Abstract
Cells sustain high rates of actin filament elongation by maintaining a large pool of actin monomers above the critical concentration for polymerization. Profilin–actin complexes constitute the largest fraction of polymerization-competent actin monomers. Filament elongation factors such as Ena/VASP and formin catalyze the transition of profilin–actin from the cellular pool onto the barbed end of growing filaments. The molecular bases of this process are poorly understood. Here we present structural and energetic evidence for two consecutive steps of the elongation mechanism: the recruitment of profilin–actin by the last poly-Pro segment of vasodilator-stimulated phosphoprotein (VASP) and the binding of profilin–actin simultaneously to this poly-Pro and to the G-actin-binding (GAB) domain of VASP. The actin monomer bound at the GAB domain is proposed to be in position to join the barbed end of the growing filament concurrently with the release of profilin.
Keywords: actin, cytoskeleton dynamics, filament elongation, profilin
Introduction
The enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) family is implicated in diverse cellular processes involving dynamic actin assembly, such as fibroblast migration, axon guidance and the movement of the bacterial pathogen Listeria monocytogenes (Krause et al, 2003). Ena/VASP proteins share a tripartite domain organization (Figure 1), consisting of N- and C-terminal Ena/VASP homology 1 and 2 (EVH1 and EVH2) domains and a central Pro-rich region. The EVH1 domain targets Ena/VASP proteins to focal adhesions, filopodia and lamellipodia by binding to target proteins containing the consensus sequence ‘FPPPP', such as vinculin (Brindle et al, 1996; Reinhard et al, 1996), lamellipodin (Krause et al, 2004), zyxin (Reinhard et al, 1995b), migfilin (Zhang et al, 2006) and palladin (Boukhelifa et al, 2004). The Pro-rich region of Ena/VASP binds profilin and the SH3 and WW domains of various signaling and scaffolding proteins, including Abl, Src and IRSp53 (Gertler et al, 1995; Ahern-Djamali et al, 1999; Krugmann et al, 2001). The EVH2 domain comprises globular and filamentous actin-binding sites (GAB and FAB), as well as a C-terminal coiled-coil (CC) region that mediates VASP tetramerization (Bachmann et al, 1999; Walders-Harbeck et al, 2002).
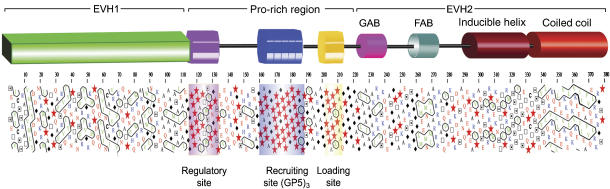
Figure 1.
Modular organization of VASP. The diagram shown on the upper part of the figure represents the modular organization of the human VASP sequence (UniProt P50552) based on the distribution of clusters of hydrophobic amino acids determined with the program HCA (Callebaut et al, 1997). Symbols are as follows: Pro,  ; Gly, ⧫; Thr, □ and Ser,
; Gly, ⧫; Thr, □ and Ser,  . Amino acids are colored according to their chemical characteristics: green, hydrophobic; red, negatively charged; blue, positively charged; black, Ala and Cys. The EVH1 and CC domains, which are stably folded and whose structures have been determined (Prehoda et al, 1999; Kuhnel et al, 2004), are characterized by a higher density of clusters of hydrophobic amino acids (contoured black). In contrast, the extended Pro-rich region (amino acids 116–213) lacks hydrophobic clusters, and is predicted to be mostly unfolded. Within this region, we identify three distinct groups of Pro residues, which are present in all members of the Ena/VASP family: regulatory site (116–135), recruiting site (160–194) and loading site (201–211). Finally, the region preceding the CC domain has the typical HCA pattern of an inducible α-helix.
. Amino acids are colored according to their chemical characteristics: green, hydrophobic; red, negatively charged; blue, positively charged; black, Ala and Cys. The EVH1 and CC domains, which are stably folded and whose structures have been determined (Prehoda et al, 1999; Kuhnel et al, 2004), are characterized by a higher density of clusters of hydrophobic amino acids (contoured black). In contrast, the extended Pro-rich region (amino acids 116–213) lacks hydrophobic clusters, and is predicted to be mostly unfolded. Within this region, we identify three distinct groups of Pro residues, which are present in all members of the Ena/VASP family: regulatory site (116–135), recruiting site (160–194) and loading site (201–211). Finally, the region preceding the CC domain has the typical HCA pattern of an inducible α-helix.
Ena/VASP functions primarily as an actin filament elongation factor, whereas it is a relatively weak nucleator (Bear et al, 2002; Barzik et al, 2005). Thus, depletion of Ena/VASP produces shorter and more densely branched filament networks, whereas its overexpression causes the opposite effect (Skoble et al, 2001; Bear et al, 2002). Since profilin–actin constitutes the major pool of polymerization-competent actin in eukaryotic cells (Pollard and Borisy, 2003), filament elongation by Ena/VASP will depend upon its ability to ‘process' profilin–actin complexes for their incorporation onto the barbed end of growing filaments. Consistent with this notion, the recruitment of profilin–actin by the central Pro-rich region of VASP enhances Listeria motility (Chakraborty et al, 1995; Kang et al, 1997; Geese et al, 2000, 2002; Auerbuch et al, 2003; Grenklo et al, 2003). Furthermore, a covalently crosslinked profilin–actin complex, which cannot function as a source for filament elongation, markedly reduces Listeria motility, and this effect depends on the ability of profilin to interact with the poly-Pro region of Ena/VASP (Grenklo et al, 2003). Profilin also appears to enhance actin polymerization by increasing the anti-capping activity of VASP (Barzik et al, 2005). The role of profilin–actin in filament elongation is not unique to Ena/VASP. Most formins also recruit profilin–actin via their Pro-rich FH1 domain, which increases the rate of filament elongation (Kovar et al, 2006). Despite the importance of profilin–actin in filament elongation, the molecular bases for this function are poorly understood. Here, we investigate the energetics and structural bases for the interactions of profilin–actin with the last poly-Pro and the poly-Pro-GAB regions of human VASP. Based on the results, we propose a model for the recruitment and processing of profilin–actin complexes during filament elongation by Ena/VASP.
