Abstract
Mammalian H/ACA RNPs are essential for ribosome biogenesis, premessenger RNA splicing, and telomere maintenance. These RNPs consist of four core proteins and one RNA, but it is not known how they assemble. By interrogating the site of H/ACA RNA transcription, we dissected their biogenesis in single cells and delineated the role of the non-core protein NAF1 in the process. NAF1 and all of the core proteins except GAR1 are recruited to the site of transcription. NAF1 binds one of the core proteins, NAP57, and shuttles between nucleus and cytoplasm. Both proteins are essential for stable H/ACA RNA accumulation. NAF1 and GAR1 bind NAP57 competitively, suggesting a sequential interaction. Our analyses indicate that NAF1 binds NAP57 and escorts it to the nascent H/ACA RNA and that GAR1 then replaces NAF1 to yield mature H/ACA RNPs in Cajal bodies and nucleoli.
Introduction
The 100–200 H/ACA RNPs of each mammalian cell affect several basic functions, such as protein synthesis, gene expression, and chromosome stability. They do so by catalyzing site-specific pseudouridylation of ribosomal and spliceosomal small nuclear RNAs, by processing ribosomal RNAs, and by stabilizing telomerase RNA. In addition to C/D RNPs, H/ACA RNPs form one of the two major classes of small nucleolar RNPs (snoRNPs) functioning in nucleoli and Cajal bodies. They consist of four evolutionarily conserved core proteins and one H/ACA RNA (Smith and Steitz, 1997; Kiss, 2001; Filipowicz and Pogacic, 2002; Decatur and Fournier, 2003; Meier, 2005).
The four core proteins—the pseudouridylase NAP57 (dyskerin; Cbf5p in yeast), GAR1, NHP2, and NOP10—are capable of forming a protein-only complex (Wang and Meier, 2004). Specifically, NOP10 and GAR1 bind independently to NAP57, whereas docking of NHP2 onto NAP57 is mediated by NOP10, thereby forming the core trimer NAP57–NOP10–NHP2 (noncovalent interactions are indicated with an en-dash). These interactions are conserved in yeast and archaeal H/ACA RNPs (Henras et al., 2004; Baker et al., 2005; Charpentier et al., 2005). Stable expression of H/ACA RNAs requires incorporation into RNPs, in particular, association with the core trimer, as genetic depletion of each of its proteins causes a specific loss of H/ACA RNAs in yeast (Henras et al., 1998; Lafontaine et al., 1998; Watkins et al., 1998; Dez et al., 2001). Therefore, assembly of H/ACA RNPs may follow the simple but unusual pathway of protein-only particle formation followed by association with an H/ACA RNA. In this manner, functional H/ACA RNPs can be assembled in archaea (Baker et al., 2005; Charpentier et al., 2005) but not in eukaryotes, where additional factors may be required (Wang and Meier, 2004).
Indeed, two proteins involved in H/ACA RNP biogenesis in yeast, the nuclear assembly factor Naf1p and Shq1p, have been identified based on their interaction with Cbf5p (yeast NAP57) and Nhp2p (Dez et al., 2002; Fatica et al., 2002; Yang et al., 2002). Although neither Naf1p nor Shq1p is part of mature H/ACA RNPs, their genetic depletion leads to a specific loss of H/ACA RNAs. While this study was in progress, chromatin immunoprecipitation (ChIP) analyses in yeast showed cotranscriptional recruitment of Naf1p and Cbf5p to H/ACA RNA genes (Ballarino et al., 2005; Yang et al., 2005). Although Naf1p and Shq1p have putative homologues in mammals, insects, nematodes, and plants, none have been detected in the sequenced genomes of archaea, suggesting a eukaryotic-specific function (unpublished data; Fatica et al., 2002).
In this study we demonstrate how human Naf1p (NAF1) is involved in H/ACA RNP assembly. We show this using a unique combination of biochemical and single-cell labeling approaches. Because most mammalian snoRNAs are processed from introns (Lestrade and Weber, 2006), we designed cell lines that expressed the H/ACA snoRNA E3 from the intron of an inducible pre-mRNA reporter construct stably integrated into their genomes. In this manner, we were able to monitor and control the transcription of a specific H/ACA RNA in a single cell and to delineate the stepwise assembly of H/ACA RNPs in the cell.
Results
Human NAF1 and GAR1 bind NAP57 competitively
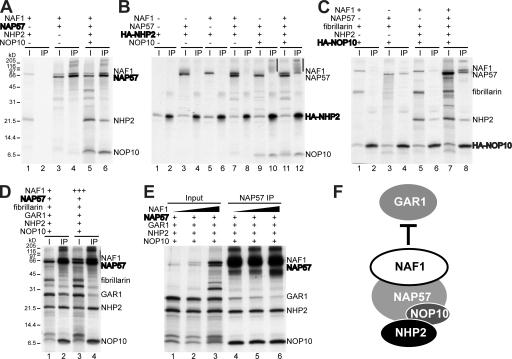
Because yeast Naf1p interacted with some of the H/ACA core proteins, we tested whether NAF1 did as well and, if so, with which ones (Ito et al., 2001; Dez et al., 2002; Fatica et al., 2002; Ho et al., 2002; Yang et al., 2002). For this purpose, we used an in vitro–translation/immunoprecipitation assay that we had previously used to dissect the molecular architecture of mammalian H/ACA RNPs (Wang and Meier, 2004). Specifically, NAF1 was in vitro translated in the presence of various combinations of H/ACA core proteins (Fig. 1, I lanes). Subsequently, one of the proteins was immunoprecipitated and the precipitates analyzed for coprecipitating proteins (Fig. 1, IP lanes).
Figure 1.
Interactions of in vitro–translated and immunoprecipitated H/ACA core proteins and NAF1. Fluorographs of the indicated proteins transcribed/translated from individual cDNAs in rabbit reticulocyte lysate in the presence of [35S]methionine and separated by SDS-PAGE before (I) and after immunoprecipitation (IP). The input lanes (I) contained 10% of the proteins used for IP. In each panel, the precipitated proteins are indicated in bold. The antibodies used for IP were directed against NAP57 (A, D, and E) or the HA epitope (B and C). 15% Tricine SDS-PAGE was used for analysis. The migration position and relative molecular mass of marker proteins is indicated on the left, and the mobility of the translated proteins is marked on the right. For unknown reasons and as previously observed (Wang and Meier, 2004), immunoprecipitated NAP57 migrated anomalously on occasion (vertical bars in B). In some experiments, the amount of cDNA encoding one of the proteins was increased to such an extent that the corresponding translated protein at least doubled in abundance (D and E). Fibrillarin was added as a negative control (C and D). (F) Schematic depiction of the mutually exclusive binding of NAF1 and GAR1 to NAP57 in the context of the core trimer.
