Abstract
During cell division, kinetochores form the primary chromosomal attachment sites for spindle microtubules. We previously identified a network of 10 interacting kinetochore proteins conserved between Caenorhabditis elegans and humans. In this study, we investigate three proteins in the human network (hDsn1Q9H410, hNnf1PMF1, and hNsl1DC31). Using coexpression in bacteria and fractionation of mitotic extracts, we demonstrate that these proteins form a stable complex with the conserved kinetochore component hMis12. Human or chicken cells depleted of Mis12 complex subunits are delayed in mitosis with misaligned chromosomes and defects in chromosome biorientation. Aligned chromosomes exhibited reduced centromere stretch and diminished kinetochore microtubule bundles. Consistent with this, localization of the outer plate constituent Ndc80HEC1 was severely reduced. The checkpoint protein BubR1, the fibrous corona component centromere protein (CENP) E, and the inner kinetochore proteins CENP-A and CENP-H also failed to accumulate to wild-type levels in depleted cells. These results indicate that a four-subunit Mis12 complex plays an essential role in chromosome segregation in vertebrates and contributes to mitotic kinetochore assembly.
Introduction
Proper segregation of chromosomes during mitosis is essential for the accurate transmission of genetic material. Each chromatid of a replicated chromosome assembles a kinetochore, which forms a dynamic interface with microtubules of the mitotic spindle (Fukagawa, 2004). To facilitate chromosome segregation, sister kinetochores must attach to and regulate the assembly properties of microtubules emanating from opposing spindle poles. This process, called chromosome biorientation, requires the integrated activities of multiple kinetochore proteins. New insights have come from the discovery that many kinetochore proteins and their associated complexes are widely conserved (Chan et al., 2005). For example, purifications from Caenorhabditis elegans, humans, and fission yeast have identified a protein network that includes Mis12, KNL-1Spc105, and the Ndc80 complex (Cheeseman et al., 2004; Obuse et al., 2004; Liu et al., 2005).
Work in C. elegans has demonstrated that depletion of a functional class of proteins that includes MIS-12 disrupts kinetochore assembly, resulting in alignment and segregation defects (Cheeseman et al., 2004). In fungi, Mis12Mtw1 functions in a complex with Nnf1, Nsl1, and Dsn1 (for review see Chan et al., 2005). Initial studies of the Mis12 orthologue in human cells suggested that it is required for proper chromosome alignment and segregation (Goshima et al., 2003). Here, we present functional analyses of three human kinetochore proteins (hNsl1DC31, hNnf1PMF1, and hDsn1Q9H410) that are part of the conserved network. These three proteins function in a discrete complex with hMis12 at inner kinetochores and are required for proper chromosome alignment and cell cycle progression as a result of a central role in kinetochore assembly.
Results and discussion
hMis12, hDsn1, hNnf1, and hNsl1 form a discrete complex
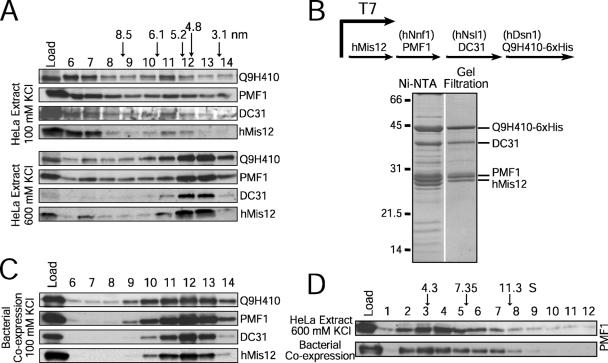
Tandem affinity purifications of hMis12 from human cells previously isolated a network of 10 interacting kinetochore proteins including hMis12, hKNL-1, the Ndc80 complex, Zwint, and three additional proteins: Q9H410, PMF1, and DC31 (Cheeseman et al., 2004; Obuse et al., 2004). In contrast, purification of Mis12Mtw1 from budding yeast identified a complex with just three other interacting proteins (Dsn1, Nsl1, and Nnf1; for review see Chan et al., 2005). This disparity suggested that hMis12 and the three additional proteins might form a stable subcomplex analogous to the budding yeast complex. To test this possibility, we generated antibodies (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200509158/DC1), fractionated mitotic HeLa extracts using gel filtration chromatography, and analyzed their migration relative to hMis12 (Fig. 1 A). Under low salt conditions, hMis12, PMF1, Q9H410, and DC31 were distributed throughout the column fractions (Fig. 1 A). Under higher salt conditions, the majority of all four proteins cofractionated with a Stokes radius of ∼4.6 nm.
Figure 1.