Results
The Pro-rich region of Ena/VASP consists of three distinct poly-Pro sites
To better understand the modular organization of VASP, and in particular that of its Pro-rich region, we analyzed the distribution of clusters of hydrophobic amino acids using the program HCA (Callebaut et al, 1997) (Figure 1). Predictably, the EVH1 and CC regions, whose crystal structures have been determined (Prehoda et al, 1999; Kuhnel et al, 2004), are characterized by a higher density of clusters of hydrophobic amino acids. Two small clusters, displaying a characteristic helical pattern (Callebaut et al, 1997), correspond to the GAB and FAB domains. The extended Pro-rich region located between the EVH1 and GAB domains lacks hydrophobic clusters and is therefore predicted to be mostly unfolded. Although this region is more divergent, we identify three distinct groups of proline residues that are present in all members of the Ena/VASP family (Figure 1). The first group, amino acids 116–135 (human VASP sequence numbers), consists of a mixture of proline and hydrophobic amino acids, a signature feature of SH3- and WW-binding sequences. Thus, programs such as iSPOT (Brannetti and Helmer-Citterich, 2003) and SCANSITE (Obenauer et al, 2003) predict that this poly-Pro site binds the SH3 domains of a number of signaling proteins, including Abl, Nck, Crk and cortactin. The interactions of Ena/VASP with some of these proteins have been demonstrated (Gertler et al, 1995; Sparks et al, 1996; Coppolino et al, 2001), although not always mapped specifically to this site. Because this site does not correspond to a canonical profilin-binding sequence (Perelroizen et al, 1994; Petrella et al, 1996; Kang et al, 1997), and since it is most likely involved in regulation, we identify it as the ‘regulatory' poly-Pro site. The second group of proline residues (amino acids 160–194) comprises three repeats of the specific profilin-binding sequence GPPPPP (or GP5), and has therefore the potential to bind multiple profilin–actin complexes (Mahoney et al, 1999). For this reason, we identify this site as the ‘recruiting' poly-Pro site. Most of the studies on the profilin–poly-Pro interaction have focused on this GP5 region or plain poly-Pro sequences (Perelroizen et al, 1994; Reinhard et al, 1995a; Petrella et al, 1996; Kang et al, 1997; Lambrechts et al, 1997; Jonckheere et al, 1999). The third group of proline residues is located between the recruiting poly-Pro site and the GAB domain, and is flanked on both sides by short flexible linkers rich in Gly residues. Because of its location, immediately N-terminal to the GAB domain, and its apparent role in delivering profilin–actin from the poly-Pro region to the GAB domain (see below), we identify this site as the ‘loading' poly-Pro site. The loading site is highly conserved among all members of the Ena/VASP family, and always presents a hydrophobic amino acid at the penultimate position, most commonly Leu (Supplementary Figure 1). The loading poly-Pro site and the GAB domain are the central focus of this study.
Binding of profilin and profilin–actin to the loading poly-Pro site of VASP
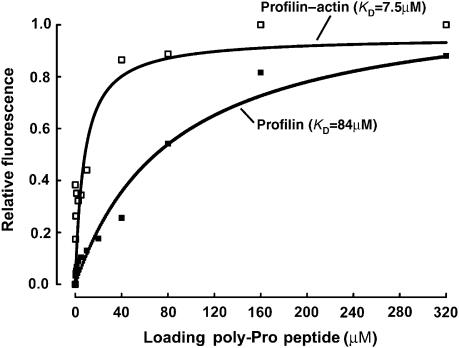
As mentioned above, various studies have investigated the binding of profilin to plain poly-Pro sequences and GP5 sequences of the kind found in the recruiting site of VASP (Perelroizen et al, 1994; Reinhard et al, 1995a; Petrella et al, 1996; Kang et al, 1997; Lambrechts et al, 1997; Jonckheere et al, 1999). In particular, profilin has been reported to bind the GP5 sequence of VASP with relatively low affinity (KD of 84–250 μM) (Petrella et al, 1996; Kang et al, 1997). However, neither the binding of profilin to the highly conserved loading poly-Pro site nor the binding of profilin–actin to any poly-Pro sequence has yet been investigated. Here, we used two different methods, isothermal titration calorimetry (ITC) and intrinsic tryptophan fluorescence, to measure the binding affinities of both profilin and profilin–actin for a 16-aa synthetic peptide (198GAGGGPPPAPPLPAAQ213) containing the loading poly-Pro site of human VASP (Figure 2; Supplementary Figure 2). Both methods consistently showed that the binding of profilin–actin for this peptide in high ionic strength buffer is 6- to 11-fold higher affinity than that of profilin alone (KD of 7.5–8 μM for profilin–actin vs 50–84 μM for profilin alone). Although not specifically addressed here, similar results were obtained for a GP5 triplet peptide, resulting in a KD of 23 μM for profilin–actin vs 230 μM for profilin alone (data not shown). We also found that the affinity of profilin for actin in G-buffer increases twofold when profilin is bound to the loading poly-Pro peptide of VASP (KD of 0.8 vs 1.6 μM) (Supplementary Figure 3, and the legend). Together, these results suggest that poly-Pro sequences bind preferentially profilin–actin than profilin alone. This finding may have important functional implications, since profilin–actin constitutes the major pool of polymerization-competent actin in cells, and the affinities determined here for poly-Pro sequences fall within the intracellular range of concentrations of profilin–actin (5–40 μM) (Condeelis, 1993; Stossel, 1993; Kang et al, 1999; Dickinson and Purich, 2002; Dickinson et al, 2002; Pollard and Borisy, 2003).
Figure 2.
Binding of profilin and profilin–actin to the loading poly-Pro site of VASP. Binding of the VASP peptide (198GAGGGPPP APPLPAAQ213) produces a significant change in the intrinsic tryptophan fluorescence of profilin alone (▪) and profilin–actin (□). In this experiment, the concentration of profilin and profilin–actin was 5 μM and the concentration of the VASP peptide varied for 0–320 μM (see Materials and methods). Each data point corresponds to the average of five independent measurements. Least-square fitting of the data, using a single-site binding model, resulted in dissociation constant (KD) estimates of 84 and 7.5 μM for profilin and profilin–actin, respectively.
Structure of profilin–actin bound to the loading poly-Pro site of human VASP
Crystal structures of profilin bound to decameric and pentadecameric L-Pro peptides have been determined (Mahoney et al, 1997, 1999). These structures revealed that profilin, like SH3 domains (Feng et al, 1994), binds plain poly-Pro sequences in two distinct backbone orientations. It was proposed at the time that non-Pro residues in profilin ligands may dictate the actual polarity and register of binding (Mahoney et al, 1999). These two factors, polarity and register, become particularly important if there is, as we speculate, a mechanism whereby profilin–actin complexes are transferred directly from the last poly-Pro site to the GAB domain of Ena/VASP during filament elongation. To test this hypothesis, we determined the structure of the ternary complex of profilin–actin bound to the 16-aa peptide corresponding to the loading poly-Pro site of human VASP. In the structure, determined to 1.8-Å resolution (Table I), actin and the VASP peptide bind on opposite sides of the profilin molecule (Figure 3A; Supplementary Figure 4A). The conformation of skeletal α-actin is very similar to that of β-actin in the structure of its complex with profilin determined previously to 2.55-Å resolution (Schutt et al, 1993). In particular, in both structures the nucleotide cleft is in the closed conformation, different from the open cleft conformation suggested by another study (Chik et al, 1996). Compared to other actin structures, the two profilin–actin complexes also display a slightly closed target-binding cleft, consisting of the cleft between actin subdomains 1 and 3 (Dominguez, 2004). This effect, which appears to be directly linked to the binding of profilin, may explain the increased affinity of the GAB domain for profilin–actin compared with actin alone (Chereau and Dominguez, 2006).
