Abstract
The Ras superfamily is comprised of at least four large families of regulatory guanosine triphosphate–binding proteins, including the Arfs. The Arf family includes three different groups of proteins: the Arfs, Arf-like (Arls), and SARs. Several Arf family members have been very highly conserved throughout eukaryotic evolution and have orthologues in evolutionally diverse species. The different means by which Arf family members have been identified have resulted in an inconsistent and confusing array of names. This confusion is further compounded by differences in nomenclature between different species. We propose a more consistent nomenclature for the human members of the Arf family that may also serve as a guide for nomenclature in other species.
Arf family history: Arfs, Arls, SARs, and other members
Arfs.
Arf was first discovered, purified, and functionally defined as the protein cofactor required for cholera toxin–catalyzed ADP ribosylation of the stimulatory regulatory subunit (Gs) of adenylyl cyclase (Enomoto and Gill, 1980; Kahn and Gilman, 1984) and, shortly thereafter, was shown to be a GTP-binding protein (Kahn and Gilman, 1986). Use of the acronym Arf is currently preferred to ADP ribosylation factor, as only Arf1–6 shares the cofactor activity for cholera toxin and because ADP ribosylation does not appear to be involved in any aspect of the normal cellular actions of any member of the family. The use of all capital letters (e.g., ARF1) refers specifically to the human gene or protein, whereas when only the first letter is capitalized (e.g., Arf1), it may refer to the protein from more than one species, an activity, or a group of proteins. Since their discovery, they have been found to be ubiquitous regulators of membrane traffic and phospholipid metabolism in eukaryotic cells (for reviews and discussion of Arf actions see Nie et al., 2003; Burd et al., 2004; Kahn, 2004). Arfs are soluble proteins that translocate onto membranes in concert with their activation, or GTP binding. The biological actions of Arfs are thought to occur on membranes and to result from their specific interactions with a large number of effectors that include coat complexes (COPI, AP-1, and AP-3), adaptor proteins (GGA1-3 and MINT1-3/X11α-γ/APBA1-3), lipid-modifying enzymes (PLD1, phosphatidylinositol (4,5)-kinase, and phosphatidylinositol (4)-kinase), and others. Arf proteins are activated by guanosine diphosphate (GDP) to GTP exchange, which is stimulated by the Sec7 domain of Arf guanine nucleotide exchange factors, and their activity is terminated upon the hydrolysis of GTP, which is stimulated by interaction with an Arf GTPase-activating protein.
Cloning and sequencing of the first Arf family member (Sewell and Kahn, 1988) led directly to the realization that Arfs are closely related to both the Ras and heterotrimeric G protein α subunit families of GTPases, and all are thought to have arisen from a common ancestor. The very high degree of conservation of Arf sequences in eukaryotes (74% between human and yeast) was also noted early on and has allowed the ready identification of orthologues in every examined eukaryote, including Giardia lamblia, which lack Ras and G protein α subunits (Murtagh et al., 1992).
Cloning by low stringency hybridization and chance led to the identification of additional members of the Arf family in a wide array of eukaryotic species. The number of mammalian Arfs grew to six by 1992 (Tsuchiya et al., 1991) and were named in their order of discovery (Price et al., 1988; Bobak et al., 1989; Kahn et al., 1991; Lee et al., 1992). The first confusion in the nomenclature was that the current human ARF4 was originally published with the name ARF2 (Kahn, et al., 1991). In fact, humans appear to have lost the ARF2 orthologue, which is present in other mammals (including rats, mice, and cows). The combination of protein sequence comparisons and intron/exon boundaries of Arf genes led to further classification of the six mammalian Arfs into classes: class I (ARF1–3 are >96% identical), class II (ARF4 and ARF5 are 90% identical to each other and 80% identical to the other Arfs), and class III (ARF6 is 64–69% identical to the other Arfs). Phylogenetic analyses support the conclusion that the three classes of Arf diverged early, as flies and worms have single representatives of each of the three classes, and the number of genes/proteins in class I and II were later expanded in vertebrates.
Arls.
