots of 2- to 4-day sporulation cultures were treated with zymolyase for 10 min at room temperature. Digested asci were spread on thin plates and dissected with a MSM System Series 300 micromanipulator (Singer). After 4 and 7 days of incubation, plates were photographed in a GelDoc1000 (Bio-Rad) and colonies were counted. Spore viability was assessed in YPD (1% yeast extract, 2% peptone, 2% glucose) with geneticin (200 mg/l).
Mating assays
Haploid control and ambiguous cells were obtained by sporulation as described above and their mating type was assessed by PCR as described (Huxley et al, 1990). Cells with opposite mating type and distinct auxotrophic markers (His+ or Ura+) were crossed. To do this, exponentially growing cells were counted in a Neubauer chamber and the volumes corresponding to 106 cells of each culture were mixed. Serial dilutions of each mating reaction were spotted in MM–His–Ura+geneticin, and after 3–4 days incubation at 30°C, plates were photographed as above.
DNA content analysis
DNA content of cells was determined using flow cytometry, and fluorescence imaging of DAPI-stained cells was carried out with an Axio Imager.Z1 microscope (Zeiss), as described (Fortuna et al, 2000).
Transcriptome analysis
DNA microarray analyses were performed with six independent cultures for each strain and hybridized against the reference in dye-swap (three control strain cultures labelled Cy5 and three labelled Cy3), in a total of six microarrays for each mutant strain. RNA isolation, labelling and hybridization were carried out as described (van de Peppel et al, 2003). Slides were scanned using the Agilent G2565AA DNA microarray scanner and raw data were extracted with Imagene 4.0 (Biodiscovery). After print-tip Lowess normalization (Yang and Speed, 2002), data visualization and analysis were performed using GeneSpring (Silicon Genetics) and SAM (Tusher et al, 2001). Comparison of GeneSpring data (P<0.05 by Student's t-test, fold change >1.6) and SAM analysis (Δ=2.15; false discovery rate=0.001) resulted in a common set of 170 significant genes, from which 81 were selected based on an average fold change of more than 1.6 in both mutant strains.
Proteome analysis
Proteins were in vivo labelled with [35S]methionine and extracted and separated by 2D-PAGE as described (Boucherie et al, 1995; Boucherie and Monribot, 2005). Radioactive protein spots were detected using a PhosphorImager (MolecularDynamics) and data analysis was performed with the ImageMaster software (Amersham Pharmacia Biotech).
Proteasome activity assay
Proteasome activity was determined essentially as described, using the fluorogenic peptide succinyl-leucine–leucine–valine–tyrosine-MCA (s-LLVY-MCA) as substrate (Demasi et al, 2003). Exponentially growing cells were harvested by centrifugation and disrupted with glass beads on a MiniBeadBeater (Biospec Products), in lysis buffer (10 mM K–Hepes, 10 mM KCl, 1.5 mM MgCl2). Activity was determined for 100 μg of protein extract in assay buffer (10 mM Tris–Cl, pH 8, 20 mM KCl 1 M, 5 mM MgCl2) with 5 μM of s-LLVY-MCA. After a 60-min incubation at 37°C, fluorescence emission was measured at 435 nm (excitation at 365 nm) with a Perkin Elmer Luminescence Spectrometer (LS 50B).
Trehalose and glycogen accumulation
Trehalose and glycogen were quantified as described (Parrou and Francois, 1997). Exponentially growing or heat-shocked cells were harvested by centrifugation, lysed in 0.25 M Na2CO3 at 95°C for 4 h and incubated overnight either with trehalase (Sigma) or amyloglucosidase (MERCK). Glucose present in the supernatant, from trehalose or glycogen breakdown, was determined using the glucose oxidase/peroxidase kit (Sigma).
Stress plate assays
Integrated cells were sporulated as described above, and spore viability was assessed in YPD, MM (0.67% yeast nitrogen base, 2% glucose, 0.2% drop-out mix containing all amino acids), SD (0.67% yeast nitrogen base without amino acids and ammonium sulphate, 2% glucose, 100 μg/ml of the required amino acids), MM+CdCl2 (100 μM) or MM+H2O2 (1.5 mM).
Accession numbers
MIAME compliant protocols and datasets in MAGE–ML are accessible from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with the following accession numbers: E-TABM-196.
