Abstract
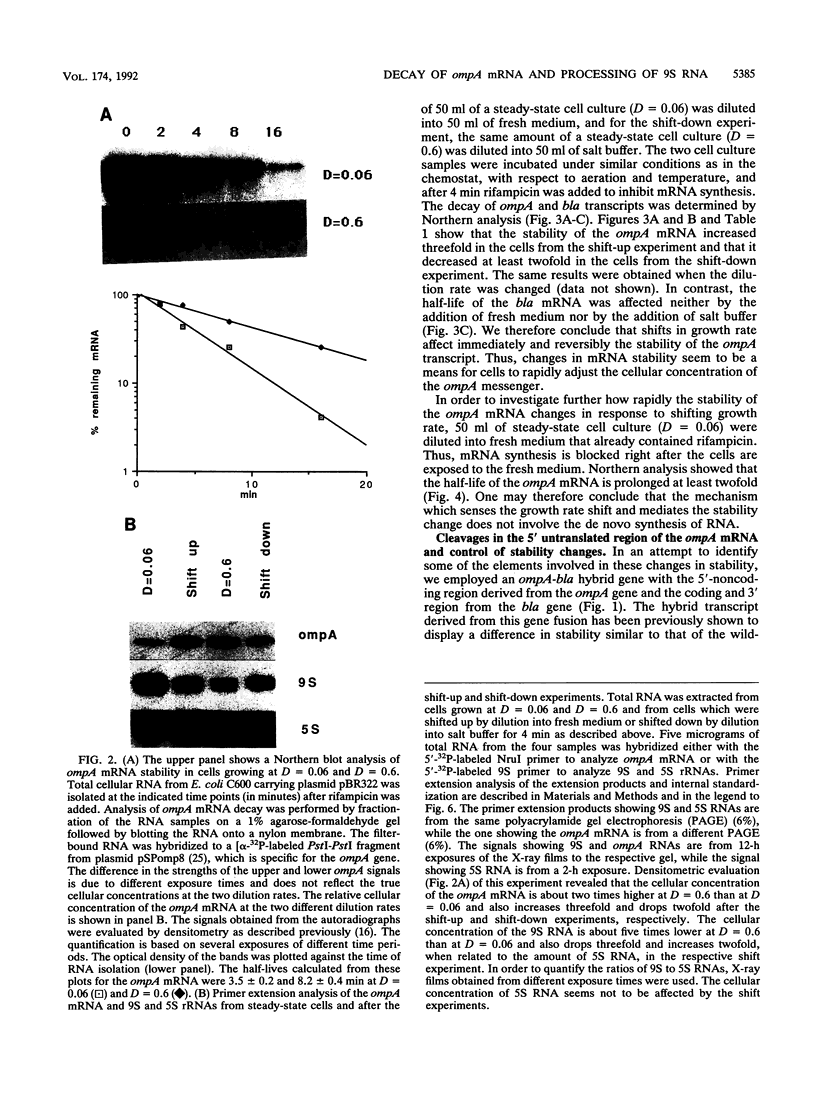
By growing Escherichia coli in continuous cultures at various growth rates, we provide definitive evidence that the stability of the ompA mRNA is growth rate dependent. Shifting fast-growing cells into physiological salt buffer led to an immediately increased rate of ompA mRNA decay and to an instantly decreased rate of 9S RNA conversion into 5S rRNA. Shifting slowly growing cells into fresh medium had the opposite effect for each of the two RNA species. The observed regulatory patterns underline the need of cells to adjust the output of ompA and 9S RNAs in response to growth rate changes. At all growth rates and throughout all shift experiments, the half-life of bla mRNA was constant. A stabilization of the ompA transcript was even observed when slowly growing cells were shifted into fresh medium already containing the transcriptional inhibitor rifampicin. A hybrid bla transcript with the 5' untranslated region from the ompA gene behaved similarly to the wild-type ompA messenger in response to a shift in growth rate. In agreement with this result, we found that the same type of 5' cleavages as have been previously shown to initiate the decay of the ompA transcript seem to be involved in stability regulation. In E. coli the degradation of mRNA has been shown to depend on the ams/rne gene. This gene controls the stability-related cleavages in the ompA transcript, catabolic processes, and the cleavages which process the 9S rRNA into 5S RNA, an anabolic process. We discuss these results with respect to the ams/rne gene and the related nuclease activities that control the ompA and 9S RNA cleavages.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics. 1978 Dec;90(4):659–671. doi: 10.1093/genetics/90.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P., Kushner S. R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Dubnau D. Induced mRNA stability in Bacillus subtilis. Proc Natl Acad Sci U S A. 1987 Jan;84(2):498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Byström A. S., von Gabain A., Björk G. R. Differentially expressed trmD ribosomal protein operon of Escherichia coli is transcribed as a single polycistronic mRNA species. J Mol Biol. 1989 Aug 20;208(4):575–586. doi: 10.1016/0022-2836(89)90149-6. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Klug G. Endonucleolytic degradation of puf mRNA in Rhodobacter capsulatus is influenced by oxygen. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1765–1769. doi: 10.1073/pnas.88.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Peters R., Bernheimer H., Berendsen W. Influence of cultural conditions and mutations on the composition of the outer membrane proteins of Escherichia coli. Mol Gen Genet. 1976 Sep 23;147(3):251–262. doi: 10.1007/BF00582876. [DOI] [PubMed] [Google Scholar]