The initial criteria for naming new Arfs were functional, and only those proteins that could (1) serve as cofactors for cholera toxin, (2) rescue the lethal arf1 − arf2 − deletion in Saccharomyces cerevisiae, and (3) directly activate PLD were given the name Arf. Thus, with the chance cloning of an essential gene in Drosophila melanogaster that encoded a protein closely related to the Arfs (50–60% identity) but lacking in these activities, it was named arflike (Tamkun et al., 1991). When orthologues were found in several other species, the name was changed to Arf-like 1 (ARL1) in those species (Kahn et al., 1992; Breiner et al., 1996; Lowe et al., 1996). Note that although the name Arf still denotes a protein with one or more specific functions or activities, the term Arl does not. The term Arl indicates only that the protein is structurally related to Arfs. Thus, the Arls are not a coherent group either functionally or phylogenetically.
PCR amplification with degenerate oligonucleotide primers (Clark et al., 1993; Schurmann et al., 1994) revealed the existence of a large number of mammalian cDNAs encoding closely related proteins. The next to be cloned and sequenced were ARL2 (Clark et al., 1993), ARL3 (Cavenagh et al., 1994), ARL4 (Schurmann et al., 1994), and ARL5 (Breiner et al., 1996). Each of the encoded proteins has a glycine at position 2, the site of N-myristoylation in all Arf proteins. Note that although ARL2 and ARL3 have the NH2-terminal glycine, they appear not to be substrates for N-myristoyltransferases.
Around this time, a protein with similar percent identities to the Arf and Arls was found, but it lacked the NH2-terminal glycine, was membrane associated, and displayed distinctive nucleotide handling properties (Schurmann et al., 1995). Thus, it was given the name Arf-related protein 1 (ARFRP1) to distinguish it from the Arls and Arfs. We realize today that this was unfortunate, as several of the more recently identified Arls also have functions and biochemical properties that are quite divergent from Arfs.
SARs.
SAR1 was among the earliest members of the Arf family sequenced, and it came out of genetic screens in the yeast S. cerevisiae as a suppressor of sec12(ts) (Nakano and Muramatsu, 1989). Its name is derived from its identification as a secretion-associated and Ras-related protein. Cloning of the mammalian orthologues revealed the presence of two closely related (90% identity) proteins/genes (Kuge et al., 1994). With <30% identity to Arfs or Arls, the SAR proteins are only slightly closer in sequence to Arfs than to other families of GTPases, but they also share considerable functional relatedness to Arfs in that they act through the recruitment of coat proteins or complexes to initiate vesicle budding. SARs lack the other aforementioned Arf activities.
Additional domains.
An interesting variation is found in ARD1/tripartite motif 23 (TRIM23), a 64-kD protein that possesses a ∼20-kD domain at its COOH terminus with 60% identity to Arfs (Mishima et al., 1993). Originally named based on the presence of the Arf domain, ARD1 is also a member of the TRIM family, from which it obtained its current name, TRIM23. A large extension is also seen in ARL13B, a protein of 428 residues that contains an Arl domain at its NH2 terminus (Chiang et al., 2004; Fan et al., 2004). Although the NH2-terminal portion of TRIM23 may possess GTPase-activating protein activity toward its own Arf domain (Vitale et al., 1996) and E3 ubiquitin ligase activity (Vichi et al., 2005), the COOH-terminal portion of ARL13B has no defined domains or functions to date.