Analysis of the autoradiograms showed that NAF1 efficiently coprecipitated with NAP57 alone (Fig. 1 A, lane 4) and in the presence of the core trimer (lane 6) but not in the absence of NAP57 (lane 2). Neither NAF1 nor NAP57 alone or together precipitated with HA-NHP2 (Fig. 1 B, lanes 4, 6, and 8). However, addition of NOP10, which mediates the interaction between NHP2 and NAP57, caused NAP57 (Fig. 1 B, lane 10) and associated NAF1 to precipitate with HA-NHP2 (lane 12). Although NAP57 efficiently coprecipitated with HA-NOP10 (Fig. 1 C, lane 4), NAF1 did not (lane 2). However, when NHP2 was included with HA-NOP10, some NAF1 did coprecipitate (Fig. 1 C, lane 6), suggesting that the association of NHP2 with NOP10 exposed a minor NAF1 interaction domain. As in the case of HA-NHP2, further addition of NAP57 to form the core trimer led to the efficient coprecipitation of NAF1 (Fig. 1 C, lane 8). In contrast, the box C/D core protein fibrillarin, added as a negative control, was not precipitated in any combination (Fig. 1 C). Collectively, these findings demonstrated that, similar to its yeast counterpart (Dez et al., 2002; Fatica et al., 2002; Yang et al., 2002), human NAF1 specifically interacted with NAP57 alone or in the context of the core trimer (Fig. 1 F).
Even in the context of the full complement of core proteins, NAF1 efficiently precipitated with NAP57 (Fig. 1 D, lane 2). Moreover, when the amount of NAF1 was increased in the input (Fig. 1 D, compare lanes 1 and 3), the amount of GAR1 relative to that of NHP2 and NOP10 was reduced in the precipitate (Fig. 1 D, compare lanes 2 and 4). This result indicated that NAF1 competed with GAR1 for NAP57 binding. Indeed, increasing the amount of NAF1 in the input while keeping the amount of the H/ACA core proteins the same (Fig. 1 E, lanes 1–3) resulted in a concomitant reduction of GAR1 relative to NHP2 and NOP10 in NAP57 precipitates (lanes 4–6). NAF1 may bind more tightly than GAR1 to NAP57 because in this system GAR1 expression was insufficient for converse competition and because of the previously observed inconsistent NAP57 association of GAR1 (Wang and Meier, 2004). Given the presence of a homologous domain in NAF1 and GAR1 (Fatica et al., 2002), it is plausible that this domain of each protein binds to the same site in NAP57. Collectively, NAF1 and GAR1 bind NAP57 in the context of the core trimer, but their binding sites overlap (or are identical), precluding simultaneous interaction (Fig. 1 F). This competition between NAF1 and GAR1 suggests a sequential interaction with NAP57.
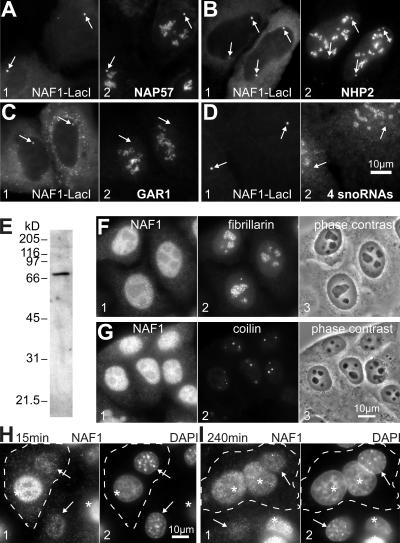
NAF1 binds NAP57 in the cell nucleus
Although these biochemical analyses clearly showed that NAF1 interacted with NAP57, it was imperative to ascertain that this interaction occurred in the biological context of the cell nucleus. Toward this end, we developed a novel nuclear tethering assay. For this purpose, NAF1 was fused to the lac repressor, LacI, which binds to lac operator sequences that were stably integrated at a single chromosomal locus of the human U2OS osteosarcoma cell line (2-6-3; Janicki et al., 2004). Transient transfection of NAF1-LacI into the 2-6-3 U2OS cells resulted in the tethering and concentration of NAF1-LacI at one site in the nucleus, easily detectable by indirect immunofluorescence with anti-NAF1 or -LacI antibodies (Fig. 2, A–D, panel 1, arrows). Using double immunofluorescence, we tested what endogenous proteins NAF1-LacI recruited to its tethering site. NAP57 accumulated above its nucleolar levels at the sites of NAF1-LacI concentration, indicating that NAF1 also bound to NAP57 in the nucleus (Fig. 2 A, panel 2). Similarly, NHP2 was observed at the tethering site, apparently recruited to NAP57 via NOP10 (Fig. 2 B). Despite its ability to interact with NAP57 (Wang and Meier, 2004), GAR1 was not concentrated at the nuclear site of the NAP57–NAF1-LacI complex (Fig. 2 C), consistent with our finding that GAR1 and NAF1 binding to NAP57 was mutually exclusive (Fig. 1, D and E). All observed interactions were due to the NAF1 moiety of the LacI fusion protein, as LacI alone failed to recruit any of the proteins tested (unpublished data). These results supported our findings from the in vitro–translation/immunoprecipitation approach (Fig. 1 F).
Figure 2.
In vivo interactions, localization, and shuttling of human NAF1. (A–D) Tethering of transfected NAF1-LacI to lac repressor repeats stably integrated into the genome of U2OS cells (arrows) and double labeled for LacI (panel 1) and endogenous H/ACA RNP components (panel 2). Double fluorescence with antibodies to NAP57 (A), NHP2 (B), GAR1 (C), and by FISH with probes to the H/ACA RNAs E3, ACA8, ACA18, and ACA25 (D). (E) Western blot of whole cell extracts probed with affinity-purified NAF1 peptide antibodies and developed by enhanced chemiluminescence. (F and G) Indirect double immunofluorescence on fixed and permeabilized HeLa cells with anti-NAF1 (panel 1) and anti-fibrillarin (F, panel 2) or anti-coilin antibodies (G, panel 2). The corresponding phase-contrast image is shown in panel 3. Note the absence of NAF1 from nucleoli (fibrillarin stain) and its lack of enrichment in Cajal bodies (coilin stain). (H and I) Heterokaryon assay. Double fluorescence of fixed individual (bottom) and fused cells (outlined) 15 min (H) and 4 h (I) after fusion stained for NAF1 (panel 1) and DNA (DAPI; panel 2). Note that NAF1 antibodies strongly label individual HeLa (asterisks) but not NIH3T3 nuclei (arrows), whereas both types of nuclei stain equally at intermediate levels only 4 h after fusion, indicating equilibration, i.e., shuttling of NAF1 (I, panel 1).
Although the absence of GAR1 suggested that NAF1-LacI failed to recruit intact H/ACA RNPs, we confirmed this by probing for the presence of four H/ACA RNAs—E3, ACA8, ACA 18, and ACA25. By using simultaneous FISH detection of the four snoRNAs, no or only negligible amounts of H/ACA RNAs were detected at the sites of NAF1-LacI concentration (Fig. 2 D). Moreover, Nopp140, which binds mature H/ACA RNPs (Meier and Blobel, 1992; Yang et al., 2000; Wang et al., 2002), failed to be recruited by NAF1-LacI (unpublished data). Therefore, NAF1 in the nucleus interacted with NAP57 alone and/or in the context of the core trimer NAP57–NOP10–NHP2 but not with mature H/ACA RNPs.