Table 1.
Crystallographic data and refinement statistics
| Profilin–actin– poly-Pro | Profilin–actin–poly- Pro-GAB | |
|---|---|---|
| Diffraction statistics | ||
| Space group | P 21 21 21 | C 2 |
| Cell parameters | ||
| a, b, c (Å) | 37.47, 75.46, 180.74 | 119.09, 56.59, 75.23 |
| α, β, γ (deg) | 90.0, 90.0, 90.0 | 90.0, 104.75, 90.0 |
| Resolution | ||
| Total (Å) | 47.09–1.8 | 35.05–1.5 |
| Outer shell (Å) | 1.86–1.8 | 1.55–1.5 |
| Completeness (%) | 98.1 (97.7) | 99.1 (98.2) |
| Redundancy | 7.0 (5.9) | 8.3 (7.5) |
| Unique reflections | 47 041 (4579) | 76 002 (7370) |
| R-mergea (%) | 7.6 (34.5) | 7.1 (29.4) |
| Average I/σ | 28.1 (5.7) | 32.44 (7.8) |
| Refinement statistics | ||
| R-factorb (%) | 15.8 | 15.6 |
| R-freec (%) | 20.1 | 19.0 |
| R.m.s. deviations | ||
| Bond length (Å) | 0.013 | 0.015 |
| Bond angles (deg) | 1.402 | 1.701 |
| Average B-factor | ||
| All atoms (Å2) | 14.92 | 10.84 |
| Actin/profilin/ VASP (Å2) | 13.18/14.26/28.82 | 9.13/7.17/12.34 |
| Solvent atoms (Å2) | 23.29 | 22.5 |
| PDB code | 2PAV | 2PBD |
| GAB, G-actin-binding domain; VASP, vasodilator-stimulated phosphoprotein. | ||
| Values in parentheses correspond to highest-resolution shell. | ||
| aR-merge=∑(I−〈I〉)/∑I; I and 〈I〉 are the intensity and the mean value of all the measurements for an individual reflection. | ||
| bR-factor=∑∣Fo−Fc∣/∑∣Fo∣; Fo and Fc are the observed and calculated structure factors. | ||
| cR-free, R-factor calculated for a randomly selected subset of the reflections (5%) that were omitted during the refinement. | ||
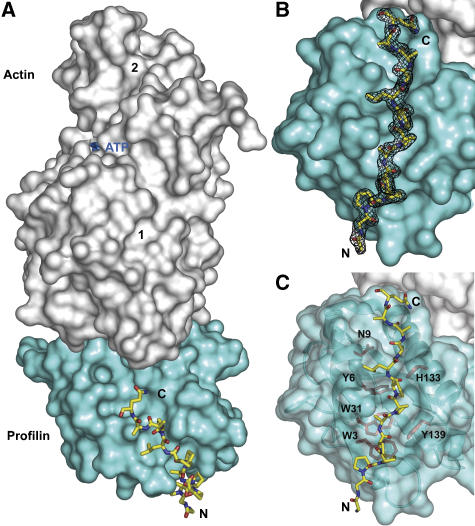
Figure 3.
Crystal structure at 1.8-Å resolution of the ternary complex of profilin–actin with the loading poly-Pro site of human VASP. (A) General view of the structure (actin, gray; profilin, cyan; VASP peptide, all-atom representation). The view is rotated ∼90° relative to the ‘classical' orientation of actin shown in Supplementary Figure 3A. (B) Electron density map around the VASP peptide, contoured at 1.0σ. (C) Illustration of the main interactions of the VASP peptide with profilin amino acids (colored red, under a transparent surface representation of profilin).
One noticeable difference between the two profilin–actin structures occurs in the DNase I-binding loop, which is disordered in the ternary complex studied here, but was visualized in the original structure of profilin–β-actin (Schutt et al, 1993). Also, the C-terminal Phe 375 of actin is rotated around the main chain backbone ∼180° between the two structures. This amino acid is at the center of the profilin–actin interface, an area that is strictly conserved between α- and β-actin, and which is very well defined in the electron density map (Supplementary Figure 4B). The conformation of profilin is also very similar in the two structures, except for the binding interface of the VASP peptide (Figure 3C), where the side chains of residues His 133, Ser 137 and Tyr 139 adopt different rotamer orientations. The N-terminus of profilin also forms part of the binding interface and moves in the current structure toward the VASP peptide, making contacts with it.
The first three amino acids of the loading poly-Pro peptide are disordered in the structure, whereas the remaining 13 (Gly 201–Gln 213) are all very well defined in the electron density map (Figure 3B). The orientation of the VASP peptide is both unambiguous and unique, with the C-terminal end of the peptide pointing toward actin (Figure 3A). The interactions with profilin are limited to the eight amino acids between Pro 203 and Pro 210, and include stacking and hydrogen bonding interactions with profilin residues Trp 3, Asn 9, Trp 31, His 133 and Tyr 139 (Figure 3C). Profilin Tyr 6 also plays a critical role in the interaction, making both a hydrogen-bonding contact with a main-chain atom and a hydrophobic stacking interaction with Leu 209 of the peptide.
The conformation of the VASP peptide is significantly different from the two backbone conformations of the decameric L-Pro peptide, but somewhat similar to that of the pentadecameric L-Pro peptide studied previously (Mahoney et al, 1997; Mahoney et al, 1999) (Supplementary Figure 5). However, important differences occur, which correlate with substitutions of some of the Pro residues in the all-Pro pentadecameric peptide by non-Pro residues in VASP. Thus, although the N-terminal portions of the two peptides superimpose well, the chains diverge slightly after Ala 206, and this effect becomes more pronounced after Leu 209 of the VASP peptide. In about 50% of Ena/VASP sequences, Ala 206 is replaced by Pro (Supplementary Figure 1) and this substitution is unlikely to have any major effect in the general conformation of the complex. However, Leu 209 is strictly conserved and is also found in WASP (Supplementary Figure 1). The role of this Leu residue appears to be twofold; it determines the orientation of the C-terminal portion of the loading poly-Pro region (toward actin), and also determines the register and polarity of the interaction. As we demonstrate next, these factors are in turn critical in defining the position of the GAB domain C-terminal to the loading poly-Pro site so that it can bind to actin.