Defining the Arf family
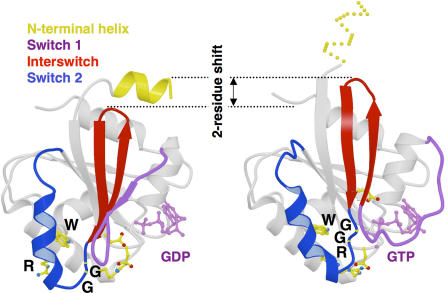
As the discussion above suggests, there are no shared functions or activities that justify grouping Arf, Arl, and SAR proteins into a family with a common nomenclature. Similarities in protein sequences within the Arf family were first identified by alignment and phylogenetic analyses and were shown to provide distinct signatures that allowed differentiation from Ras, G protein α subunits, and other GTPases. These include an NH2-terminal extension, a glycine acceptor for myristate at position 2, an aspartate at position 26 (in contrast to the glycine 12 of Ras that carries oncogenic potential), and other residues that are very highly conserved within the family. These early observations were put on more solid functional footing when they were found to map to unique elements in their three-dimensional structures, which allow for the GDP/GTP switch to be coupled with interaction signals opposite to the nucleotide-binding site (for review see Pasqualato et al., 2002). The prominent feature of this unique nucleotide switch is a nonconventional GDP-bound form in which the two β strands that connect the nucleotide-sensitive switch 1 and 2 regions (also called the interswitch) are retracted in the protein core and must undergo a two-residue shift to reach the active conformation (Fig. 1). However, the interswitch cannot do so unless the NH2-terminal helical extension, which caps the interswitch and locks it in the retracted conformation, has been displaced. In the case of ARF1, biochemical studies have established that this requires the interaction of the NH2 terminus with membranes, thus allowing the nucleotide-binding site to detect and respond to remote protein–membrane interactions (Antonny et al., 1997). Like Arf proteins, each Sar has an NH2-terminal amphipathic helix that functions as a structural GDP/GTP switch to anchor the GTP-bound form to membranes of the endoplasmic reticulum (Huang et al., 2001; Bi et al., 2002). Furthermore, membrane insertion of this NH2-terminal helix was recently shown to initiate membrane bending at the early stages of COPII coat assembly and to be subsequently required for the completion of COPII vesicle fission (Lee et al., 2005).
Figure 1.
The structural “air de famille.” In Arfs, Arls, and SAR proteins, the interswitch toggles from an unusual retracted conformation in the GDP-bound form that is fastened by the NH2-terminal helix to an exposed conformation in the GTP-bound form that is stabilized by the W/GG/R signature (shown here for ARF6-GDP and ARF6-GTP). This large conformational change, which involves a two-residue β-strand register shift in the core of the G domain, allows the nucleotide-binding site to detect remote interactions taking place at the NH2 terminus (reproduced from Pasqualato et al., 2001 with permission).
Structural analysis of ARF1 and ARF6 GDP/GTP cycles and their comparison with those of small GTP-binding proteins whose interswitch does not toggle identified three structural determinants for this movement: a helical NH2-terminal extension that fastens the retracted, GDP-bound interswitch; a shorter interswitch that can retract completely; and a sequence signature (wDvGGqXXXRxxW) that provides both flexibility for the movement (GG) and hydrogen bonds for stabilization of the active conformation (R/W). These characteristics are present in all Arf and most Arl sequences, which, therefore, are predicted to have the ability to undergo the interswitch toggle to detect interactions opposite to the nucleotide-binding site, whatever their nature, and propagate them to this site (Pasqualato et al., 2002).
These structural criteria for unifying Arf and Arl proteins as a family have since been supported by various structures of GDP-bound Arf and Arl proteins (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200512057/DC1). It should be noted, however, that one subgroup, ARL4, has a long interswitch that may have lost the ability to toggle, whereas structures of NH2-terminally truncated ARL8A and ARL8B bound to GDP have a GTP-like conformation. This suggests that truncation of the NH2 terminus is sufficient in this family to destabilize the retracted interswitch or that these proteins have lost their ability to undergo the interswitch toggle. Recent work on ARL3 suggests that proteins interacting with the NH2 terminus could also work as the displacing factor as an alternative to membranes (Behnia et al., 2004; Setty et al., 2004).
Arf family nomenclature
Table I contains information on proposed and previous names as well as other information on the human ARF family members. EST and genomic sequencing resulted in the identification of subsequent Arf-like proteins, and these proteins/genes were often misnamed or named multiple times by different research groups. Some of these names suggest relationships that are misleading, and some are called Arfs despite (presumably) lacking any Arf activities. In many cases, no functional data are yet available for the most recently identified Arf family members. One protein has been referred to by four different names, and some proteins/genes were named by curators of databases responding to specific requests in a manner that disagreed with common usage by researchers in the field. The confusion is magnified when species differences are considered (e.g., yeast Arl3 is the orthologue of ARFRP1).
Table I.