Human NAF1 is a shuttling nucleoplasmic protein
Although NAF1-LacI bound to the lac operator sequences in the nucleus, excessive expression led to its accumulation in the cytoplasm (Fig. 2, B and C, panel 1). Therefore, to investigate the localization of endogenous NAF1, we raised polyclonal antibodies against a synthetic NAF1 peptide. On Western blots of whole cell extracts, affinity-purified NAF1 antibodies recognized a single protein band that migrated at the same position as in vitro–translated NAF1 (compare Fig. 2 E and Fig. 1 A, lane 1). By indirect immunofluorescence, they yielded a nucleoplasmic staining pattern excluding nucleoli (the sites of mature snoRNPs, which were double labeled with antibodies to the C/D snoRNP protein fibrillarin; Fig. 2 F). Furthermore, NAF1 was not enriched in Cajal bodies, another site of mature snoRNPs, identified by coilin immunofluorescence (Fig. 2 G).
Given the nuclear and cytoplasmic localization of the NAF1-LacI fusion protein, we tested whether NAF1 shuttled between the nucleus and cytoplasm using a heterokaryon approach (Borer et al., 1989). For this purpose, we fused human HeLa and mouse NIH3T3 cells (Fig. 2, H and I, outlined cells), whose nuclei can be distinguished by a difference in DNA staining (Fig. 2, H and I, panel 2, asterisks and arrows). Because the NAF1 antibodies barely recognized the mouse protein while brightly staining the HeLa nuclei (Fig. 2, H and I, panel 1), we could test whether NAF1 equilibrated between the nuclei of different origin in fused cells. To inhibit de novo protein synthesis, the fusions were performed in the presence of cycloheximide. Although NAF1 barely redistributed between the different nuclei right after fusion, it fully equilibrated 4 h later (Fig. 2, compare panel 1 in H and I). Specifically, reduced staining in the HeLa nuclei and increased staining in the 3T3 nuclei of the fused cells indicated equilibration of HeLa NAF1 between the nuclei of different origin, i.e., shuttling of NAF1.
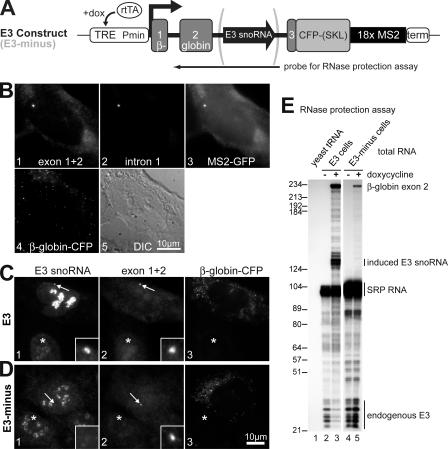
Cell line with inducible and observable H/ACA RNA transcription
So far, our data have indicated that NAF1, although absent from mature H/ACA RNPs, bound NAP57 and the core trimer. Based on the shuttling of NAF1, such an interaction could occur in the cytoplasm followed by nuclear translocation for association with H/ACA RNAs. We reasoned that the most likely sites for nuclear RNP assembly were the sites of H/ACA RNA transcription. To test our hypothesis, we developed a unique cell system. Our four requirements for this system were (1) the inducible transcription of a specific H/ACA RNA, (2) the visualization of its transcription site, (3) the processing of the H/ACA RNA from an intron, and (4) the generation of a functional mRNA. Specifically, two cell lines were generated (based on the U2OS cell line) that had tandem arrays of the following constructs stably integrated at a single locus in their genome. Both constructs consisted of the human β-globin gene with three exons, two introns, and a short 5′ untranslated region (UTR; Fig. 3 A; Kiss and Filipowicz, 1995; Darzacq et al., 2002). Exon 3 was truncated and fused in-frame to CFP containing the peroxisomal targeting tripeptide Ser-Lys-Leu (SKL) at its COOH terminus (Tsukamoto et al., 2000); thus, fluorescent peroxisomes resulted from the transcription of a functional mRNA. The 3′ UTR encompassed 18 MS2 stem loop repeats, which provided tight binding sites for the MS2 coat protein when transcribed (Janicki et al., 2004; Shav-Tal et al., 2004a), thereby allowing the detection of the transcription locus with a MS2-GFP fusion protein. Most important, the rat H/ACA snoRNA E3 was placed in the second intron of the β-globin gene of the first construct (Fig. 3 A). Expression of the gene was induced by doxycycline through the transactivator rtTA (reverse tetracycline-responsive transcriptional activator), which was stably integrated in these U2OS cells (Fig. 3 A). This cell line was termed E3. A control cell line that contained the same construct but without the E3 snoRNA was named E3-minus.
Figure 3.
Stable human U2OS cell lines expressing rat H/ACA RNA E3 from an inducible promoter. (A) Schematic of the construct that was stably integrated into the genome of the E3 and E3-minus cell lines. Expression was driven by a minimal CMV promoter (Pmin) under the control of the tetracycline response element (TRE) and was induced in the presence of doxycycline (dox) by the transactivator rtTA. The construct contained the polyadenylation/cleavage signal and transcriptional terminator (term) of the bovine growth hormone. The size of the probe used in the RNase protection assay (E) is indicated underneath. (B) Fluorescent micrographs of an E3 cell induced for transgene expression and assayed by FISH with Cy3-labeled probes to exon 1 and 2 (panel 1) and a Cy5-labeled probe to intron 1 (panel 2) and by direct fluorescence for MS2-GFP (panel 3) and for β-globin–CFP (panel 4). Panel 5 depicts the corresponding differential interference contrast image of the cell. (C and D) Triple fluorescence by FISH with probes to the induced rat E3 snoRNA (panel 1), which cross-hybridizes with the endogenous human E3 in uninduced cells (asterisks), and to exon 1 and 2 for identification of the transcription sites (panel 2), and by direct fluorescence of β-globin–CFP in peroxisomes (panel 3) to differentiate the induced cells from the uninduced cells. Insets (width = 2.6 μm) show a magnification of the transcription sites (arrows). (E) RNase protection assay. Autoradiograph of the sequencing gel used to separate the fragments protected from digestion by RNase A and T1. Yeast tRNA (lane 1) and total RNA from E3 (lanes 2 and 3) or E3-minus cells (lanes 4 and 5) isolated before (lanes 2 and 4) or 24 h after (lanes 3 and 5) induction with doxycycline were hybridized to the radiolabeled probe indicated in A. In addition, a separate probe corresponding to 100 nucleotides of SRP RNA was included in all samples as a control. The migrating positions of the protected fragments are indicated on the right. Note that because the probe corresponded to the rat E3 snoRNA of the integrated constructs, it protected only the smaller fragments of the endogenous human E3 snoRNA, which differed in 13 nucleotides from that of the rat.
After doxycycline induction of the transgene, proper expression and processing of the RNAs in our cell lines was confirmed by fluorescence microscopy and RNase protection assays. First, using FISH with DNA probes for exon 1 and 2 of the β-globin construct, the exons were detected as a single spot in the nucleoplasm and in mature β-globin–CFP mRNPs in the cytoplasm (Fig. 3 B, panel 1). Hybridization to the same nucleoplasmic spot with a probe for intron 1 confirmed its identity as a transcription site (Fig. 3 B, panel 2). Alternatively and for potential live imaging studies, the transcription sites and cytoplasmic mRNPs were detected by transiently transfected MS2-GFP (Fig. 3 B, panel 3). The presence of the mRNA and its 3′ UTR in the cytoplasm suggested that it was full-length and that the β-globin–CFP pre-mRNA was correctly spliced and exported. This was corroborated by the visualization of its translation product, β-globin–CFP, in peroxisomes where it localized by virtue of its COOH-terminal SKL targeting sequence (Fig. 3 B, panel 4). Importantly, in the absence of doxycycline and transgene transcription, no transcription sites, mRNPs, or β-globin–CFP were detected (unpublished data). The presence of a single transcription site per nucleus suggested that tandem arrays of our constructs integrated at a single site in the genome of these U2OS cells (Shav-Tal et al., 2004b; Darzacq et al., 2005). Indeed, FISH on metaphase spreads corroborated a single insertion site (unpublished data).