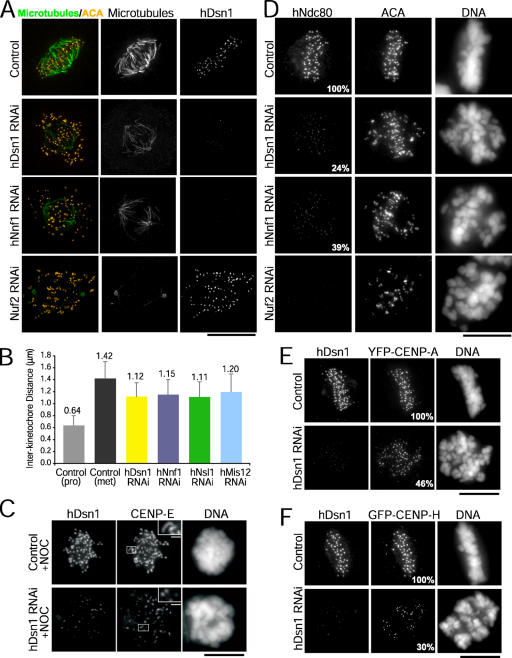
hMis12, hDsn1, hNnf1, and hNsl1 form a discrete complex. (A) Immunoblots of mitotic HeLa extracts fractionated on a Superose 6 gel filtration column at 100 or 600 mM KCl. Stokes radii of standards: thyroglobulin (8.5 nm), ferritin (6.1 nm), catalase (5.2 nm), aldolase (4.8 nm), and ovalbumin (3.1 nm). (B) Coexpression of hMis12, PMF1, DC31, and Q9H410-6xHis using a polycistronic system in bacteria. Coomassie-stained peak fractions are shown. (C) Migration of the recombinant complex on a Superose 6 gel filtration column. (D) Comigration of the purified recombinant complex and the endogenous complex on a 5–20% sucrose gradient. PMF1 blots are shown for brevity. Standards are BSA (4.3 S), aldolase (7.35 S), and catalase (11.3 S).
To determine whether the stable association of hMis12, PMF1, Q9H410, and DC31 accounted for this cofractionation, we coexpressed all four proteins in bacteria using a polycistronic system (Fig. 1 B). The four proteins copurified to near homogeneity through nickel affinity and gel filtration chromatography, indicating that they form a stable complex (Fig. 1 B). The recombinant complex cofractionated as a single peak nearly coincident with that observed in HeLa extracts at 600 mM KCl (Fig. 1, C and D). We estimate that the Svedberg coefficient (S) value is ∼5.8 (Fig. 1 D), resulting in a native molecular mass of ∼112 kD. The combined subunit molecular mass is 119.7 kD, suggesting that the complex contains a single molecule of each protein. We conclude that hMis12, PMF1, DC31, and Q9H410 are the human equivalent of the Mis12 complex in budding yeast. Nnf1 and PMF1 show weak sequence similarity (unpublished data), and the position of coiled coils and limited regions of identity suggest that Q9H410 and DC31 are analogous to Dsn1 and Nsl1 (Obuse et al., 2004). Therefore, we refer to these four proteins as the hMis12 complex and to the three new subunits as hDsn1 (Q9H410), hNnf1 (PMF1), and hNsl1 (DC31).
The hMis12 complex localizes to inner kinetochores
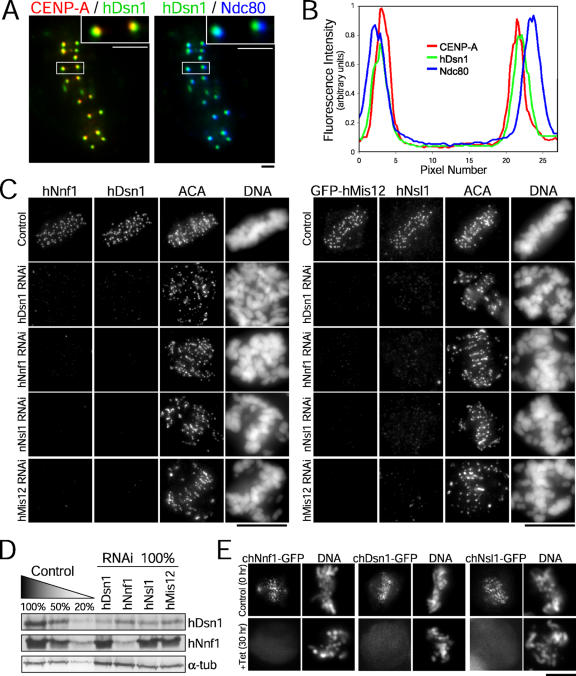
Immunolocalization studies demonstrated that the three new hMis12 complex subunits localize coincidently with centromere protein (CENP) A at inner kinetochores and internally to Ndc80 at outer kinetochores (Fig. 2, A and B), which is consistent with previous studies of hMis12 (Goshima et al., 2003; Obuse et al., 2004). Punctate nuclear localization was observed in only a subset of interphase cells, suggesting that the hMis12 complex is not constitutively present at centromeres throughout the cell cycle (not depicted). All of the hMis12 complex proteins localized to kinetochores at constant levels throughout mitosis, which is consistent with this complex being a stable component of the mitotic inner kinetochore (not depicted).