The Arf family GTPases: summary of names, identifiers, and NH2-terminal sequences
| Accepted symbols |
Proposed new names | Previous HGNC symbol |
Former common name |
Other names or information |
Accession number (protein) |
Length (aa) |
NH2-terminal sequence |
Gene locus ID |
Human locus |
|---|---|---|---|---|---|---|---|---|---|
| ARF1 | ARF1 | ARF1 | NP_001649 | 181 | MGNIFANLFKGL | 375 | 1q42 | ||
| ARF3 | ARF3 | ARF3 | NP_001650 | 181 | MGNIFGNLLKSL | 377 | 12q13 | ||
| ARF4 | ARF4 | ARF4 | NP_001651 | 180 | MGLTISSLFSRL | 378 | 3p21.2-p21.1 | ||
| ARF5 | ARF5 | ARF5 | NP_001653 | 180 | MGLTVSALFSRI | 381 | 7q31.3 | ||
| ARF6 | ARF6 | ARF6 | NP_001654 | 175 | MGKVLSKIFGNK | 382 | 14q21.3 | ||
| ARL1 | ARL1 | ARL1 | ARFL1 | NP_001168 | 181 | MGGFFSSIFSSL | 400 | 7q13 | |
| ARL2 | ARL2 | ARL2 | ARFL2 | NP_001658 | 184 | MGLLTILKKMKQ | 402 | 11q13 | |
| ARL3 | ARL3 | ARL3 | ARFL3 | NP_004302 | 182 | MGLLSILRKLKS | 403 | 10q23.3 | |
| ARL4Ab | ADP ribosylation factorlike 4A | ARL4c | ARL4 | NP_005729 | 200 | MGNGLSDQTSIL | 10124 | 7p21-p15.3 | |
| ARL4Cb | ADP ribosylation factorlike 4C | ARL7c | ARL7 | LAK | NP_005728 | 192 | MGNISSNISAFQ | 10123 | 2q37.1 |
| ARL4Db | ADP ribosylation factorlike 4D | ARF4Lc | ARL9 | ARL6/ARF4L/ ARL5/ARL4L |
NP_001652 | 201 | MGNHLTEMAPTA | 379 | 17q12-q21 |
| ARL5Ab | ADP ribosylation factorlike 5A | ARL5c | ARL5 | NP_036229 | 179 | MGILFTRIWRLF | 26225 | 2q23.3 | |
| ARL5Bb | ADP ribosylation factorlike 5B | ARL8c | ARL5B | ARL8/similar to ARL5/ARL5-like |
NP_848930 | 179 | MGLIFAKLWSLF | 221079 | 10p12.31 |
| ARL5Cb | ADP ribosylation factorlike 5C | ARL12c | ARL12 | XP_372668 | 179a | MGQLIAKLMSIF | 390790 | 17q12 | |
| ARL6 | ARL6 | ARL6 | BBS3 | NP_115522 | 186 | MGLLDRLSVLLG | 84100 | 3q11.2 | |
| ARL8Ab | ADP ribosylation factorlike 8A | ARL10Bc | ARL8A | ARL10B/GIE2 | NP_620150 | 186 | MIALFNKLLDWF | 127829 | 1q32.1 |
| ARL8Bb | ADP ribosylation factorlike 8B | ARL10Cc | ARL8B | ARL10C/GIE1 | NP_060654 | 186 | MLALISRLLDWF | 55207 | 3p26.1 |
| ARL9 | ARL9 | NP_996802 | 123/ 265a |
MEFLEIGGSK/ MERGKVKKKE |
132946 | 4q12 | |||
| ARL10b | ADP ribosylation factorlike 10 | ARL10Ac | ARL10A | NP_775935 | 244 | MAPRPLGPLVLA | 285598 | 5q35.2 | |
| ARL11 | ARL11 | ARL11 | ARLTS1 | NP_612459 | 196 | MGSVNSRGHKAE | 115761 | 13q14.2 | |
| ARL13Ab | ADP ribosylation factorlike 13A | ARL13c | dJ341D10.2 | NP_001013008 | 297 | MFRLLSSCCSCL | 392509 | Xq22.1 | |
| ARL13Bb | ADP ribosylation factorlike 13B | ARL2L1c | DKFZp761H079 | NP_878899 | 428 | MFSLMASCCGWF | 200894 | 3q11.2 | |
| ARL14b | ADP ribosylation factorlike 14 | ARF7c | ARL10 | ARF7 | NP_079323 | 192 | MGSLGSKNPQTK | 80117 | 3q25.33 |
| ARL15b | ADP ribosylation factorlike 15 | ARFRP2c | FLJ20051 | NP_061960 | 204 | MSDLRITEAFLY | 54622 | 5p15.2 | |
| ARL16b | ADP ribosylation factorlike 16 | LOC339231 | XP_290777 | 173a | MCLLLGATGVGK | 339231 | 17q25.3 | ||
| ARFRP1 | ARFRP1 | ARFRP1 | Arp, Arp1 | NP_003215 | 201 | MYTLLSGLYKYM | 10139 | 20q13.3 | |
| SAR1Ab | SARA1c | SAR1A | HsSara1 | NP_064535 | 198 | MSFIFEWIYNGF | 56681 | 10q22.1 | |
| SAR1Bb | SARA2c | SAR1B | HsSara2 | NP_057187 | 198 | MSFIFDWIYSGF | 51128 | 5q31.1 | |
| TRIM23 | TRIM23 | ARD1 (α) | ARFD1, RNF46 | NP_001647 | 574 | MATLWNKLGAG | 373 | 5q12.3 |
Because of previous usage and to avoid confusion, the new assignments result in there being no gene/protein named ARL7 or ARL12. See Table S2 for a list of earlier names and references in which earlier names were used (available at http://www.jcb.org/cgi/content/full/jcb.200512057/DC1).