In E3 cells, the snoRNA produced from the transgene was detected at the transcription site by FISH with two probes complementary to the rat E3 snoRNA (Fig. 3 C, panels 1 and 2, arrows and insets). Additionally, E3 accumulated in nucleoli in a granular pattern typical for snoRNPs, suggesting its proper incorporation into H/ACA RNPs (Fig. 3 C, panel 1). Induced cells were routinely identified by the β-globin–CFP protein product of the transgene in peroxisomes (Fig. 3 C, panel 3). Because the endogenous human E3 snoRNA differed in 13 nucleotides from the induced rat E3, hybridization to endogenous E3 was weaker (Fig. 3 C, panel 1, asterisk). In the E3-minus control cell line, no E3 was observed at the transcription site (Fig. 3 D, panels 1 and 2, arrows and insets) and no enhanced fluorescence occurred in nucleoli of induced versus uninduced cells (Fig. 3 D, panel 1, asterisk). The transgene, however, was properly expressed as attested by the β-globin–CFP signal in peroxisomes (Fig. 3 D, panel 3).
As a final quality test of our stable cell lines, we investigated the length of the induced snoRNA and the regulation of gene expression using RNase protection assays. Specifically, a radiolabeled RNA probe was synthesized that spanned the intronic snoRNA and exon 2 of β-globin (Fig. 3 A). When incubated with total RNA from the cell lines, fragments from this probe hybridizing to the stable target RNAs were protected from digestion by RNase A and T1 and migrated at their expected positions on a sequencing gel (Fig. 3 E, lanes 2–5). However, hybridization to yeast tRNA yielded no protected fragments (Fig. 3 E, lane 1). After induction by doxycycline, prominent bands of full-length E3 snoRNA and exon 2 were protected by total RNA from E3 cells (Fig. 3 E, lane 3) but only exon 2 by RNA from E3-minus cells (lane 5). The generation of full-length E3 snoRNA indicated its proper processing and incorporation into H/ACA RNPs. Full-length exon 2 represented correctly spliced globin-CFP mRNA. Transcription of the transgene was tightly regulated because no E3 and exon 2 were detected in the absence of doxycycline (Fig. 3 E, lanes 2 and 4). Endogenous E3 snoRNA was present independent of induction of the transgene (Fig. 3 E, lanes 2–5). Note that only smaller fragments of the probe were protected by partial hybridization to endogenous human E3 snoRNA because of slight sequence differences to the rat E3 of the transgene. As a control, a separate probe was included that protected a 100-nt fragment of the unrelated RNA of the signal recognition particle (SRP). Equal amounts of SRP RNA were protected by total RNA from induced and uninduced cells but not by tRNA (Fig. 3 E). These data indicated that our transgene was correctly induced, transcribed, and spliced and that the rat E3 snoRNA was properly processed and assembled into stable H/ACA RNPs.
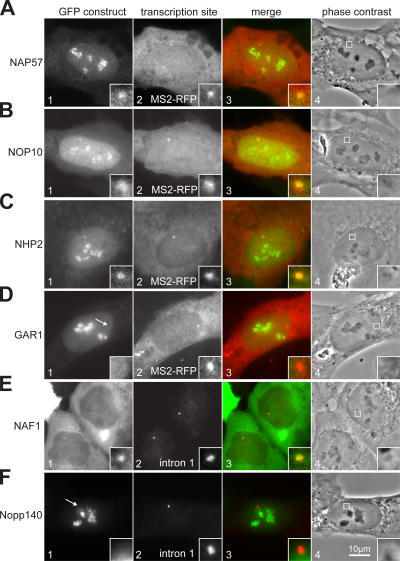
Visualization of the H/ACA core trimer and NAF1 at the site of snoRNA transcription
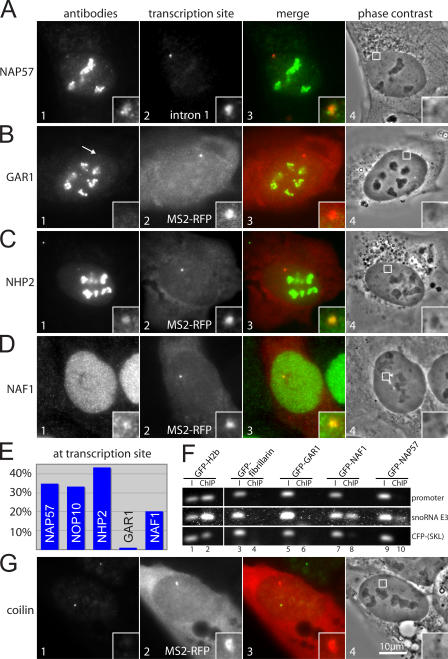
To test which proteins associated with the nascent H/ACA RNA at its site of transcription, we transfected GFP-tagged constructs into the E3 cell line and induced expression of the transgene. Localization at the transcription site was monitored by double fluorescence with cotransfected MS2-RFP or by FISH with the intron 1 probe (Fig. 4, panel 2). All core proteins localized to nucleoli in their characteristic granular pattern, suggesting their incorporation into H/ACA RNPs (Fig. 4, A–D, panel 1). In addition, the proteins of the core trimer, NAP57-, NHP2-, and NOP10-GFP, were identified at the site of H/ACA RNA transcription, indicating an early association of the core trimer with the nascent RNA (Fig. 4, A–C, insets). However, the fourth core protein, GAR1-GFP, was consistently absent from H/ACA RNA transcription sites (Fig. 4 D, arrow and insets). The putative assembly factor, NAF1-GFP, concentrated at the transcription sites but was excluded from nucleoli (Fig. 4 E). Overexpressed NAF1-GFP accumulated in the cytoplasm like NAF1-LacI (Fig. 4 E and Fig. 2, B and C, panel 1). These observations suggested an early involvement of NAF1 in H/ACA RNP biogenesis, followed by GAR1 replacement during or after release of the maturing particles from the site of transcription. Indeed, the absence from transcription sites of GFP-Nopp140, a chaperone associated with functional H/ACA RNPs in nucleoli (Wang et al., 2002), supported the lack of mature particles at the site of transcription (Fig. 4 F). Additionally, the C/D snoRNP protein GFP-fibrillarin, although localizing properly to nucleoli, was absent from the transcription site (unpublished data). Importantly, none of the proteins was observed at the transcription site of E3-minus cells, confirming that all colocalization was due to the nascent H/ACA RNA.
Figure 4.