Figure 2.
Kinetochore localization of the hMis12 complex is interdependent. (A) HeLa cells stained for CENP-A (red), hDsn1 (green), and Ndc80 (blue). Insets show higher magnification of the boxed pair of sister kinetochores. (B) Corresponding fluorescence intensity profiles in a line scan. (C) HeLa cells were transfected with the indicated siRNAs and stained for DNA, centromeres (ACA), and the indicated proteins. (D) HeLa cells were harvested after the transfection of siRNAs, and serial dilutions of control and depleted extracts were immunoblotted for hDsn1 and hNnf1. α-Tubulin was used as a loading control. (E) Chicken Mis12 loss-of-function mutant cells stably expressing Dsn1-GFP, Nsl1-GFP, and Nnf1-GFP constructs were fixed and analyzed at 0 (control) and 30 h (knockdown) after the addition of tet. Bars (A), 1 μm; (C and E), 10 μm.
Kinetochore localization of hMis12 complex subunits is interdependent
To analyze the consequences of hMis12 complex inhibition, we transfected HeLa cells with gene-specific siRNAs. Quantification of kinetochore fluorescence intensities indicated that hMis12 complex constituents could be depleted by 73–93% (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200509158/DC1). Because of variable depletion levels from cell to cell in a population, immunoblot analyses indicated that hDsn1 and hNnf1 were depleted by >80–90%, whereas hMis12 and hNsl1 were depleted by ∼50% (Fig. S1). To generate an alternative genetic means to examine vertebrate Mis12 depletion, we also obtained a conditional loss-of-function mutant for Mis12 in chicken DT40 cells (Fig. S2). Cell growth was sustained in chMis12-deleted cells by the expression of chMis12 cDNA under the control of a tetracycline (tet)-repressible promoter. chMis12 protein was significantly reduced 18 h after the addition of tet and was undetectable by 24 h (Fig. S2).
RNAi-mediated depletion of hMis12, hDsn1, hNnf1, or hNsl1 resulted in a dramatic reduction in the kinetochore levels of the other three proteins (Fig. 2 C). This reduction was caused by alterations in both kinetochore targeting and, in some cases, stability of the assayed protein (Fig. 2 D). Controlled depletion of chMis12 also resulted in the delocalization of chDsn1, chNnf1, and chNsl1 (Fig. 2 E). Thus, the vertebrate Mis12 complex proteins are interdependent for kinetochore localization and exhibit interdependent stability, which is consistent with their function as a complex at inner kinetochores.
The hMis12 complex is required for chromosome alignment and segregation
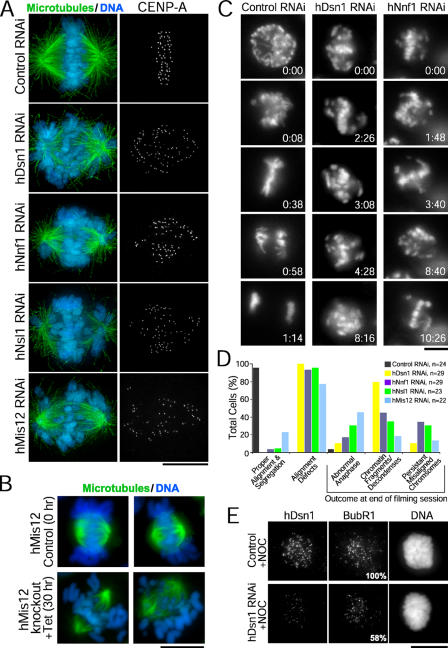
Reduced levels of Mis12 complex proteins resulted in chromosome alignment defects in both human and chicken cells, although spindle bipolarity was not perturbed (Fig. 3, A and B). To analyze mitotic chromosome dynamics, we transfected siRNAs into HeLa cells stably expressing YFP-histone H2B (Kops et al., 2004) and performed live imaging. Cells depleted of hDsn1, hNnf1, hNsl1, or hMis12 remained in mitosis for a mean of 8 h, whereas control cells completed mitosis in 1 h (Fig. 3, C and D; and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200509158/DC1). Previous studies indicated that chromosome misalignment also led to a mitotic delay after hDsn1 depletion (Obuse et al., 2004) but not after hMis12 depletion (Goshima et al., 2003). Although chromosomes were observed making multiple attempts to congress, they never fully aligned, and some cells underwent aberrant segregation or exhibited abnormal chromatin morphology (Fig. 3, C and D; and Videos 2 and 3). Similarly, the depletion of chMis12 resulted in a significant accumulation of cells with unaligned chromosomes (Fig. S2). Kinetochore levels of the spindle checkpoint protein BubR1 were reduced by 42% in depleted HeLa cells treated with nocodazole (Fig. 3 E and Table S1). These findings demonstrate that the Mis12 complex is required for normal chromosome alignment and segregation. Although the spindle checkpoint is active after hMis12 complex inhibition, the decreased BubR1 levels and occasional chromosome missegregation indicate that the checkpoint may be partially compromised.