- Lundberg U., Nilsson G., von Gabain A. The differential stability of the Escherichia coli ompA and bla mRNA at various growth rates is not correlated to the efficiency of translation. Gene. 1988 Dec 10;72(1-2):141–149. doi: 10.1016/0378-1119(88)90136-9. [DOI] [PubMed] [Google Scholar]
- Lundberg U., von Gabain A., Melefors O. Cleavages in the 5' region of the ompA and bla mRNA control stability: studies with an E. coli mutant altering mRNA stability and a novel endoribonuclease. EMBO J. 1990 Sep;9(9):2731–2741. doi: 10.1002/j.1460-2075.1990.tb07460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol Microbiol. 1991 Apr;5(4):857–864. doi: 10.1111/j.1365-2958.1991.tb00759.x. [DOI] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Melin L., Rutberg L., von Gabain A. Transcriptional and posttranscriptional control of the Bacillus subtilis succinate dehydrogenase operon. J Bacteriol. 1989 Apr;171(4):2110–2115. doi: 10.1128/jb.171.4.2110-2115.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J., Schottel J. L. A novel transcriptional response by the cat gene during slow growth of Escherichia coli. J Bacteriol. 1991 Jun;173(11):3523–3530. doi: 10.1128/jb.173.11.3523-3530.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem. 1979 Nov 10;254(21):11154–11159. [PubMed] [Google Scholar]
- Mudd E. A., Krisch H. M., Higgins C. F. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990 Dec;4(12):2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature. 1984 Nov 1;312(5989):75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Lundberg U., von Gabain A. In vivo and in vitro identity of site specific cleavages in the 5' non-coding region of ompA and bla mRNA in Escherichia coli. EMBO J. 1988 Jul;7(7):2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. Functional mRNA half lives in E. coli. Mol Gen Genet. 1978 Nov 9;166(3):329–336. doi: 10.1007/BF00267626. [DOI] [PubMed] [Google Scholar]
- Resnekov O., Rutberg L., von Gabain A. Changes in the stability of specific mRNA species in response to growth stage in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8355–8359. doi: 10.1073/pnas.87.21.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M. K., Singh B., Ray B. K., Apirion D. Maturation of 5-S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur J Biochem. 1983 Mar 1;131(1):119–127. doi: 10.1111/j.1432-1033.1983.tb07238.x. [DOI] [PubMed] [Google Scholar]
- Sandler P., Weisblum B. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J Mol Biol. 1988 Oct 20;203(4):905–915. doi: 10.1016/0022-2836(88)90116-7. [DOI] [PubMed] [Google Scholar]
- Taraseviciene L., Miczak A., Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991 Apr;5(4):851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]