Colocalization of GFP constructs to the H/ACA RNA transcription site. (A) Double fluorescence of an E3 cell cotransfected with NAP57-GFP (panel 1) and MS2-RFP (panel 2) and induced for transgene expression. Panel 3 shows a merge of the two in pseudocolor. Panel 4 depicts the corresponding phase-contrast image outlining, with a box the area containing the transcription site, which is enlarged in the insets. (B–F) Same as A, except transfected with GFP fused to the proteins indicated on the left. H/ACA RNA transcription sites in E and F were detected by FISH with the probe to intron 1. Note the failure of GAR1-GFP and GFP-Nopp140 to colocalize to the transcription site (arrows). The width of the insets corresponds to 2.6 μm.
The results with the transfected constructs were corroborated with endogenous proteins by indirect immunofluorescence and quantitated. In the same way as the transiently transfected constructs and in addition to their known location in nucleoli and nucleoplasm, endogenous NAP57, NHP2, and NAF1, but not GAR1, were concentrated at the E3 transcription site (Fig. 5, A–D). For quantitation, the presence of each protein, transfected and endogenous, at an E3 transcription site was scored. The identifications of each factor at the site of transcription from at least 100 cells are given as a percentage (Fig. 5 E). These data documented the presence of NAP57, NOP10, NHP2, and NAF1 at the E3 transcription site but a lack of association of GAR1. The <100% coincidence of the localizing factors with the transcription site was likely caused by variable stages of H/ACA RNA transcription and the fact that uncertain identifications were discounted. Nevertheless, the quantitation supported our conclusions and proved the images in Fig. 4 and Fig. 5 (A–D) to be truly representative.
Figure 5.
Colocalization of endogenous proteins with the H/ACA RNA transcription site. (A) Double fluorescence of a fixed and permeabilized E3 cell after induction of the transgene and immunostained for endogenous NAP57 (panel 1) and for the H/ACA RNA transcription site by FISH with the probe to intron 1 (panel 2). Panel 3 shows a merge of the two in pseudocolor. Panel 4 depicts the corresponding phase-contrast image outlining, with a box indicating the area containing the transcription site, which is enlarged in the insets. (B–D) Same as in A, except immunostained for the endogenous proteins indicated on the left, and the transcription site was identified by the fluorescence of transfected MS2-RFP (panel 2). (E) Bar diagram of the percentage of cells, with the indicated proteins observed at the site of H/ACA RNA transcription. The following numbers of cells with a transcription site (in parentheses) were counted for each protein: NAP57 (150); NOP10 (101); NHP2 (101); GAR1 (104); and NAF1 (159). Questionable colocalizations were discounted. (F) ChIPs of the GFP-fusion constructs indicated on top after their transient transfection into E3 cells and induction of the transgene. Immunoprecipitations were performed with GFP antibodies. Ethidium bromide stain of DNA amplified by PCR, with primers to the regions of the transgene indicated on the right and separated on agarose gels. Amplifications of the input (I; odd lanes) and the ChIPs (even lanes) are shown. (G) Same as in B and C, except stained for the Cajal body marker protein coilin.
To confirm the recruitment of NAP57 and NAF1 to the snoRNA transcription site by an independent method, we used ChIP on our induced cell lines transiently transfected with GFP-tagged constructs. Using PCR, we tested for the presence of three DNA fragments in the inputs and ChIPs: the promoter region, the rat snoRNA E3, and the CFP-SKL of the transgene. The presence of the correct fragment in the inputs was confirmed in all cases (Fig. 5 F, odd lanes). Immunoprecipitation with anti-GFP antibodies detected an association of the positive control, the core histone H2b, with all fragments (Fig. 5 F, lane 2). Although GAR1 and the control C/D snoRNP protein fibrillarin were not associated with any of the three fragments (Fig. 5 F, lanes 4 and 6), NAP57 and NAF1 precipitated the fragment corresponding to snoRNA E3 but not the other two (lanes 8 and 10). These results indicated that NAP57 and NAF1 were recruited specifically during transcription of H/ACA snoRNA E3 but not before or after. Altogether, the ChIP data supported our single-cell fluorescence localizations.
For the following three reasons, we also investigated whether Cajal bodies coincided with H/ACA RNA transcription sites. First, Cajal bodies have been implicated as sites of H/ACA RNP–mediated pseudouridylation (Darzacq et al., 2002). Second, the transcription of other small nuclear RNAs, e.g., U2, had been physically associated with Cajal bodies (Frey et al., 1999; Smith and Lawrence, 2000). Third, we wanted to verify that the observed H/ACA RNA transcription sites were unique entities and distinct from Cajal bodies. This was to be expected because GAR1 and Nopp140, both verified Cajal body constituents (Meier and Blobel, 1994; Pogacic et al., 2000), were not present at transcription sites (Fig. 4, B and F; and Fig. 5 B) and because only 27% of our U2OS cells contained Cajal bodies. Indeed, most Cajal bodies, identified by their marker protein coilin, were distinct from the snoRNA E3 transcription site (Fig. 5 G). Nevertheless, in a small number of instances (18 out of 245 cells with both Cajal bodies and transcription sites), an association of the two was observed, suggesting that, as in the case of U2 snRNA, there may be some relationship between active snoRNA genes and Cajal bodies.
NAF1 is essential for stable H/ACA RNA accumulation and is recruited to the transcription site in a NAP57-dependent fashion
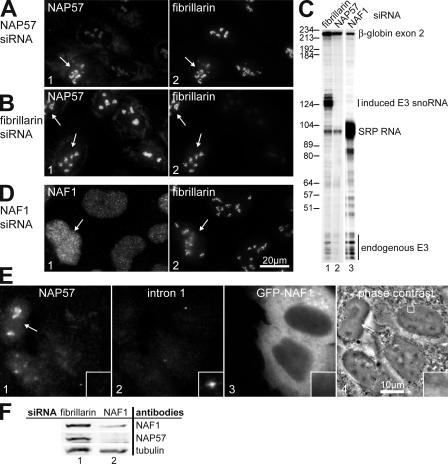
Having established that both NAP57 and NAF1 were recruited to the site of H/ACA RNA transcription, we used a siRNA approach to investigate how the appearance of the two proteins related to each other. For this purpose, E3 cells were transiently transfected with NAP57 siRNA for 72 h and the knockdown confirmed by indirect immunofluorescence. NAP57 was knocked down in the nucleoli of >50% of cells, whereas the levels of fibrillarin in the same nucleoli remained unaffected (Fig. 6 A, compare panels 1 and 2). Similarly, knockdown of fibrillarin left the amount and distribution of NAP57 unaffected (Fig. 6 B, compare panels 1 and 2). As judged by comparison of fluorescence intensities of nucleoli in silenced versus unsilenced cells (Fig. 6, A and B, arrows), NAP57 and fibrillarin were knocked down by ∼60 and ∼80%, respectively.
Figure 6.
NAP57 and NAF1 RNA interference. (A) Double immunofluorescence of NAP57 (panel 1) and fibrillarin (panel 2) in E3 cells 3 d after transfection with NAP57 siRNA. Note that NAP57 is not knocked down in all cells (arrow). (B) Same as A, except the cells were transfected with fibrillarin siRNA. (C) RNase protection assay as in Fig. 3 E on RNA from E3 cells transfected with siRNAs directed against the mRNAs of the proteins indicated on top and induced for transgene expression. Note that NAP57 and NAF1, but not fibrillarin, siRNAs abolished specifically the accumulation of induced rat E3 snoRNA but not of the exon of the gene from which it was expressed. (D) Same as A, except the cells were transfected with NAF1 siRNA. (E) NAF1 is not present at the H/ACA RNA transcription site in cells knocked down for NAP57. Triple fluorescence of endogenous NAP57 (panel 1), FISH with intron 1 probe marking the transcription site (panel 2), and transfected GFP-NAF1 (panel 3) in cells transfected with NAP57 siRNA. Panel 4 shows a phase-contrast image of the same cells and outlines the area of the inset. The arrow points to an unsilenced cell. The width of inset is 2.6 μm. (F) Western blots of lysates from cells treated with siRNA to fibrillarin (lane 1) and NAF1 (lane 2), probed with antibodies to the antigens indicated on the right, and scanned on an infrared imaging system.