Figure 3.
The hMis12 complex is required for chromosome alignment and segregation. (A) After transfection of the indicated siRNAs, HeLa cells stably expressing YFP–CENP-A were stained for tubulin (green), DNA (blue), and CENP-A. (B) Chicken DT40 cells were stained for tubulin (green) and DNA (blue) at 0 (control) or 30 h (knockdown) after tet addition. (C) HeLa cells stably expressing YFP-histone H2B were imaged by time-lapse fluorescence microscopy after transfection of the indicated siRNAs; time is given in hours/minutes. (D) Summary of live imaging analysis in HeLa cells. (E) After siRNA transfection, HeLa cells were treated with nocodazole (NOC) and stained for hDsn1 and BubR1. Values indicating the mean kinetochore fluorescence intensities are given relative to controls. Bars, 10 μm.
The hMis12 complex is required for chromosome biorientation
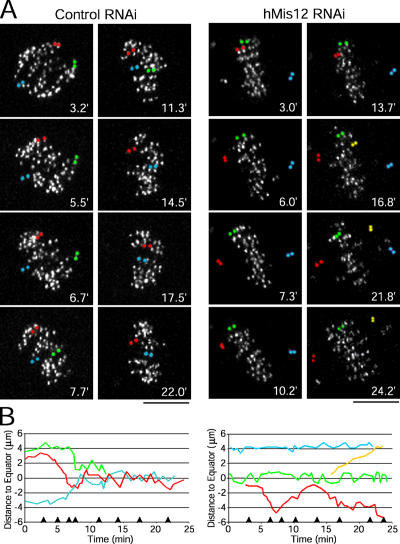
To characterize the chromosome alignment defects after hMis12 depletion, we performed RNAi in HeLa cells stably expressing YFP-labeled CENP-A (Kops et al., 2004) and imaged kinetochore motility at high temporal resolution. In control cells, paired sister kinetochores congressed to the metaphase plate and oscillated until anaphase onset (Fig. 4, A and B; and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200509158/DC1). In depleted cells, some kinetochore pairs remained at the spindle poles for the duration of the analysis. Strikingly, sister kinetochores were frequently observed moving back and forth between spindle poles and the metaphase plate (Fig. 4, A and B; and Video 5). These defects were observed in every cell depleted of any subunit of the hMis12 complex (Videos 5–8). In total, these findings indicate that upon inhibition of the hMis12 complex, sister kinetochores are defective in both the establishment and maintenance of bioriented kinetochore microtubule attachments, which is consistent with previous studies in fungi and C. elegans (Goshima and Yanagida, 2000; Pinsky et al., 2003; Scharfenberger et al., 2003; Cheeseman et al., 2004).
Figure 4.
The hMis12 complex is required for chromosome biorientation. (A) HeLa cells stably expressing YFP–CENP-A were imaged after transfection of the indicated siRNAs; time is given in minutes. Paired sister kinetochores were pseudocolored to illustrate their dynamics. (B) Distance of pseudocolored sister kinetochores in A to the metaphase plate over time for the control cell (left) and hMis12-depleted cell (right). Arrowheads on the x axis indicate the time of the panels in A. Bars, 5 μm.
The hMis12 complex is required for kinetochore fiber formation
These defects in biorientation prompted us to analyze the stability of kinetochore microtubule attachments. Cells were incubated in ice-cold media before fixation to depolymerize all microtubules except stable kinetochore fibers (Inoué, 1964). Compared with controls, the kinetochore fibers in depleted cells were significantly diminished (Fig. 5 A). In addition, chromosomes located near the spindle poles lacked any associated microtubule staining, indicating that no stable end-on kinetochore microtubule attachments were present on these chromosome populations (Fig. 5 A). Overall, the reduction in kinetochore fiber stability following inhibition of the hMis12 complex was less severe than that following inhibition of the Ndc80 complex (Fig. 5 A; DeLuca et al., 2005).
Figure 5.
The hMis12 complex is required for kinetochore fiber formation and kinetochore assembly. (A) After the indicated perturbations, HeLa cells were incubated in ice-cold media for 10 min and stained for tubulin (green), centromeres (ACA, yellow), DNA, and hDsn1. (B) Interkinetochore distances measured in transfected HeLas. Error bars represent SD. (C) After siRNA transfection, HeLa cells were treated with nocodazole and stained for hDsn1 and CENP-E. Insets show boxed sister kinetochore pairs at higher magnification. (D) After the indicated perturbations, HeLa cells were stained for Ndc80, centromeres (ACA), DNA, and hDsn1 (not depicted). (E and F) After the depletion of hDsn1, HeLa cells stably expressing YFP–CENP-A (E) or GFP–CENP-H (F) were stained for hDsn1 and YFP. (D–F) The mean kinetochore fluorescence intensities, which are expressed as percentages relative to controls, are indicated in the bottom right of panels. Bars, 10 μm; (insets), 1 μm.