The sequence currently in the database is predicted to be incorrect, and our corrected information was used herein.
New names approved by the HGNC.
Previous names that were recently changed.
The need for a generally agreed upon nomenclature for the ARF family has become acute as a result of increasing confusion and interest in their study. It is not possible today to propose a completely consistent nomenclature, as there are simply too many studies with some of the earlier discovered proteins (e.g., ARFRP1 should be an ARL).
The nomenclature developed and described in this article builds on previous efforts to describe phylogenetic relationships and bring consistency to nomenclature (Pasqualato et al., 2002; Li et al., 2004; Logsdon and Kahn, 2004). It is the result of many discussions between researchers in the field and with the HUGO Genome Nomenclature Committee (HGNC) and has been widely circulated to Arf family researchers. We describe the presence in the human proteome of 29 members of the Arf family and a system for naming newly identified proteins in human or other species. The use of letter suffixes is reserved for those groups of proteins within the family that share higher percent identities and are, therefore, likely to share some level of functional redundancy. One exception to this is the ARL13A and ARL13B proteins, which have been given a common number based upon phylogenetic evidence. The consensus nomenclature for the Arf family is shown in Table I along with previous names and unique gene/protein identifying information. Note that in three cases (ARL5C, ARL9, and ARL16), the intron/exon boundary predictions in the database are thought to be incorrect (based upon comparisons with sequences in other species), resulting in differences in the predicted protein sequences. In these cases, we use our corrected sequences for comparisons and provide the predicted protein sequences of the human proteins (see supplemental material, available at http://www.jcb.org/cgi/content/full/jcb.200512057/DC1). In addition, there is one case (ARL9) in which it appears that alternative splicing yields two different proteins, one of which is truncated and predicted to be unable to bind nucleotides, so both are provided in the supplemental protein sequence material.
We also identify several gene sequences that have questionable EST/mRNA support and are likely pseudogenes derived from members of the Arf family. These genes, which are annotated by the HGNC, are therefore not included as Arf family members and are listed, along with their identifiers, in Table II. It is expected that additional pseudogenes will be found and added to this list over time. We also note some uncertainty as to whether ARL5C in Table I is a transcribed gene, as it may lack part of the consensus GTP-binding signature depending on which predicted protein sequence is used.
Table II.