Structure of profilin–actin bound to the loading poly-Pro-GAB region of VASP
We had previously shown that profilin–actin binds the GAB domain of VASP with higher affinity than actin alone, leading to the proposal that profilin–actin complexes might transition from the loading poly-Pro site to the GAB domain during filament elongation (Chereau and Dominguez, 2006). We had also proposed that the GAB domain is related to WH2 and would therefore be expected to bind in the cleft between actin subdomains 1 and 3 (Chereau et al, 2005). However, an important question remained; the structures of WH2–actin indicated that the binding sites for profilin and WH2 partially overlap and a conformational change would be necessary if the two were to bind actin simultaneously (Chereau et al, 2005). To test this hypothesis we determined the 1.5-Å resolution crystal structure of the ternary complex of profilin—actin, with a 43-aa peptide comprising the loading poly-Pro and GAB domains of human VASP (residues Gly 202–Ser 244) (Figure 4A). The length of the VASP peptide was defined on the basis of the structures of the loading poly-Pro region described above (Figure 3), and that of the WH2 of WASP bound to actin (Chereau et al, 2005), ensuring that none of the amino acids involved in interactions with profilin and actin were excluded.
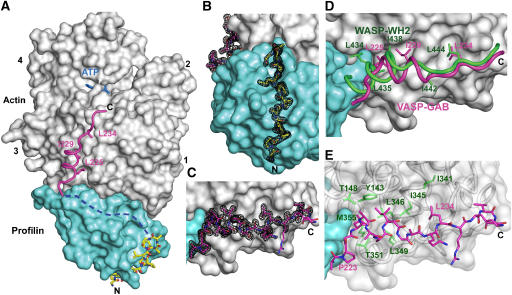
Figure 4.
Crystal structure at 1.5-Å resolution of the ternary complex of profilin–actin with the loading poly-Pro-GAB region of human VASP. (A) General view of the structure (actin, gray; profilin, cyan). The two portions of the peptide visualized in the electron density map are colored differently (corresponding to the colors used in Figures 1 and 3): poly-Pro, all-atom representation with carbon atoms in yellow and GAB in magenta. The linker between these two sites is missing in the structure (represented by a discontinuous blue line), but can be modeled along the proposed path (see also Supplementary Movie 6). (B, C) Electron density map contoured at 1.0σ around the poly-Pro and GAB, respectively. (D) Superimpositions of the structures of the GAB of VASP (magenta) and the WH2 of WASP (green) (Chereau et al, 2005). Only the side chains of hydrophobic amino acids that interact with actin are shown. Note that the helix of the GAB is both shifted forward and rotated ∼45° relative to that of WH2, which is at least in part due to the presence of profilin in the current structure. The LKKT portions of the two structures, however, superimpose well. (E) Illustration of the main interactions of the GAB (magenta) with hydrophobic amino acids (green) of the cleft between actin subdomains 1 and 3.
This ternary complex crystallized in a different space group than that of the loading poly-Pro site (Table I), yet the profilin–actin portions of the two structures are very similar. Differences in three loops of actin (amino acids 199–203, 241–247 and 322–329) can all be attributed to different crystal packing contacts. The electron density map is generally very well defined, and includes two portions of the VASP peptide, amino acids Gly 202–Ala 212 (loading poly-Pro site) and Ala 222–Ser 238 (GAB domain) (Figures 4B and C). The conformation and interactions of the loading poly-Pro site are very similar to those described above (Figures 3C and 4B). The only noticeable difference is that in this structure the loop Tyr 24–Pro 28 of profilin moves toward the peptide and makes one additional hydrogen-bonding contact with it.
As anticipated, the GAB domain binds roughly at the same site as the WH2 of WASP (Figure 4D). Both domains consist of an N-terminal helix that binds in the cleft between actin subdomains 1 and 3, and a C-terminal extended region that binds along the actin surface, climbing toward the pointed end of the actin monomer. Based on this observation, the GAB domain of Ena/VASP can be unambiguously classified as a WH2 domain. However, comparison of the structure with those of WH2 and Tβ domains determined in the absence of profilin (Hertzog et al, 2004; Chereau et al, 2005; Lee et al, 2007) also reveals important differences. Thus, the helix of the GAB domain is rotated ∼45° and shifted forward half a helical turn (Figure 4D). These changes add to a displacement of the GAB domain away from profilin, whereas profilin is unaffected. It is unclear if these changes can all be attributed to the presence of profilin, since the sequence of the GAB domain contains some important differences compared to classical WH2 domains. In particular, the helix of the GAB domain is only two turns long, interrupted at the N-terminus by a combination of Gly and Pro residues, whereas in most WH2s the helix is at least one turn longer and would be expected to conflict with profilin (Figure 4D). Despite these differences the interaction of the helix of the GAB domain presents the same hydrophobic character as the helices of other actin-binding proteins that commonly bind in this cleft (Dominguez, 2004). Thus, VASP residues Leu 225 and Ile 229, on the hydrophobic side of this helix, face the hydrophobic cleft in actin (Figure 4E).
C-terminal to the helix of the GAB domain is the sequence 234LRKV237. This sequence, conserved in all members of the Ena/VASP family (Supplementary Figure 1), corresponds to the so-called ‘LKKT' motif found in a number of actin-binding proteins, including thymosin β (Hertzog et al, 2004), gelsolin (Irobi et al, 2003) and WH2 (Paunola et al, 2002; Chereau et al, 2005). The interactions of the LKKT motif with actin are conserved in these proteins as well as in the GAB domain of VASP. The most important element of this interaction is the binding of VASP residue Leu 234 in a hydrophobic pocket on the actin surface, surrounded by actin residues Ile 341 and Ile 345 (Figure 4E). In the current structure, as well as in the structure of the WH2 of WASP (Chereau et al, 2005), the amino acids C-terminal to the LKKT motif are disordered and do not appear to interact with actin.
Also disordered in the structure is the linker between the poly-Pro and GAB domains, not a surprising result given the high content of Gly residues in this linker (213QGPGGGGAG221). Since the VASP peptide is not observed as a continuous chain, the possibility exists that two different peptides are bound. However, this is unlikely since the complex was crystallized using an actin–profilin peptide stoichiometry of 1:1.1:1.1 (see Materials and methods), and no traces of density are observed in the 1.5-Å resolution map that would correspond to the significant unbound portions of two peptides. Another possibility is domain swapping of the VASP peptide between symmetry related molecules, although again there is no direct evidence of this in the electron density map. Nevertheless, none of these possibilities change the fact that profilin–actin binds the poly-Pro and GAB domains simultaneously in the structure. Furthermore, the missing amino acids of the linker can be easily modeled along a groove at the profilin–actin interface, so that they span the distance between the two sites (Supplementary Movie 6). Note also that GAB is followed by FAB, which tethers Ena/VASP to the elongating filament, thereby directing the incorporation of profilin–actin from the poly-Pro-GAB onto the barbed end of a specific filament (Figure 5).