As an additional assessment of kinetochore microtubule attachments, we measured the distance between sister kinetochores, which correlates with the tension exerted across the centromere (Waters et al., 1996). We restricted our analysis to aligned bioriented chromosomes with end-on attachments. We found that the mean interkinetochore distance was decreased from 1.42 μm in control metaphase cells to 1.11–1.20 μm in depleted cells (P < 0.001; n > 100 chromosomes from five cells per condition; Fig. 5 B). This 30–40% reduction in centromere stretch relative to the rest length of 0.64 μm (measured in control prophase cells) suggests that the net force produced by kinetochore microtubule attachments is diminished upon depletion of hMis12 complex proteins.
The hMis12 complex contributes to outer kinetochore assembly
Our previous study in C. elegans suggested that MIS-12 regulates outer kinetochore assembly (Cheeseman et al., 2004). Thus, we examined localization of the Ndc80 complex, which physically associates with the Mis12 complex (Cheeseman et al., 2004; Obuse et al., 2004) and is a major constituent of the outer plate (DeLuca et al., 2005). In cells with greatly reduced levels of Mis12 complex subunits, the kinetochore localization of endogenous Ndc80 (Fig. 5 D) and stably expressed GFP-hNuf2 (not depicted) was significantly lower than in control cells. Measurements of kinetochore fluorescence intensity indicated that after the depletion of any of the four Mis12 complex components, Ndc80 targeting was reduced a mean of 61–82% (Table S1). Chicken DT40 cells depleted of Mis12 also showed a significant reduction in kinetochore-localized Ndc80 (Fig. S2). These results suggest that the outer kinetochore is disrupted in cells depleted of the Mis12 complex. In support of this idea, CENP-E targeting was diminished in depleted HeLa cells, and expansion of the fibrous corona into crescent-shaped collars, which is normally observed upon nocodazole treatment, did not occur (Fig. 5 C). Because CENP-E and the Ndc80 complex both contribute to the formation of stable, bioriented kinetochore microtubule attachments, the role of the hMis12 complex in chromosome biorientation may be explained, in part, by its requirement for the recruitment of outer kinetochore components.
The hMis12 complex contributes to inner kinetochore assembly
To determine whether inner kinetochore assembly is also affected in cells depleted of hMis12 complex subunits, we quantified the levels of CENP-A and CENP-H, two DNA-proximal kinetochore components (Chan et al., 2005). We found that the levels of both were reduced in cells depleted of hDsn1: CENP-A was decreased by 49–54%, and CENP-H was decreased by 70% (Fig. 5, E and F; and Table S1). Previous data indicated that CENP-A was unaffected, whereas CENP-H was decreased upon the depletion of hMis12 (Goshima et al., 2003). Because the targeting of CENP-H requires CENP-A (Regnier et al., 2005), the reduction in CENP-H could be explained, in part, by the lower CENP-A levels we observed. The functional consequence of this CENP-A reduction is unclear. Reduction of CENP-A to ∼30% by direct siRNA does not prevent mitotic progression and does not cause chromosome alignment and segregation defects as severe as those seen in hMis12 complex–inhibited cells (Black, B., D. Foltz, and D. Cleveland, personal communication). Consequently, we favor the idea that outer kinetochore defects are primarily responsible for the dramatic phenotypes observed in hMis12 complex depletions. It should also be noted that hMis12 was previously suggested to localize to kinetochores in a manner that is partially independent of CENP-A (Goshima et al., 2003). Exploring the role of the hMis12 complex in this key early step of kinetochore assembly is an important future direction.
The hMis12 complex plays a central role in human kinetochore assembly
The data presented in this study demonstrate conservation of a four-subunit Mis12 complex in human cells. Functional analyses revealed defects in chromosome alignment and segregation, kinetochore fiber stability, and chromosome biorientation. These defects are likely explained by a central role for this complex in kinetochore assembly. Both inner and outer kinetochore protein targeting is defective in cells depleted of the hMis12 complex. The reduction in CENP-A levels may reflect a role for the hMis12 complex in CENP-A loading/stabilization or could be an indirect consequence of defective chromosome segregation in a prior cell division. The effect on Ndc80 complex targeting is likely direct given the conserved physical interaction between the Mis12 and Ndc80 complexes. In general, the defects in chromosome movement and kinetochore fiber formation in hMis12 complex–inhibited cells were less severe than those observed upon the direct depletion of the Ndc80 complex. This difference could arise from technical variations in RNAi efficacy; however, studies in fungi and C. elegans have revealed that the Ndc80 complex functions mostly downstream of the Mis12 complex, yet its inhibition results in more severe kinetochore microtubule attachment and chromosome segregation defects (Goshima et al., 1999; Janke et al., 2001; Nabetani et al., 2001; De Wulf et al., 2003; Pinsky et al., 2003; Scharfenberger et al., 2003; Westermann et al., 2003; Cheeseman et al., 2004). The similar trend in human cell studies (DeLuca et al., 2005; this study) leads us to speculate that additional mechanisms contribute to Ndc80 complex function in chromosome segregation. For example, Ndc80 complex targeting is also mediated by CENP-H (Hori et al., 2003), which was still present at ∼30% of wild-type levels in hMis12 complex–depleted cells.