Pseudogenes of the Arf family in the human genome
| New symbol |
New name | Other names/ information |
Chromosome location |
Accession number |
Gene locus ID |
|---|---|---|---|---|---|
| ARF1P1 | ADP ribosylation factor 1 pseudogene 1 | 7q21.3 | XM_498225 | 442334 | |
| ARF1P2 | ADP ribosylation factor 1 pseudogene 2 | ARL17A | 17q21.31 | NM_016632.1 | 51326 |
| ARF4P | ADP ribosylation factor 4 pseudogene | 9q34 | NG_001075 | 380 | |
| ARF4P2 | ADP ribosylation factor 4 pseudogene 2 | 20q13.33 | NG_001031 | 170485 | |
| ARF4P3 | ADP ribosylation factor 4 pseudogene 3 | 13q32.3 | XM_372496 | 390423 | |
| ARL4P | ADP ribosylation factorlike 4 pseudogene | ARL4B | 10q21.2 | XM_370560 | 387684 |
| ARL4P2 | ADP ribosylation factorlike 4 pseudogene 2 | ARL4B | 4p14 | NG_005394 | 152709 |
| SAR1P1 | SAR1 gene homologue (S. cerevisiae) pseudogene 1 | SARAP | 6p21 | AL035402 | 387048 |
| SAR1P2 | SAR1 gene homologue (S. cerevisiae) pseudogene 2 | SARA1P | 10q26.2 | AC026226 | 641312 |
| SAR1P3 | SAR1 gene homologue (S. cerevisiae) pseudogene 3 | 4q27 | XP_293671 | 344988 |
This is a list of the HGNC-recognized pseudogenes from the Arf family along with their locations in the genome, accession numbers, and gene identifiers.
Finally, we note that although the large majority of Arf family members appear to have very broad and perhaps ubiquitous tissue expression patterns, a few are far more restricted in their expression. Thus, it is expected that further additions and perhaps even deletions will be needed to keep the nomenclature of this family current and as consistent as possible. To ensure that new family members are assigned unique symbols, we strongly encourage authors to consult the HGNC before publishing any new names for members of this gene/protein family. This is a confidential service provided by the HGNC that will help prevent future confusion from arising. We also suggest that curators and researchers focusing on other organisms use the information provided in this article as much as possible to simplify and clarify the nomenclature across species.
Other researchers supporting the use of this nomenclature include: Bruno Antonny, Bill Balch, Vytas Bankaitis, Gary Bokoch, Juan Bonifacino, Chris Burd, Jim Casanova, Tamara Caspary, Dany Cassel, Rick Cerione, Pierre Chardin, Philippe Chavrier, Shamshad Cockcroft, Peter Cullen, Ivan de Curtis, Maria Antonella De Matteis, Julie Donaldson, Cryslin D'Souza-Schorey, John Exton, Victor Faundez, Jim Goldenring, Jean Gruenberg, Alan Hall, Fuchu He, Wangjin Hong, Victor Hsu, Mary Hunzicker-Dunn, Trevor Jackson, Cathy Jackson, Hans Joost, Toshi Katada, Fang-jen Lee, Michel Leroux, Jennifer Lippincott-Schwartz, John Logsdon, Alberto Luini, Vivek Malhotra, Ed Manser, Tobias Meyer, Paul Melancon, Joel Moss, Aki Nakano, Kazu Nakayama, Tommy Nilsson, Susanne Pfeffer, Richard Premont, Paul Randazzo, Anne Ridley, Scotty Robinson, Anne Rosenwald, Craig Roy, Hisataka Sabe, Randy Schekman, Nava Segev, Val Sheffield, Phil Stahl, Elizabeth Sztul, Chris Turner, Anne Theibert, Martha Vaughan, Kanamarlapudi Venkateswarlu, Fred Wittinghofer, Keqiang Ye, and Marino Zerial.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM68029 and GM67226 to R.A. Kahn), the Association pour la Recerche contre la Cancer (to J. Cherfils), and the French Research Ministry (ACI-BCMS to J. Cherfils).
Abbreviations used in this paper: GDP, guanosine diphosphate; TRIM, tripartite motif.