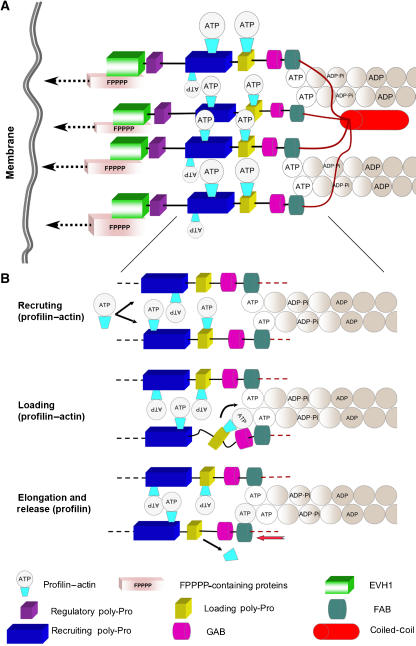
Figure 5.
Model of filament elongation by Ena/VASP. (A) Ena/VASP tetramers are recruited to the sites of active filament assembly via interactions of the EVH1 domain with FPPPP-containing scaffolding proteins such as zyxin (Reinhard et al, 1995b), vinculin (Brindle et al, 1996; Reinhard et al, 1996), lamellipodin (Krause et al, 2004), migfilin (Zhang et al, 2006) and palladin (Boukhelifa et al, 2004). These proteins are connected to the plasma membrane directly or via interactions with membrane-binding scaffolding proteins (indicated by dashed arrows). VASP tetramerization and tethering via the EVH1 domain are likely important for processivity in cells (see Discussion). The recruiting poly-Pro site contains multiple profilin–actin binding modules, which may serve to increase the local concentration of actin monomers for assembly. It is unclear if each subunit of Ena/VASP supports the elongation of one filament or, as shown in the figure, two subunits per filament are required (one per each long-pitch helix). (B) Close view of the elongation machinery showing three steps of the elongation mechanism (indicated by black arrows): (1) recruitment of profilin–actin to the poly-Pro regions, (2) loading of profilin–actin from the last poly-Pro site onto the GAB domain and (3) filament elongation (red arrow) resulting from the addition of the actin subunit at the barbed end and the release of profilin.
Discussion
Two monomeric actin-binding proteins, profilin and thymosin-β4, contribute to maintaining a large fraction (∼50%) of the cellular actin in the unpolymerized pool at a concentration (∼20–100 μM) that is 200- to 1000-fold higher than the critical concentration for barbed-end polymerization (∼0.1 μM) (Pollard and Borisy, 2003). While free ATP–actin is present in low amounts (0.1–1 μM) and thymosin-β4–actin is polymerization incompetent, profilin–actin (present at 5–40 μM intracellular concentration) constitutes the main source of actin monomers for polymerization (Dickinson and Purich, 2002; Dickinson et al, 2002). Multiple properties allow profilin to play such a central role in filament assembly (dos Remedios et al, 2003; Witke, 2004; Yarmola and Bubb, 2006); (1) profilin catalyzes the exchange of ADP for ATP on actin, which replenishes the pool of ATP–actin monomers ready for polymerization, (2) it inhibits filament nucleation, which explains in part the need for nucleation promoting factors, (3) profilin-bound actin monomers cannot add to pointed ends, but can elongate filament barbed ends at approximately the same rate as free actin monomers, (4) profilin competes effectively with thymosin-β4 for actin, which is due to its greater affinity for actin than thymosin-β4 and high dissociation constants of their respective complexes with actin, and (5) profilin binds poly-Pro sequences in various cytoskeletal proteins, including the Arp2/3 complex activator WASP and the elongation factors Ena/VASP and formin. Thus, recruitment of profilin–actin increases the elongation rates of formin (Kovar et al, 2006) and Ena/VASP-dependent Listeria motility (Chakraborty et al, 1995; Kang et al, 1997; Geese et al, 2000, 2002; Machner et al, 2001; Auerbuch et al, 2003; Grenklo et al, 2003).
How filament elongation factors process profilin–actin for its incorporation at the barbed end of the filament has remained a mystery. A view commonly held is that the role of the Pro-rich region of Ena/VASP (as well as formin) is to increase the local concentration of profilin–actin available for assembly (Kang et al, 1997; Geese et al, 2000; Machner et al, 2001; Dickinson and Purich, 2002; Grenklo et al, 2003; Kovar et al, 2006). This work provides evidence for a more active role of the Pro-rich region of Ena/VASP in elongation, by directing the transition of profilin–actin to the GAB domain, from where the actin monomer can join the barbed end of the growing filament (Figure 5). Thus, we have shown here that the Pro-rich region of Ena/VASP has a complex organization, consisting of three discrete modules: the regulatory, recruiting and loading poly-Pro sites (Figure 1). While the binding of profilin to poly-Pro sequences of the kind found in the recruiting site had been previously studied (Petrella et al, 1996; Kang et al, 1997), the loading site had not yet been investigated. This site is particularly important since it is highly conserved among all the members of the Ena/VASP family and WASP, and is connected to the GAB domain by a short Gly-rich linker (Supplementary Figure 1).
We found that the loading poly-Pro site binds profilin–actin with significantly higher affinity than profilin alone (Figure 2; Supplementary Figure 2). The implication of this finding is that Ena/VASP has the ability to discriminate between polymerization-competent profilin–actin complexes and profilin alone, which could jam the elongation machinery. We had previously reported that the GAB domain also has higher affinity for profilin–actin than actin alone (Chereau and Dominguez, 2006). Therefore, both the loading poly-Pro and GAB domains have a clear preference for profilin–actin. However, the two sites must cooperate, and not compete, for the transfer of profilin–actin from the cellular pool onto the barbed end of the elongating filament. To test this hypothesis, we determined the high-resolution structures of profilin–actin bound to the loading poly-Pro (Figure 3) and poly-Pro-GAB of human VASP (Figure 4). Among the main results of the structures are the following: (1) the conformation of profilin–α-actin in the two structures is very similar to that of profilin–β-actin determined previously (Schutt et al, 1993), (2) the presence of non-Pro amino acids in the loading poly-Pro site, and in particular Leu 209 that is conserved in Ena/VASP and WASP, define the register, polarity and orientation of binding, (3) these factors are in turn essential in determining the position of the GAB domain, so that it can access its binding site on actin within the profilin–actin complex, (4) the GAB domain is related to the WH2 of WASP and makes similar interactions with actin, (5) the GAB domain can bind actin concomitantly with profilin, which requires significant repositioning compared with WH2 domains crystallized in the absence of profilin, and most importantly (6) the loading poly-Pro and GAB domains of Ena/VASP do not compete for profilin–actin, which they can bind simultaneously (Figure 4).