To test the impact of NAP57 and fibrillarin knockdown on the expression of the transgene, E3 cells were transfected with siRNAs and induced with doxycycline before total RNA isolation and RNase protection analysis. In this manner, fibrillarin knockdown had no impact on the expression and stability of the E3 snoRNA and the globin-CFP mRNA (Fig. 6 C, lane 1). However, NAP57 knockdown abolished production of stable E3 snoRNA but left globin-CFP mRNA induction unaffected (Fig. 6 C, lane 2). The impact of NAP57 knockdown on endogenous E3 snoRNA was less severe, reflecting the increased half-life of the snoRNA once incorporated into mature RNPs. In separate experiments, knockdown of NAF1 also caused a specific loss of induced E3 snoRNA but not of exon 2 (Fig. 6 C, lane 3), even though NAF1 was only reduced by ∼50% (Fig. 6 D). These data indicated that reduced levels of NAP57 and NAF1 abolished packaging of newly synthesized H/ACA RNA into stable RNPs.
Because both NAP57 and NAF1 proved essential for H/ACA RNP biogenesis, we tested whether their recruitment to nascent snoRNA was interdependent. For this purpose, NAF1-GFP was transfected into E3 cells knocked down for NAP57 but induced for transgene expression. Surprisingly, in NAP57-silenced cells, NAF1-GFP was never detected at E3 snoRNA transcription sites identified by FISH with the intron 1 probe (Fig. 6 E). However, in control fibrillarin knocked down cells, NAF1-GFP was observed at snoRNA transcription sites (unpublished data). Therefore, recruitment of NAF1 to nascent H/ACA RNAs was NAP57 dependent.
Finally, we tested whether NAP57 was also dependent on NAF1. Because of the partial knockdown, individual cells reduced in NAF1 were difficult to unambiguously identify by indirect immunofluorescence. Therefore, Western blotting was used to compare the amount of the two proteins in whole cell lysates (Fig. 6 F). After NAF1 knockdown, NAP57 levels were reduced compared with those of tubulin and of control cells knocked down for fibrillarin (Fig. 6 F, compare lanes 1 and 2). Thus, NAP57 levels were dependent on NAF1.
Discussion
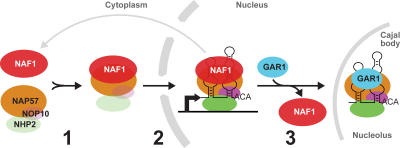
The capability to detect and image discrete regions in the cell nucleus at which specific complexes assemble, in conjunction with detailed biochemical analysis of these same processes, is a powerful tool for the unraveling of intricate interactions taking place during the generation of molecular complexes. Following this approach, we dissected the spatiotemporal assembly of human H/ACA RNPs using newly established cell lines that allow induction and visualization of H/ACA RNA transcription. Our data support the following three-step model of human H/ACA RNP biogenesis (Fig. 7). First, NAF1 stabilizes newly synthesized NAP57 before integration into the RNP. NOP10 and NHP2 may associate with NAP57 at this point to form the core trimer because the NAF1–NAP57 interaction is unimpeded by the presence of NOP10 and NHP2. Second, the NAF1–NAP57–(NOP10–NHP2) complex is recruited to the site of H/ACA RNA transcription. Third, GAR1 replaces NAF1 in the maturing particle either at the transcription site triggering its release or during transit through the nucleoplasm to the site of function in nucleoli and Cajal bodies. Therefore, assembly of these simple five-component RNPs displays a surprising spatiotemporal complexity that serves as paradigm for the assembly of larger RNPs. For example, ribosome assembly requires an unexpected number of nonribosomal proteins (Granneman and Baserga, 2004).
Figure 7.
Three-step model of human H/ACA RNP assembly. NAF1 associates with NAP57 in the cytoplasm (1). The complex is recruited to the site of H/ACA RNA transcription, as are NOP10 and NHP2 (2). GAR1 replaces NAF1 to yield mature H/ACA RNPs in nucleoli and Cajal bodies (3). Released NAF1 recycles across the nuclear envelope. NOP10 and NHP2 may already bind to NAP57 in the cytoplasm.
A general concept that emerges from our studies is that newly synthesized, free NAP57 is always paired with NAF1. Specifically, the two proteins are interdependent (Fig. 6). The need for such a stabilizing activity is supported by the fact that mammalian NAP57, when expressed in bacteria or insect cells, precipitates and is insoluble (unpublished data). The shuttling of the normally nuclear NAF1 is consistent with its association with NAP57 already in the cytoplasm followed by their coimport (Fig. 2 I). If free NAP57 and NAF1 are normally associated, then not only may NAF1 recruitment to the H/ACA RNA transcription site be dependent on NAP57 but also vice versa. For example, NAF1 may target NAP57 to nascent H/ACA snoRNAs by binding to the phosphorylated COOH-terminal domain (CTD) of RNA polymerase II and remain there through the interaction of NAP57 with the snoRNA (Fig. 7). Such a mechanism is supported by findings with yeast Naf1p, which binds to the CTD (Ito et al., 2001; Fatica et al., 2002; Ho et al., 2002). Two recent reports using ChIP analysis showed that Naf1p and Cbf5p (yeast NAP57) are also recruited cotranscriptionally in yeast; however, it remains to be resolved whether phosphorylated CTD is required or not (Ballarino et al., 2005; Yang et al., 2005). Regardless, our dissection of the role of human NAF1 in H/ACA RNP biogenesis suggests that it is the orthologue of yeast Naf1p and that it functions as a NAP57 chaperone.
The last step of H/ACA RNP biogenesis, the replacement of NAF1 by GAR1, may occur in Cajal bodies, possibly aided by the survival of motor neurons (SMN) protein. The following three observations support such a scenario. First, SMN, all snoRNP core proteins, and most of their RNAs tested have been detected in Cajal bodies (Gall, 2000; Kiss, 2004). Second, SMN, which assembles the Sm proteins onto spliceosomal U snRNAs, interacts with GAR1, raising the possibility of its involvement also in H/ACA RNP biogenesis (Pellizzoni et al., 2001; Whitehead et al., 2002). Third, fibrillarin, the glycine and arginine domain–containing protein of C/D snoRNPs, also interacts with SMN and also is involved in a late RNP assembly step (Lafontaine and Tollervey, 2000; Jones et al., 2001; Pellizzoni et al., 2001; Watkins et al., 2002). Therefore, and in analogy to the C/D snoRNP U3 (Verheggen et al., 2002), H/ACA RNP biogenesis may be added to the many functions of Cajal bodies. The occurrence of the final step of H/ACA RNP maturation in Cajal bodies is further attractive because it would preclude the presence of functional H/ACA RNPs in the nucleoplasm, where they might endanger nontarget RNAs by pseudouridylation.