References
- Antonny, B., S. Beraud-Dufour, P. Chardin, and M. Chabre. 1997. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 36:4675–4684. [DOI] [PubMed] [Google Scholar]
- Behnia, R., B. Panic, J.R. Whyte, and S. Munro. 2004. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 6:405–413. [DOI] [PubMed] [Google Scholar]
- Bi, X., R.A. Corpina, and J. Goldberg. 2002. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 419:271–277. [DOI] [PubMed] [Google Scholar]
- Bobak, D.A., M.S. Nightingale, J.J. Murtagh, S.R. Price, J. Moss, and M. Vaughan. 1989. Molecular cloning, characterization, and expression of human ADP- ribosylation factors: two guanine nucleotide-dependent activators of cholera toxin. Proc. Natl. Acad. Sci. USA. 86:6101–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiner, M., A. Schurmann, W. Becker, and H.G. Joost. 1996. Cloning of a novel member (ARL5) of the ARF-family of Ras-related GTPases. Biochim. Biophys. Acta. 1308:1–6. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., T.I. Strochlic, and S.R. Gangi Setty. 2004. Arf-like GTPases: not so Arf-like after all. Trends Cell Biol. 14:687–694. [DOI] [PubMed] [Google Scholar]
- Cavenagh, M.M., M. Breiner, A. Schurmann, A.G. Rosenwald, T. Terui, C. Zhang, P.A. Randazzo, M. Adams, H.G. Joost, and R.A. Kahn. 1994. ADP-ribosylation factor (ARF)-like 3, a new member of the ARF family of GTP-binding proteins cloned from human and rat tissues. J. Biol. Chem. 269:18937–18942. [PubMed] [Google Scholar]
- Chiang, A.P., D. Nishimura, C. Searby, K. Elbedour, R. Carmi, A.L. Ferguson, J. Secrist, T. Braun, T. Casavant, E.M. Stone, and V.C. Sheffield. 2004. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). Am. J. Hum. Genet. 75:475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, J., L. Moore, A. Krasinskas, J. Way, J. Battey, J. Tamkun, and R.A. Kahn. 1993. Selective amplification of additional members of the ADP-ribosylation factor (ARF) family: cloning of additional human and Drosophila ARF- like genes. Proc. Natl. Acad. Sci. USA. 90:8952–8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto, K., and D.M. Gill. 1980. Cholera toxin activation of adenylate cyclase. Roles of nucleoside triphosphates and a macromolecular factor in the ADP ribosylation of the GTP-dependent regulatory component. J. Biol. Chem. 255:1252–1258. [PubMed] [Google Scholar]
- Fan, Y., M.A. Esmail, S.J. Ansley, O.E. Blacque, K. Boroevich, A.J. Ross, S.J. Moore, J.L. Badano, H. May-Simera, D.S. Compton, et al. 2004. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 36:989–993. [DOI] [PubMed] [Google Scholar]
- Huang, M., J.T. Weissman, S. Beraud-Dufour, P. Luan, C. Wang, W. Chen, M. Aridor, I.A. Wilson, and W.E. Balch. 2001. Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export. J. Cell Biol. 155:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, R.A. 2003. ARF Family GTPases. Kluwer Academic Publishers, Boston. 350 pp.
- Kahn, R.A., and A.G. Gilman. 1984. Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J. Biol. Chem. 259:6228–6234. [PubMed] [Google Scholar]
- Kahn, R.A., and A.G. Gilman. 1986. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J. Biol. Chem. 261:7906–7911. [PubMed] [Google Scholar]
- Kahn, R.A., F.G. Kern, J. Clark, E.P. Gelmann, and C. Rulka. 1991. Human ADP-ribosylation factors. A functionally conserved family of GTP- binding proteins. J. Biol. Chem. 266:2606–2614. [PubMed] [Google Scholar]
- Kahn, R.A., P. Randazzo, T. Serafini, O. Weiss, C. Rulka, J. Clark, M. Amherdt, P. Roller, L. Orci, and J.E. Rothman. 1992. The amino terminus of ADP-ribosylation factor (ARF) is a critical determinant of ARF activities and is a potent and specific inhibitor of protein transport. J. Biol. Chem. 267:13039–13046. [PubMed] [Google Scholar]
- Kuge, O., C. Dascher, L. Orci, T. Rowe, M. Amherdt, H. Plutner, M. Ravazzola, G. Tanigawa, J.E. Rothman, and W.E. Balch. 1994. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J. Cell Biol. 125:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, F.J., J. Moss, and M. Vaughan. 1992. Human and Giardia ADP-ribosylation factors (ARFs) complement ARF function in Saccharomyces cerevisiae. J. Biol. Chem. 267:24441–24445. [PubMed] [Google Scholar]
- Lee, M.C., L. Orci, S. Hamamoto, E. Futai, M. Ravazzola, and R. Schekman. 2005. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 122:605–617. [DOI] [PubMed] [Google Scholar]
- Li, Y., W.G. Kelly, J.M. Logsdon Jr., A.M. Schurko, B.D. Harfe, K.L. Hill-Harfe, and R.A. Kahn. 2004. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 18:1834–1850. [DOI] [PubMed] [Google Scholar]
- Logsdon, J.J.M., and R.A. Kahn. 2004. The Arf family tree. In Arf Family GTPases. R.A. Kahn, editor. Kluwer Academic Publishers, Boston. 1–21.