The structures do not provide clear insights as to why the loading poly-Pro binds more tightly to profilin–actin (Figure 2; Supplementary Figure 2). However, compared with other actin structures, profilin produces a slight closure of the cleft between subdomains 1 and 3, known as the target-binding cleft (Dominguez, 2004), which may explain the increased affinity of profilin–actin for the GAB domain observed previously (Chereau and Dominguez, 2006). Another possibility is the mutual stabilization of profilin and actin within their complex, making the binding of both the GAB and poly-Pro domains more favorable. This latter effect appears to work both ways, since the affinity of actin for profilin also increases when profilin is bound to the loading poly-Pro peptide of VASP (Supplementary Figure 3).
What is the next step after profilin–actin has been recruited to the loading poly-Pro and GAB domains? We would like to suggest that the actin monomer bound to the GAB domain is already in contact with the barbed end of the FAB-tethered filament (Figure 5B). This idea is based strictly on structural constraints imposed by the short length of the linker between the GAB and FAB domains (Supplementary Figure 1), independently of where on F-actin the latter binds. However, as noted before (Chereau and Dominguez, 2006), the FAB domain is related to the C region of WASP and the two appear to be related to WH2. In other words, the GAB-FAB of Ena/VASP is related to the WH2-C of WASP, a relationship that extends at both ends to the loading poly-Pro and, to a lesser extent, to the acidic region (Supplementary Figure 1). WH2 domains often occur in tandem repeats (Paunola et al, 2002) and, if this proposal is correct, GAB-FAB and WH2-C constitute specialized forms of tandem WH2 domains, involved in filament elongation and Arp2/3 activation, respectively. FAB would then be expected to bind in the cleft between subdomains 1 and 3 of the last actin subunit at the barbed end of the elongating filament. The transition of an actin monomer from the GAB domain to the barbed end of the elongating filament would then trigger a series of events, including the release of profilin and the stepping of Ena/VASP (Figure 5B).
What is the driving force of these events? The mechanism proposed here is generally in agreement with the direct-transfer polymerization model of Dickinson and Purich (2002) (Dickinson et al, 2002). According to this model, ATP hydrolysis by actin is the driving force for the processive stepping of actin polymerization-based motors. However, the precise role of ATP hydrolysis in elongation remains controversial. While it has been suggested that ATP hydrolysis is required for profilin dissociation (Romero et al, 2007), other experiments show that filament elongation rates can exceed the ATP hydrolysis rate by at least 20-fold (Blanchoin and Pollard, 2002). Furthermore, phosphate dissociation after hydrolysis is slow (Carlier and Pantaloni, 1986), implying that a significant fraction of the filament consists of ADP-Pi-actin, which is considered to be structurally and functionally indistinguishable from ATP–actin (Pollard and Borisy, 2003). Therefore, based on the evidence to date, it is unclear whether ATP hydrolysis or a nucleotide-independent conformational change due to the G- to F-actin transition triggers profilin dissociation and Ena/VASP stepping.
How does this mechanism apply to all four subunits of the full-length Ena/VASP tetramer? Although the core of the elongation machinery consists of the poly-Pro-GAB-FAB region (Figure 5), it is very likely that all the domains of Ena/VASP are necessary for processive elongation in cells (the exception may be the motility of Listeria, which has evolved different recruitment mechanisms). Ena/VASP tetramers are recruited to sites of rapid assembly by interactions of the EVH1 domains with FPPPP-containing proteins such as zyxin (Reinhard et al, 1995b), vinculin (Brindle et al, 1996; Reinhard et al, 1996), lamellipodin (Krause et al, 2004), migfilin (Zhang et al, 2006) and palladin (Boukhelifa et al, 2004). Filaments associated with Ena/VASP tend to be unbranched, longer and form crosslinked bundles (Skoble et al, 2001; Bear et al, 2002; Svitkina et al, 2003). A tetramer may be more efficient at generating such actin bundles, in particular if the subunits of Ena/VASP work cooperatively, both within and between neighboring tetramers. But more importantly, tetramerization via the CC domain and tethering via the EVH1 domain may enable Ena/VASP to hold on to a particular filament during processive stepping, by sequentially allowing each subunit to release and advance while the others remain attached. Consistent with this idea, recent evidence shows that all the domains of Ena/VASP are required for proper localization and continued polymerization at filopodial tips (Applewhite et al, 2007). In particular, the GAB domain was found to play a key role in maintaining filopodial tip localization, which is consistent with recent evidence that the WH2 of N-WASP transiently tethers actin filament to the membrane (Co et al, 2007).
Many questions will require further investigation. For instance, it is unclear whether a single subunit of Ena/VASP can support the elongation of a whole filament, or whether two subunits per filament are required (one for each of the long-pitch helices of the filament). It is also possible that cycles of elongation by Ena/VASP alternate with short periods of depolymerization, or backward steps. Finally, we have stressed here the striking resemblance between the loading poly-Pro-GAB-FAB of VASP and the equivalent region in WASP (Supplementary Figure 1). Some of the results obtained here may therefore be applicable to WASP-Arp2/3-mediated branch nucleation. However, there also are important differences, notably that the helix of WH2 is longer than that of the GAB domain (Figure 4D), which may preclude the transfer of profilin–actin from the last poly-Pro of WASP to WH2.
Materials and methods
Preparation of proteins and peptides
The cDNA encoding for human profilin I was purchased from ATCC, amplified by PCR and inserted into vector pET29/T7 (Novagen). BL21(DE3) competent cells (Invitrogen) were transformed with this constructs and grown in LB medium at 37°C until the OD at 600 nm reached a value of 0.6. Expression was induced by the addition of 1 mM isopropyl-β-D-1-thiogalactopyranoside and carried out for 4 h at 37°C. Cells were harvested by centrifugation, resuspended in 10 mM Tris pH 7.5, 100 mM glycine, 100 mM NaCl, 1 mM DTT and lysed using a French press. The soluble lysate was purified on an affinity poly-L-proline sepharose column. Profilin was eluted from the column using 30% (v/v) DMSO, dialysed against 10 mM Tris pH 7.5, 50 mM NaCl and concentrated to ∼14 mg/ml. Actin was prepared from rabbit skeletal muscle as described (Graceffa and Dominguez, 2003). Peptides corresponding to human VASP residues 198–213 (loading poly-Pro) and 202–244 (loading poly-Pro and GAB domain) were synthesized on an ABI431 peptide synthesizer and purified by HPLC (see Supplementary Figure 1 for sequence information). The concentrations of the peptides were determined by amino-acid analysis (Dana-Farber Cancer Institute, Boston, MA, USA).