Materials and methods
Cell culture and siRNA transfection
HeLa cells and clonal lines derived from HeLa cells stably expressing GFP-Mis12 (Cheeseman et al., 2004), YFP–CENP-A (a gift from D. Foltz, Ludwig Institute for Cancer Research [LICR], La Jolla, CA), and YFP-histone H2B (a gift from J. Shah, Harvard Medical School, Boston, MA; Kops et al., 2004), or GFP–CENP-H and GFP-hNuf2 (this study) were maintained in DME supplemented with 10% FBS, penicillin/streptomycin, and l-glutamine (Invitrogen) at 37°C in a humidified atmosphere with 5% CO2. Cells were plated on 12- or 22-mm glass coverslips coated with poly-l-lysine (Sigma-Aldrich) for immunostaining or 35-mm glass bottom microwell dishes (MatTek) for time-lapse imaging. Predesigned siRNAs targeting hNsl1DC31 (Ambion), hDsn1Q9H410, hNnf1PMF1, hMis12 (Dharmacon), hNuf2 (a gift from J. DeLuca, University of North Carolina, Chapel Hill, NC; DeLuca et al., 2005), or nonspecific control siRNAs (Dharmacon) were transfected according to manufacturer's directions using Oligofectamine and serum-free OptiMEM (Invitrogen). FBS was added to 10% to the transfection reaction after incubation on cells for 5–6 h. Cells were assayed 48 h after transfection for hDsn1, hNnf1, or hNsl1 and 72 h after transfection for hMis12.
Chicken Mis12 target disruption constructs for each gene were generated such that genomic fragments encoding an entire coding region in a single exon were replaced with a histidinol or puromycin resistance cassette under control of the β-actin promoter. Target constructs were transfected with a Gene Pulser II electroporator (Bio-Rad Laboratories). Chicken DT40 cells were cultured and transfected as described previously (Fukagawa et al., 2001, 2004). All DT40 cells were cultured at 38°C in DME supplemented with 10% FCS, 1% chicken serum, and penicillin/streptomycin. To repress the expression of the tet-responsive transgenes, tet (Sigma-Aldrich) was added to the culture medium to a final concentration of 2 μg/ml. Chicken homologues of Dsn1, Nsl1, and Nnf1 were cloned and fused with GFP under control of the cytomegalovirus promoter. These fusion constructs were transfected into chMis12 conditional knockout cells to examine their localization.
Protein expression, chromatography, and antibody production
For gel filtration and sucrose gradient analysis, HeLa cells were grown until they were ∼80% confluent, and 100 ng/ml nocodazole was added for 14 h to enrich for mitotic cells. Cells were harvested and washed into lysis buffer (50 mM Hepes, pH 7.4, 1 mM EGTA, 1 mM MgCl2, 100 mM KCl, and 10% glycerol). Cell extract was prepared as described previously (Cheeseman et al., 2004) and fractionated on either a Superose 6 gel filtration column or a 5–20% sucrose gradient at 50 krpm for 8 h in either lysis buffer or lysis buffer with a total of 600 mM KCl. Coexpression of Mis12 complex proteins was conducted by cloning the cDNAs for each protein into pST39 (a gift from S. Tan, Pennsylvania State University, University Park, PA; Tan, 2001). Escherichia coli carrying the plasmid were induced with 0.1 mM IPTG, and the proteins were purified using nickel affinity chromatography. The eluate from the nickel column was further fractionated on gel filtration and sucrose gradients as described above. Affinity-purified rabbit polyclonal antibodies were generated against hDsn1 (residues 181–356), hNsl1 (residues 22–281), hNnf1 (full length), hMis12 (residues 105–205), chMis12 (full length), and chNdc80 (residues 465–640) as described previously (Desai et al., 2003).