- Lowe, S.L., S.H. Wong, and W. Hong. 1996. The mammalian ARF-like protein 1 (Arl1) is associated with the Golgi complex. J. Cell Sci. 109:209–220. [DOI] [PubMed] [Google Scholar]
- Mishima, K., M. Tsuchiya, M.S. Nightingale, J. Moss, and M. Vaughan. 1993. ARD 1, a 64-kDa guanine nucleotide-binding protein with a carboxyl-terminal ADP-ribosylation factor domain. J. Biol. Chem. 268:8801–8807. [PubMed] [Google Scholar]
- Murtagh, J.J., Jr., M.R. Mowatt, C.M. Lee, F.J. Lee, K. Mishima, T.E. Nash, J. Moss, and M. Vaughan. 1992. Guanine nucleotide-binding proteins in the intestinal parasite Giardia lamblia. Isolation of a gene encoding an approximately 20-kDa ADP-ribosylation factor. J. Biol. Chem. 267:9654–9662. [PubMed] [Google Scholar]
- Nakano, A., and M. Muramatsu. 1989. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 109:2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, Z., D.S. Hirsch, and P.A. Randazzo. 2003. Arf and its many interactors. Curr. Opin. Cell Biol. 15:396–404. [DOI] [PubMed] [Google Scholar]
- Pasqualato, S., J. Menetrey, M. Franco, and J. Cherfils. 2001. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato, S., L. Renault, and J. Cherfils. 2002. Arf, Arl, Arp, and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Rep. 3:1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, S.R., M. Nightingale, S.C. Tsai, K.C. Williamson, R. Adamik, H.C. Chen, J. Moss, and M. Vaughan. 1988. Guanine nucleotide-binding proteins that enhance choleragen ADP-ribosyltransferase activity: nucleotide and deduced amino acid sequence of an ADP-ribosylation factor cDNA. Proc. Natl. Acad. Sci. USA. 85:5488–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann, A., M. Breiner, W. Becker, C. Huppertz, H. Kainulainen, H. Kentrup, and H.G. Joost. 1994. Cloning of two novel ADP-ribosylation factor-like proteins and characterization of their differential expression in 3T3-L1 cells. J. Biol. Chem. 269:15683–15688. [PubMed] [Google Scholar]
- Schurmann, A., S. Massmann, and H.G. Joost. 1995. ARP is a plasma membrane-associated Ras-related GTPase with remote similarity to the family of ADP-ribosylation factors. J. Biol. Chem. 270:30657–30663. [DOI] [PubMed] [Google Scholar]
- Setty, S.R., T.I. Strochlic, A.H. Tong, C. Boone, and C.G. Burd. 2004. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat. Cell Biol. 6:414–419. [DOI] [PubMed] [Google Scholar]
- Sewell, J.L., and R.A. Kahn. 1988. Sequences of the bovine and yeast ADP-ribosylation factor and comparison to other GTP-binding proteins. Proc. Natl. Acad. Sci. USA. 85:4620–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun, J.W., R.A. Kahn, M. Kissinger, B.J. Brizuela, C. Rulka, M.P. Scott, and J.A. Kennison. 1991. The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc. Natl. Acad. Sci. USA. 88:3120–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, M., S.R. Price, S.C. Tsai, J. Moss, and M. Vaughan. 1991. Molecular identification of ADP-ribosylation factor mRNAs and their expression in mammalian cells. J. Biol. Chem. 266:2772–2777. [PubMed] [Google Scholar]
- Vichi, A., D.M. Payne, G. Pacheco-Rodriguez, J. Moss, and M. Vaughan. 2005. E3 ubiquitin ligase activity of the trifunctional ARD1 (ADP-ribosylation factor domain protein 1). Proc. Natl. Acad. Sci. USA. 102:1945–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale, N., J. Moss, and M. Vaughan. 1996. ARD1, a 64-kDa bifunctional protein containing an 18-kDa GTP-binding ADP-ribosylation factor domain and a 46-kDa GTPase-activating domain. Proc. Natl. Acad. Sci. USA. 93:1941–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.