Supplementary Material
Supplementary Figures 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure Legends
Supplementary Tables
Acknowledgments
We are thankful to Joel Arrais for help with submission of the microarray data to ArrayExpress, Cidαlia Pina-Vaz and Biocant for the use of the flow cytometry and Mass-Spectrometry laboratories, respectively. This study was supported by the Portuguese Foundation for Science and Technology through projects POCI/FEDER-SAU-MMO-55476/2004, POCI/FEDER-BIA-MIC-55466-2004, FCT/FEDER/CONC-REEQ/737/2001, an EMBO YIP Award (REF: 0134/2003) and a Human Frontier Science Program Award (REF: HFSP RGP45/2005) and the Research Institute of the University of Aveiro.
References
- Al Mamun AA, Marians KJ, Humayun MZ (2002) DNA polymerase III from Escherichia coli cells expressing mutA mistranslator tRNA is error prone. J Biol Chem 277: 46319–46327 [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA (2003) Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucherie H, Dujardin G, Kermorgant M, Monribot C, Slonimski P, Perrot M (1995) Two-dimensional protein map of Saccharomyces cerevisiae: construction of a gene–protein index. Yeast 11: 601–613 [DOI] [PubMed] [Google Scholar]
- Boucherie H, Monribot C (2005) Two-dimensional gel electrophoresis of total yeast proteins. In Yeast Protocols, Xiao W (ed), 2nd edn, pp 47–64. Totowa, NJ: Humana Press [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E, Lagniel G, Perrot M, Bussereau F, Boudsocq A, Jacquet M, Labarre J (1999) The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol Microbiol 33: 274–283 [DOI] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC (1993) An osmosensing signal transduction pathway in yeast. Science 259: 1760–1763 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR (1986) The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination' codon, TGA. EMBO J 5: 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ (1997) Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143: 303–311 [DOI] [PubMed] [Google Scholar]
- Crick FH (1968) The origin of the genetic code. J Mol Biol 38: 367–379 [DOI] [PubMed] [Google Scholar]
- Demasi M, Silva GM, Netto LE (2003) 20S proteasome from Saccharomyces cerevisiae is responsive to redox modifications and is S-glutathionylated. J Biol Chem 278: 679–685 [DOI] [PubMed] [Google Scholar]
- DiDomenico BJ, Bugaisky GE, Lindquist S (1982) The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell 31: 593–603 [DOI] [PubMed] [Google Scholar]
- Enjalbert B, Nantel A, Whiteway M (2003) Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell 14: 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuna M, Sousa MJ, Côrte-Real M, Leão C, Salvador A, Sansonetty F (2000) Cell cycle analysis of yeasts using Syber Green I. In Current Protocols in Cytometry, Robinson JP (ed), pp 11.13.1–11.13.9. New York: Wiley [DOI] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR (1999) Ploidy regulation of gene expression. Science 285: 251–254 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (1994) High efficiency transformation with lithium acetate. In Molecular Genetics of Yeast: a Practical Approach, Johnston JR (ed), pp 121–134. Oxford: IRL Press [Google Scholar]
- Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK (2002) A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296: 1462–1466 [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Bose ME, Mielke MR, Muller U, Fox CA (2000) Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154: 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I (1990) Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet 6: 236. [DOI] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S (2004) The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA 101: 7329–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Jung US, Piotrowski J, Levin DE (1995) The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev 9: 1559–1571 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Yanagida M (2004) Suppression of a mitotic mutant by tRNA-Ala anticodon mutations that produce a dominant defect in late mitosis. J Cell Sci 117: 2283–2293 [DOI] [PubMed] [Google Scholar]
- Kuge S, Jones N, Nomoto A (1997) Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J 16: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J (1995) Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158: 113–117 [DOI] [PubMed] [Google Scholar]
- Massey SE, Moura G, Beltrao P, Almeida R, Garey JR, Tuite MF, Santos MA (2003) Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res 13: 544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda I, Silva R, Santos MA (2006) Evolution of the genetic code in yeasts. Yeast 23: 203–213 [DOI] [PubMed] [Google Scholar]
- Mizzen LA, Welch WJ (1988) Characterization of the thermotolerant cell. I. Effects on protein synthesis activity and the regulation of heat-shock protein 70 expression. J Cell Biol 106: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I (2004) Reprogrammed genetic decoding in cellular gene expression. Mol Cell 13: 157–168 [DOI] [PubMed] [Google Scholar]