Immunofluorescence and drug treatments
For analysis of checkpoint proteins BubR1 and CENP-E, cells were treated with 10 μg/ml nocodazole diluted in media for 1 h before processing for immunofluorescence. For analysis of kinetochore proteins, cells were rinsed in PBS (12 mM PO4 −2, 137 mM NaCl, and 3 mM KCl, pH 7.4), extracted for 5 min in warmed PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) plus 1% Triton X-100, and fixed at room temperature for 20 min in PHEM plus 4% formaldehyde. For analysis of spindle microtubules, cells were not extracted before fixation. Treatment and fixation of cells for the cold-stable kinetochore fiber assay were performed as described previously (Lampson and Kapoor, 2005). All fixed cells were rinsed with TBS-TX (20 mM Tris, 150 mM NaCl, pH 7.5, and 0.1% Triton X-100) and blocked in AbDil (2% BSA and 0.1% NaN3 in TBS-TX) for 30–60 min. For fluorescence of microtubules, DM1α (Sigma-Aldrich) was used at 1:500. For visualization of kinetochore proteins, mouse anti-HEC1 (9G3; Abcam) was used at 1:1,000, and human anticentromere antibodies (ACAs; Antibodies, Inc.) were used at 1:200. YFP–CENP-A, GFP–CENP-H, or GFP-Mis12–expressing HeLas were counterstained with goat anti-GFP (a gift from D. Drechsel, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) at 1:500. Mouse anti–CENP-A (a gift from K. Yoda, Nagoya University, Nagoya, Japan), rabbit anti–CENP-E (a gift from D. Cleveland, LICR), and mouse anti-BubR1 (a gift from S. Taylor, University of Manchester, Manchester, UK) were used at 1:200. Polyclonal antibodies against hDsn1, hNsl1, and hNnf1 were used at 1 μg/ml. Cy2-, Cy3-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1:100, although in some cases, directly labeled antibodies against hDsn1 and hNnf1 were used without secondaries. DNA was visualized using 10 μg/ml Hoechst in TBS-TX. Coverslips were mounted using 0.5% p-phenylenediamine and 20 mM Tris-Cl, pH 8.8, in 90% glycerol. For DT40 immunofluorescence, cells were collected onto slides with a cytocentrifuge, fixed in 3% PFA in 250 mM Hepes, pH 7.4, for 15 min at room temperature, permeabilized in 0.5% NP-40 in PBS for 10 min at room temperature, rinsed three times in 0.5% BSA, and processed for indirect immunofluorescence.
Microscopy and image acquisition
Images of fixed cells were acquired on a deconvolution microscope (DeltaVision; Applied Precision) equipped with a CCD camera (CoolSNAP; Roper Scientific). 30–80 z sections were acquired at 0.2-μm steps using a 100× NA 1.3 U-planApo objective (Olympus) with 1 × 1 binning. For analysis of microtubule attachments, images were deconvolved using the DeltaVision software (Applied Precision). Measurements of the intensity of kinetochore localization were conducted on nondeconvolved images. All images for a specific experiment used identical exposure settings and scaling. For live cell imaging, medium was replaced with CO2-independent medium supplemented with 10% FBS, penicillin/streptomycin, and l-glutamine (Invitrogen) and was covered with mineral oil immediately before analysis. Cells were maintained at 35–37°C using a heated stage. Images of cells expressing YFP-histone H2B were collected on an inverted microscope (Eclipse 300; Nikon) and a 60× NA 1.4 planApo objective. Acquisition parameters, including exposure, focus, and illumination, were controlled by MetaMorph software (Universal Imaging). Single focal plane images were collected by a camera (CoolSNAP HQ; Photometrics) at 2-min intervals with an exposure time of 80 ms with 2 × 2 binning. Images of cells expressing YFP–CENP-A were collected on a spinning disc confocal (McBain Instruments) mounted on an inverted microscope (TE2000e; Nikon) using a 60× NA 1.40 planApo objective plus 1.5× auxiliary magnification. Five z sections were acquired at 1-μm steps at 10-s time intervals with exposure times of 100 ms and 2 × 2 binning. Z stacks were projected in MetaMorph. All subsequent analysis and processing of images were performed using MetaMorph software. Images of DT40 cells were collected with a cooled CCD camera (CoolSNAP HQ; Photometrics Image Point) mounted on an inverted microscope (IX71; Olympus) with a 60× NA 1.40 planApo objective lens together with a filter wheel. Images were analyzed with an IPLab software (Signal Analytics) or DeltaVision deconvolution system (Applied Precision).
Data analysis
All distance and fluorescence intensity measurements were made using MetaMorph software. Analysis of kinetochore movements was performed by measuring the distance from the center of sister kinetochores to a point on the spindle equator along the trajectory of chromosome movement. The bulk of metaphase-aligned chromosomes were used as a reference point for the spindle equator, which occasionally shifted during our analysis. Interkinetochore distances were measured using the centers of the paired CENP-A dots. Kinetochore fluorescence intensities were determined by measuring the integrated fluorescence intensity within a 7 × 7 pixel square positioned over a single kinetochore and subtracting the background intensity of a 7 × 7 pixel square positioned in a region of cytoplasm lacking kinetochores. Maximal projected images were used for these measurements. To determine significant differences between means, unpaired t tests assuming unequal variance were performed; differences were considered significant when P < 0.05.