In conclusion, we have developed a novel cell system to visualize the site of transcription of an intronic H/ACA RNA and to define factors recruited to the nascent H/ACA RNA after induction of its transcription. These stable cell lines were based on a previously established system to monitor gene expression in living cells (Janicki et al., 2004; Shav-Tal et al., 2004a) and allowed us to dissect the biogenesis of H/ACA RNA beyond what was possible with ChIP analysis (Ballarino et al., 2005; Yang et al., 2005). We have defined some roles of the H/ACA-specific factor NAF1 in RNP assembly. Nevertheless, additional factors may be involved, as in the case of C/D RNP biogenesis, where the export factors CRM1 (chromosome region maintenance 1) and PHAX (phosphorylated adaptor for RNA export) are important (Boulon et al., 2004). Our cell lines will now allow testing if these and other factors also play a role in H/ACA RNP biogenesis.
Materials and methods
Tissue culture, transfection, and cell lines
HeLa, NIH3T3, and COS-1 cells were cultured in high-glucose DME (Invitrogen) containing 10% FBS (Invitrogen). E3 and E3-minus cells were cultured in low-glucose DME (Invitrogen) containing 10% FBS. Cells were transiently transfected with expression plasmids using FuGene 6 (Roche) for 24–30 h and with siRNAs using Lipofectamine 2000 (Invitrogen) for 72 h according to manufacturer's protocol. Transcription of the transgenes was induced using 6 μg/ml doxycycline overnight.
E3 and E3-minus cells were generated by electroporating the respective constructs together with a puromycin-resistance plasmid (pPuro; a gift from C. Dugast-Darzacq, Institut Jacques Monod, Paris, France) into human U2OS tet-on cells (CLONTECH Laboratories, Inc.) and selection with 10 μg/ml puromycin. After doxycycline induction, individual colonies were screened for globin-CFP expression in peroxisomes. CFP-positive clones were tested for a detectable transcription site by FISH or transfection with MS2-GFP.
DNA/RNA constructs
The human NAF1 clone was obtained from American Type Culture Collection (clone 3447276). For transient transfections, the following plasmids were used: NAF1-GFP (pNK16; NAF1 fused to monomeric GFP in monomeric GFP–C1; Snapp et al., 2003); RFP-NAF1 (pNK31; NAF1 fused to monomeric RFP in monomeric RFP–C1; Campbell et al., 2002); NAF1 (pNK40; in pcDNA3; Invitrogen); NAP57-GFP, NOP10-GFP, NHP2-GFP, and GAR1-GFP (Pogacic et al., 2000); GFP-Nopp140 (pTM95; Dundr et al., 2004); GFP-fibrillarin (Platani et al., 2000); MS2-GFP (pNK35; the NLS was removed from and photo-activatable GFP replaced by GFP in MS2–photo-activatable GFP; Shav-Tal et al., 2004a); MS2-RFP (pNK36; same as for MS2-GFP); and LacI-NAF1 (pNK41; LacI from pAFS51 [Robinett et al., 1996] via pJRC70 [a gift from J. Chubb, University of Dundee, Dundee, UK] cloned into pNK40).
As templates for in vitro transcription/translation, the following constructs were used: NAP57, NOP10, NHP2, GAR1, HA-NOP10, and HA-NHP2 as described previously (Wang and Meier, 2004); NAF1 (pSR29; NAF1 under T7 promoter in pBSII SK+; Stratagene); and HA-NAF1 (pSR36; NAF1 with a single HA tag in pTM93; Isaacet al., 1998).
The pTet-globin-snoE3-CFP-18MS2-2 (for E3 cells) and pTet-globin-CFP-18MS2-2 (for E3-minus cells) were generated by sequentially inserting the following, mostly PCR-amplified fragments, into pCMV-globin (Darzacq et al., 2002): CFP with a COOH-terminal SKL tripeptide from pECFP-N1 (CLONTECH Laboratories, Inc.); 18 MS2 repeats from pSL-24X (Fusco et al., 2003; six repeats were lost because of recombination in Escherichia coli); Tet promoter from pTRE-2 (CLONTECH Laboratories, Inc.) replacing the CMV promoter; and, after excision of the neomycin-resistance gene, rat snoRNA E3 from pTM105 (Wang et al., 2002).
As a probe for RNase protection assays, an 883-nt fragment encompassing part of intron 1 through intron 2 after the snoRNA insertion site (Fig. 3 A) was cloned into PCR4-TOPO vector (Invitrogen), yielding pSR41. As a probe for SRP RNA, nucleotides 119–221 of the RNA were amplified from plasmid p7swt2 (Strub et al., 1991) and cloned into pBSII KS+ to yield pSR27. The probe for U19 was as published (Kiss et al., 1996).
The following siRNAs were used at a final concentration of 50 nM: siNAP57, GGACGGCAUUGAGGUCAAUTT (Ambion); a combination of siNAF1.3, GGCCUUAGAUUUUUAGUGAUTT (Ambion), and siNAF1.4, GGAUAUCGUAACAGAGAAUTT (Ambion); and sifibrillarin, GUCUUCAUUUGUCGAGGAATT (Dharmacon).
Fluorescence and immunochemical approaches
For indirect immunofluorescence, cells were fixed and permeabilized as we described previously (Isaac et al., 1998) and mounted in p-phenylenediamine–containing medium (Pringle et al., 1991). The following primary antibodies were used: polyclonal anti-NAP57 serum (RU8 at 1:200) raised in rabbits (Covance Research Products) against the synthetic peptide 20KSLPEEDVAEIQHAE34 (GenScript Corporation) of human NAP57 with an additional NH2-terminal cysteine as described previously (Meier and Blobel, 1992); anti-NAF1 IgGs (CRX6 at 1:100) raised in rabbits against the synthetic peptide 146CISSLPPVLSDGDDDLQVEKENKN168 of human NAF1 as described for the NAP57 antiserum and affinity purified over a peptide column (Meier and Blobel, 1992); anti-GAR1 IgG (p16 at 1.5 μg/ml; Dragon et al., 2000); anti-NHP2 IgG (p14 at 3 μg/ml; Pogacic et al., 2000); anti-fibrillarin IgG (D77 at 1 μg/ml; Aris and Blobel, 1988); anti-coilin ascites fluid (5P10 at 1:1,000; Almeida et al., 1998); and anti-LacI IgG (9A5 at 1 μg/ml; Upstate Biologicals). The following fluorescently labeled secondary antibodies were used at a 1:200 dilution: goat anti–rabbit IgGs (rhodamine; Chemicon); goat anti–mouse IgGs (fluorescein; Boehringer); and goat anti–rabbit F(ab′)2 fragments (Cy5; Jackson ImmunoResearch Laboratories).
Imaging was performed at room temperature using a 60×/1.4 NA planapo objective on an inverted microscope (1X 81; Olympus) containing automatic excitation and emission filter wheels connected to an air-cooled charge-coupled device camera (Sensicam QE; Roper Scientific) run by IPLab Spectrum software (Scanalytics). Images were processed for contrast and brightness using Photoshop CS2 (Adobe). Because of slight bleed-through from the fluorescein into the cyan channel in Fig. 3 B (panel 4), the fluorescein image was subtracted from the cyan image using ImageJ software (NIH).