Crystallization, data collection and structure determination
The complexes of profilin–actin with the two VASP peptides were prepared by mixing 66 μM actin in G-buffer (2 mM HEPES pH 7.4, 0.2 mM CaCl2 and 0.2 mM ATP) with 73 μM profilin (in 10 mM Tris pH 7.5, 50 mM NaCl), followed by the addition of 73 μM VASP peptide (same buffer as profilin). The complexes were stored on ice and used at this concentration during crystallization trials at 4 and 20°C. The best crystals of profilin–actin with the loading poly-Pro peptide were obtained at 4°C in 4 μl sitting drops, consisting of a 1:1 (v/v) mixture of protein solution with 200 mM sodium formate and 20% (w/v) PEG 3350 well solution. The best crystals of profilin–actin with the poly-Pro-GAB peptide were obtained at 4°C in 3 μl sitting drops, consisting of a 1:2 (v/v) mixture of the protein solution with 150 mM DL-malic acid pH 7.0 and 18% (w/v) PEG 3350 well solution. The crystals were flash-frozen in liquid nitrogen, using 20% glycerol as cryoprotectant. X-ray data sets were collected at the BioCARS beamline 14-BM-C (Advance Photon Source, Argonne, IL, USA). The data sets were indexed and scaled with program HKL-2000 (HKL Research Inc.). The structures were determined by molecular replacement using the CCP4 program AMoRe and the structure of profilin–β-actin (Schutt et al, 1993) as a search model. Model building and refinement were performed with the program Coot (Emsley and Cowtan, 2004) and CCP4 program Refmac (Table I).
Binding of VASP peptide measured by tryptophan fluorescence
Binding of the loading poly-Pro peptide of human VASP to profilin and profilin–actin was measured by the change of intrinsic tryptophan fluorescence using a Cary Eclipse Fluorescence spectrophotometer (Varian). The excitation wavelength was set to 295 nm and the emission spectra were recorded from 300 to 400 nm. The experiments were performed at 20°C in 10 mM sodium phosphate pH 7.4 and 150 mM NaCl. The concentration of profilin and profilin–actin in the cell was 5 μM. The VASP peptide was added at varying concentrations (0, 0.15, 0.30, 0.6, 1.25, 2.5, 5, 10, 20, 40, 80, 160 and 320 μM). Each data point represents the average of five independent measurements (Figure 2). Least-square fitting of the data, using a single-site binding model, resulted in dissociation constant (KD) estimates of 84 and 7.5 μM for profilin and profilin–actin, respectively.
Binding of VASP peptide measured by ITC
Binding of the VASP peptide to profilin and profilin–actin was also measured using ITC on a VP-ITC instrument (MicroCal, Northampton, MA, USA). To determine ΔH and Ka of association, the VASP peptide, at a concentration of 1 mM, was titrated in 10-μl injections, into 1.44 ml of 100 μM profilin (or 80 μM profilin–actin). The experiments were all performed at 25°C in 10 mM sodium phosphate pH 7.4 and 150 mM NaCl. The duration of each injection was 10 s, with an interval of 3 min between injections. The heat of binding was corrected for the small exothermic heat of injection, determined by injecting VASP peptides into buffer. Data were analyzed using the MicroCal's Origin program.
Supplementary Material
Supplementary Figure 1–5
Supplementary Movie 6
Acknowledgments
This work was supported by NIH grant HL086655. We thank Paul Leavis and Elizabeth Gowell for the synthesis of VASP peptides. Use of the Advanced Photon Source was supported by DOE contract no. W-31-109-Eng-38. Use of the BioCARS facilities was supported by NIH grant RR07707.
References
- Ahern-Djamali SM, Bachmann C, Hua P, Reddy SK, Kastenmeier AS, Walter U, Hoffmann FM (1999) Identification of profilin and src homology 3 domains as binding partners for Drosophila enabled. Proc Natl Acad Sci USA 96: 4977–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima SI, Svitkina TM, Gertler FB, Borisy GG (2007) Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell 18: 2579–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Loureiro JJ, Gertler FB, Theriot JA, Portnoy DA (2003) Ena/VASP proteins contribute to Listeria monocytogenes pathogenesis by controlling temporal and spatial persistence of bacterial actin-based motility. Mol Microbiol 49: 1361–1375 [DOI] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M (1999) The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem 274: 23549–23557 [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA (2005) Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem 280: 28653–28662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB (2002) Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109: 509–521 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD (2002) Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry 41: 597–602 [DOI] [PubMed] [Google Scholar]
- Boukhelifa M, Parast MM, Bear JE, Gertler FB, Otey CA (2004) Palladin is a novel binding partner for Ena/VASP family members. Cell Motil Cytoskeleton 58: 17–29 [DOI] [PubMed] [Google Scholar]
- Brannetti B, Helmer-Citterich M (2003) iSPOT: a web tool to infer the interaction specificity of families of protein modules. Nucleic Acids Res 31: 3709–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle NP, Holt MR, Davies JE, Price CJ, Critchley DR (1996) The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem J 318 (Part 3): 753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon JP (1997) Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci 53: 621–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D (1986) Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry 25: 7789–7792 [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove CJ, Jockusch BM, Reinhard M, Walter U, Wehland J (1995) A focal adhesion factor directly linking intracellularly motile Listeria monocytogenes and Listeria ivanovii to the actin-based cytoskeleton of mammalian cells. EMBO J 14: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Dominguez R (2006) Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J Struct Biol 155: 195–201 [DOI] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R (2005) Actin-bound structures of Wiskott–Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA 102: 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chik JK, Lindberg U, Schutt CE (1996) The structure of an open state of beta-actin at 2.65 Å resolution. J Mol Biol 263: 607–623 [DOI] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J (2007) Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell 128: 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J (1993) Life at the leading edge: the formation of cell protrusions. Annu Rev Cell Biol 9: 411–444 [DOI] [PubMed] [Google Scholar]
- Coppolino MG, Krause M, Hagendorff P, Monner DA, Trimble W, Grinstein S, Wehland J, Sechi AS (2001) Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcgamma receptor signalling during phagocytosis. J Cell Sci 114: 4307–4318 [DOI] [PubMed] [Google Scholar]
- Dickinson RB, Purich DL (2002) Clamped-filament elongation model for actin-based motors. Biophys J 82: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RB, Southwick FS, Purich DL (2002) A direct-transfer polymerization model explains how the multiple profilin-binding sites in the actoclampin motor promote rapid actin-based motility. Arch Biochem Biophys 406: 296–301 [DOI] [PubMed] [Google Scholar]