Online supplemental material
Fig. S1 shows immunoblots of HeLa cell lysates probed with antibodies to hDsn1, hNnf1, or hNsl1 as well as immunoblots after RNAi of each of these proteins. Fig. S2 illustrates the disruption of chMis12 in DT40 cells, the cell cycle consequences of chMis12 knockdown, and the concomitant decrease in kinetochore-localized chNdc80. Table I summarizes the quantification of kinetochore fluorescence intensities of targeted Mis12 complex subunits and CENP-A, CENP-H, Ndc80, and BubR1. Videos 1–3 show mitosis in HeLa cells expressing YFP-histone H2B after transfection of control siRNA (Video 1), siRNA targeting hDsn1 (Video 2), or siRNA targeting hNnf1 (Video 3). Videos 4–8 show mitosis in HeLa cells expressing YFP–CENP-A after transfection of control siRNA (Video 4), siRNA targeting hMis12 (Video 5), siRNA targeting hDsn1 (Video 6), siRNA targeting hNnf1 (Video 7), and siRNA targeting hNsl1 (Video 8). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200509158/DC1.
Supplementary Material
Acknowledgments
We thank P. Maddox and B. Black for critical reading of this manuscript, D. Foltz and J. Shah for YFP–CENP-A and YFP-histone H2B cells, S. Tan for reagents and advice on polycistronic expression, and J. DeLuca for discussions and hNuf2 siRNA.
This work was supported by LICR, a grant from the National Institutes of Health to A. Desai (R01GM074215-01), and by Grants-in-Aid for Scientific Research on Priority Areas cancer cell biology, cell cycle, and nuclear dynamics from the Ministry of Education, Science, Sports and Culture of Japan to T. Fukagawa. S. Kline is a fellow of the American Cancer Society, I.M. Cheeseman is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research, and A. Desai is the Connie and Bob Lurie Scholar of the Damon Runyon Cancer Research Foundation (grant DRS 38-04).
S.L. Kline and I.M. Cheeseman contributed equally to this article.
Abbreviations used in this paper: ACA, anticentromere antibody; CENP, centromere protein; tet, tetracycline.
References
- Chan, G.K., S.T. Liu, and T.J. Yen. 2005. Kinetochore structure and function. Trends Cell Biol. 15:589–598. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Niessen, S. Anderson, F. Hyndman, J.R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J.G., Y. Dong, P. Hergert, J. Strauss, J.M. Hickey, E.D. Salmon, and B.F. McEwen. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Shevchenko, A. Hyman, and K. Oegema. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf, P., A.D. McAinsh, and P.K. Sorger. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa, T. 2004. Assembly of kinetochores in vertebrate cells. Exp. Cell Res. 296:21–27. [DOI] [PubMed] [Google Scholar]
- Fukagawa, T., V. Regnier, and T. Ikemura. 2001. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 29:3796–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, Y. Takami, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784–791. [DOI] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633. [DOI] [PubMed] [Google Scholar]
- Goshima, G., S. Saitoh, and M. Yanagida. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 13:1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., T. Haraguchi, Y. Hiraoka, H. Kimura, and T. Fukagawa. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362. [DOI] [PubMed] [Google Scholar]
- Inoué, S. 1964. Organization and function of the mitotic spindle. In Primitive Motile Systems in Cell Biology. R.D. Allen and N. Kamiya, editors. Academic Press, New York. 549–598.
- Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, M.M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops, G.J., D.R. Foltz, and D.W. Cleveland. 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 101:8699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson, M.A., and T.M. Kapoor. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7:93–98. [DOI] [PubMed] [Google Scholar]
- Liu, X., I. McLeod, S. Anderson, J.R. Yates, and X. He. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24:2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani, A., T. Koujin, C. Tsutsumi, T. Haraguchi, and Y. Hiraoka. 2001. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 110:322–334. [DOI] [PubMed] [Google Scholar]
- Obuse, C., O. Iwasaki, T. Kiyomitsu, G. Goshima, Y. Toyoda, and M. Yanagida. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135–1141. [DOI] [PubMed] [Google Scholar]
- Pinsky, B.A., S.Y. Tatsutani, K.A. Collins, and S. Biggins. 2003. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell. 5:735–745. [DOI] [PubMed] [Google Scholar]
- Regnier, V., P. Vagnarelli, T. Fukagawa, T. Zerjal, E. Burns, D. Trouche, W. Earnshaw, and W. Brown. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25:3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfenberger, M., J. Ortiz, N. Grau, C. Janke, E. Schiebel, and J. Lechner. 2003. Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J. 22:6584–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21:224–234. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., R.V. Skibbens, and E.D. Salmon. 1996. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J. Cell Sci. 109:2823–2831. [DOI] [PubMed] [Google Scholar]
- Westermann, S., I.M. Cheeseman, S. Anderson, J.R. Yates III, D.G. Drubin, and G. Barnes. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.