- Nicholls S, Straffon M, Enjalbert B, Nantel A, Macaskill S, Whiteway M, Brown AJ (2004) Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot Cell 3: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba T, Andachi Y, Muto A, Osawa S (1991) CGG: an unassigned or nonsense codon in Mycoplasma capricolum. Proc Natl Acad Sci USA 88: 921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama T, Muto A, Osawa S (1990) Role of GC-biased mutation pressure on synonymous codon choice in Micrococcus luteus, a bacterium with a high genomic GC-content. Nucleic Acids Res 18: 1565–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S, Jukes TH (1989) Codon reassignment (codon capture) in evolution. J Mol Evol 28: 271–278 [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Parrou JL, Francois J (1997) A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem 248: 186–188 [DOI] [PubMed] [Google Scholar]
- Parrou JL, Teste MA, Francois J (1997) Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 143 (Part 6): 1891–1900 [DOI] [PubMed] [Google Scholar]
- Perreau VM, Keith G, Holmes WM, Przykorska A, Santos MA, Tuite MF (1999) The Candida albicans CUG-decoding ser-tRNA has an atypical anticodon stem–loop structure. J Mol Biol 293: 1039–1053 [DOI] [PubMed] [Google Scholar]
- Santos MAS, Cheesman C, Costa V, Moradas-Ferreira P, Tuite MF (1999) Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp.. Mol Microbiol 31: 937–947 [DOI] [PubMed] [Google Scholar]
- Santos MAS, Perreau VM, Tuite MF (1996) Transfer RNA structural change is a key element in the reassignment of the CUG codon in Candida albicans. EMBO J 15: 5060–5068 [PMC free article] [PubMed] [Google Scholar]
- Santos MAS, Tuite MF (1995) The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res 23: 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MAS, Ueda T, Watanabe K, Tuite MF (1997) The non-standard genetic code of Candida spp.: an evolving genetic code or a novel mechanism for adaptation? Mol Microbiol 26: 423–431 [DOI] [PubMed] [Google Scholar]
- Schultz DW, Yarus M (1994) Transfer RNA mutation and the malleability of the genetic code. J Mol Biol 235: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Silva RM, Miranda I, Moura G, Santos MA (2004) Yeast as a model organism for studying the evolution of non-standard genetic codes. Brief Funct Genomic Proteomic 3: 35–46 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Singer MA, Lindquist S (1998) Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell 1: 639–648 [DOI] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Aebersold R, Domon B (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics (in press) [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Herbert P, Lewendon A, Shaw WV, Tuite MF (1998) Missense translation errors in Saccharomyces cerevisiae. J Mol Biol 282: 13–24 [DOI] [PubMed] [Google Scholar]
- Storchova Z, Pellman D (2004) From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 5: 45–54 [DOI] [PubMed] [Google Scholar]
- Sugiyama H, Ohkuma M, Masuda Y, Park SM, Ohta A, Takagi M (1995) In vivo evidence for non-universal usage of the codon CUG in Candida maltosa. Yeast 11: 43–52 [DOI] [PubMed] [Google Scholar]
- Sun K, Coic E, Zhou Z, Durrens P, Haber JE (2002) Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes Dev 16: 2085–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ueda T, Watanabe K (1997) The ‘polysemous' codon—a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J 16: 1122–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald-Dietrich A, Giege R, Rudinger-Thirion J (2005) Evidence for the existence in mRNAs of a hairpin element responsible for ribosome dependent pyrrolysine insertion into proteins. Biochimie 87: 813–817 [DOI] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH (1999) Novel sensing mechanisms and targets for the cAMP–protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 33: 904–918 [DOI] [PubMed] [Google Scholar]
- Trotter EW, Kao CM, Berenfeld L, Botstein D, Petsko GA, Gray JV (2002) Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J Biol Chem 277: 44817–44825 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Nagata M, Imoto T (2001) Aggregation and chemical reaction in hen lysozyme caused by heating at pH 6 are depressed by osmolytes, sucrose and trehalose. J Biochem (Tokyo) 130: 491–496 [DOI] [PubMed] [Google Scholar]
- van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, Holstege FC (2003) Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep 4: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Speed T (2002) Design issues for cDNA microarray experiments. Nat Rev Genet 3: 579–588 [DOI] [PubMed] [Google Scholar]
- Zhao X, Pujol C, Soll DR, Hoyer LL (2003) Allelic variation in the contiguous loci encoding Candida albicans ALS5, ALS1 and ALS9. Microbiology 149: 2947–2960 [DOI] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406: 90–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure Legends
Supplementary Tables