- Dominguez R (2004) Actin-binding proteins—a unifying hypothesis. Trends Biochem Sci 29: 572–578 [DOI] [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ (2003) Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev 83: 433–473 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Feng S, Chen JK, Yu H, Simon JA, Schreiber SL (1994) Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3–ligand interactions. Science 266: 1241–1247 [DOI] [PubMed] [Google Scholar]
- Geese M, Loureiro JJ, Bear JE, Wehland J, Gertler FB, Sechi AS (2002) Contribution of Ena/VASP proteins to intracellular motility of Listeria requires phosphorylation and proline-rich core but not F-actin binding or multimerization. Mol Biol Cell 13: 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geese M, Schluter K, Rothkegel M, Jockusch BM, Wehland J, Sechi AS (2000) Accumulation of profilin II at the surface of Listeria is concomitant with the onset of motility and correlates with bacterial speed. J Cell Sci 113 (Part 8): 1415–1426 [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM (1995) enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev 9: 521–533 [DOI] [PubMed] [Google Scholar]
- Graceffa P, Dominguez R (2003) Crystal structure of monomeric actin in the ATP state: structural basis of nucleotide-dependent actin dynamics. J Biol Chem 278: 34172–34180 [DOI] [PubMed] [Google Scholar]
- Grenklo S, Geese M, Lindberg U, Wehland J, Karlsson R, Sechi AS (2003) A crucial role for profilin–actin in the intracellular motility of Listeria monocytogenes. EMBO Rep 4: 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF (2004) The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117: 611–623 [DOI] [PubMed] [Google Scholar]
- Irobi E, Burtnick LD, Urosev D, Narayan K, Robinson RC (2003) From the first to the second domain of gelsolin: a common path on the surface of actin? FEBS Lett 552: 86–90 [DOI] [PubMed] [Google Scholar]
- Jonckheere V, Lambrechts A, Vandekerckhove J, Ampe C (1999) Dimerization of profilin II upon binding the (GP5)3 peptide from VASP overcomes the inhibition of actin nucleation by profilin II and thymosin beta4. FEBS Lett 447: 257–263 [DOI] [PubMed] [Google Scholar]
- Kang F, Laine RO, Bubb MR, Southwick FS, Purich DL (1997) Profilin interacts with the Gly–Pro–Pro–Pro–Pro–Pro sequences of vasodilator-stimulated phosphoprotein (VASP): implications for actin-based Listeria motility. Biochemistry 36: 8384–8392 [DOI] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS (1999) Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J Biol Chem 274: 36963–36972 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD (2006) Control of the assembly of ATP- and ADP–actin by formins and profilin. Cell 124: 423–435 [DOI] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB (2003) Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol 19: 541–564 [DOI] [PubMed] [Google Scholar]
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, Yaffe MB, Boussiotis VA, Gertler FB (2004) Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell 7: 571–583 [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A (2001) Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol 11: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Kuhnel K, Jarchau T, Wolf E, Schlichting I, Walter U, Wittinghofer A, Strelkov SV (2004) The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc Natl Acad Sci USA 101: 17027–17032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C (1997) The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J 16: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R (2007) Structural basis for the actin-binding function of missing-in-metastasis. Structure 15: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Urbanke C, Barzik M, Otten S, Sechi AS, Wehland J, Heinz DW (2001) ActA from Listeria monocytogenes can interact with up to four Ena/VASP homology 1 domains simultaneously. J Biol Chem 276: 40096–40103 [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Janmey PA, Almo SC (1997) Structure of the profilin–poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol 4: 953–960 [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Rozwarski DA, Fedorov E, Fedorov AA, Almo SC (1999) Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat Struct Biol 6: 666–671 [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB (2003) Scansite 2.0: proteome protwide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunola E, Mattila PK, Lappalainen P (2002) WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett 513: 92–97 [DOI] [PubMed] [Google Scholar]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF (1994) Interaction of profilin with G-actin and poly(L-proline). Biochemistry 33: 8472–8478 [DOI] [PubMed] [Google Scholar]
- Petrella EC, Machesky LM, Kaiser DA, Pollard TD (1996) Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry 35: 16535–16543 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Lee DJ, Lim WA (1999) Structure of the enabled/VASP homology 1 domain–peptide complex: a key component in the spatial control of actin assembly. Cell 97: 471–480 [DOI] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U (1995a) The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J 14: 1583–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Jouvenal K, Tripier D, Walter U (1995b) Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein). Proc Natl Acad Sci USA 92: 7956–7960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Rudiger M, Jockusch BM, Walter U (1996) VASP interaction with vinculin: a recurring theme of interactions with proline-rich motifs. FEBS Lett 399: 103–107 [DOI] [PubMed] [Google Scholar]
- Romero S, Didry D, Larquet E, Boisset N, Pantaloni D, Carlier MF (2007) How ATP hydrolysis controls filament assembly from profilin–actin: IMPLICATION FOR FORMIN PROCESSIVITY. J Biol Chem 282: 8435–8445 [DOI] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U (1993) The structure of crystalline profilin–beta-actin. Nature 365: 810–816 [DOI] [PubMed] [Google Scholar]
- Skoble J, Auerbuch V, Goley ED, Welch MD, Portnoy DA (2001) Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch formation, and Listeria monocytogenes motility. J Cell Biol 155: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Rider JE, Hoffman NG, Fowlkes DM, Quillam LA, Kay BK (1996) Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci USA 93: 1540–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel TP (1993) On the crawling of animal cells. Science 260: 1086–1094 [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG (2003) Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol 160: 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walders-Harbeck B, Khaitlina SY, Hinssen H, Jockusch BM, Illenberger S (2002) The vasodilator-stimulated phosphoprotein promotes actin polymerisation through direct binding to monomeric actin. FEBS Lett 529: 275–280 [DOI] [PubMed] [Google Scholar]
- Witke W (2004) The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 14: 461–469 [DOI] [PubMed] [Google Scholar]
- Yarmola EG, Bubb MR (2006) Profilin: emerging concepts and lingering misconceptions. Trends Biochem Sci 31: 197–205 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tu Y, Gkretsi V, Wu C (2006) Migfilin interacts with vasodilator-stimulated phosphoprotein (VASP) and regulates VASP localization to cell–matrix adhesions and migration. J Biol Chem 281: 12397–12407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1–5
Supplementary Movie 6