FISH was performed as previously described (Chartrand et al., 2000), except that cells were grown on untreated coverslips and no dextran sulfate or vanadylribonucleoside complex was included with 40% formamide for hybridization with a 10-ng probe. Probes were synthesized and labeled with either Cy5 or Cy3 (GE Healthcare). In some cases, immunostaining was done after FISH, using the routine protocol, except that cells were fixed again but not permeabilized.
Heterokayon assays were performed essentially as described previously (Borer et al., 1989). Cell fusions were induced by the addition of 50% polyethylene glycol 8000 for 10 min, and 100 μg/ml of cycloheximide was added 1 h before and during fusion. The cells were fixed 15 min and 4 h after fusion and labeled with NAF1 antibodies followed by DNA stain with 5 μg/ml DAPI for 2 min.
For the NAF1-LacI tethering assay, the 2-6-3 U2OS cell line (Janicki et al., 2004) was transiently transfected with NAF1-LacI and analyzed by indirect double immunofluorescence with the indicated antibodies and by FISH.
In vitro–transcription/translation and immunoprecipitation assays (Wang and Meier, 2004) and Western blots (Isaac et al., 1998) were performed as described previously, except the blot in Fig. 6 F, which was probed with NAF1 rabbit serum (CRY6 at 1:100), NAP57 IgG (RU8 at 0.6 μg/ml), and tubulin monoclonal antibodies (DMA1 at 1.1 μg/ml; Sigma-Aldrich), developed with infrared conjugated goat anti–rabbit IgGs (Alexa fluor 680; Rockland Immunochemicals) and goat anti–mouse IgGs (IR800; Invitrogen), and scanned on an infrared imaging system (Odyssey; LI-COR Biosciences).
RNase protection assay and ChIP
RNase A/T1 protection analysis was performed as described previously (Goodall et al., 1990), with minor changes. Antisense probes were synthesized in the presence of α-[32P]CTP using the Maxiscript kit (Ambion) and gel purified. 1–5 μg of total RNA (prepared with TRIzol [Invitrogen]) were mixed with probe (50,000 cpm) for hybridization and digestion with 40 μg/ml RNase A (Roche) and 2 μg/ml RNase T1 (Calbiochem). Typically, half of the samples were analyzed in 6% polyacrylamide–8M urea sequencing gels followed by autoradiography.
ChIP was performed as described in detail by the Farnham Laboratory (Weinmann et al., 2001). Cells used for ChIP were transfected with the indicated GFP-tagged constructs using FuGene 6 (Roche) and induced simultaneously with 6 μg/ml doxycycline for 24 h. The GFP fusion proteins were precipitated with anti-GFP agarose beads (MBL International Corporation).
Acknowledgments
We are deeply indebted to the following people and laboratories for contributing the referenced reagents: John Aris, Maria Carmo-Fonseca, Jonathan Chubb, Claire Dugast-Darzacq, Witek Filipowicz, Tamas Kiss, Angus Lamond, Annemarie Poustka, Erik Snapp, David Spector, and Katharina Strub. We thank Cristina Montagna and Jenetta Smith of the Albert Einstein Genome Imaging Facility for metaphase spread analysis. We acknowledge the services of the Albert Einstein Analytical Imaging Facility.
This work was supported by grants from the National Institutes of Health (HL079566 [to U.T. Meier] and EB2060 [R.H. Singer]) and the American Cancer Society (RSG-01-050-01-GMC; to U.T. Meier).
X. Darzacq, N. Kittur, and S. Roy contributed equally to this paper.
X. Darzacq's present address is Centre National de la Recherche Scientifique, École Normale Supérieure, 75230 Paris, Cedex 05, France.
Y. Shav-Tal's present address is Faculty of Life Sciences, Bar-Ilan University, Ramat-Gan 52900, Israel.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; CTD, COOH-terminal domain; SKL, Ser-Lys-Leu; snoRNP, small nucleolar RNP; UTR, untranslated region; SMN, survival of motor neurons; SRP, signal recognition particle.
References
- Almeida, F., R. Saffrich, W. Ansorge, and M. Carmo-Fonseca. 1998. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 142:899–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris, J., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D.L., O.A. Youssef, M.I. Chastkofsky, D.A. Dy, R.M. Terns, and M.P. Terns. 2005. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 19:1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarino, M., M. Morlando, F. Pagano, A. Fatica, and I. Bozzoni. 2005. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:5396–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer, R.A., C.F. Lehner, H.M. Eppenberger, and E.A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 56:379–390. [DOI] [PubMed] [Google Scholar]
- Boulon, S., C. Verheggen, B.E. Jady, C. Girard, C. Pescia, C. Paul, J.K. Ospina, T. Kiss, A.G. Matera, R. Bordonne, and E. Bertrand. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 16:777–787. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier, B., S. Muller, and C. Branlant. 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 33:3133–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand, P., E. Bertrand, R.H. Singer, and R.M. Long. 2000. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 318:493–506. [DOI] [PubMed] [Google Scholar]
- Darzacq, X., B.E. Jady, C. Verheggen, A.M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 21:2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq, X., R.H. Singer, and Y. Shav-Tal. 2005. Dynamics of transcription and mRNA export. Curr. Opin. Cell Biol. 17:332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W.A., and M.J. Fournier. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278:695–698. [DOI] [PubMed] [Google Scholar]
- Dez, C., A. Henras, B. Faucon, D. Lafontaine, M. Caizergues-Ferrer, and Y. Henry. 2001. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 29:598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez, C., J. Noaillac-Depeyre, M. Caizergues-Ferrer, and Y. Henry. 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22:7053–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., V. Pogacic, and W. Filipowicz. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., M.D. Hebert, T.S. Karpova, D. Stanek, H. Xu, K.B. Shpargel, U.T. Meier, K.M. Neugebauer, A.G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., M. Dlakic, and D. Tollervey. 2002. Naf1p is a box H/ACA snoRNP assembly factor. RNA. 8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319–327. [DOI] [PubMed] [Google Scholar]
- Frey, M.R., A.D. Bailey, A.M. Weiner, and A.G. Matera. 1999. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 9:126–135. [DOI] [PubMed] [Google Scholar]
- Fusco, D., N. Accornero, B. Lavoie, S.M. Shenoy, J.M. Blanchard, R.H. Singer, and E. Bertrand. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall, J.G. 2000. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16:273–300. [DOI] [PubMed] [Google Scholar]
- Goodall, G.J., K. Wiebauer, and W. Filipowicz. 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181:148–161. [DOI] [PubMed] [Google Scholar]
- Granneman, S., and S.J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43–50. [DOI] [PubMed] [Google Scholar]
- Henras, A., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J.P. Gelugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras, A.K., R. Capeyrou, Y. Henry, and M. Caizergues-Ferrer. 2004. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA. 10:1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G.D. Bader, L. Moore, S.L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415:180–183. [DOI] [PubMed] [Google Scholar]
- Isaac, C., Y. Yang, and U.T. Meier. 1998. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 142:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 98:4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki, S.M., T. Tsukamoto, S.E. Salghetti, W.P. Tansey, R. Sachidanandam, K.V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R.H. Singer, and D.L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K.W., K. Gorzynski, C.M. Hales, U. Fischer, F. Badbanchi, R.M. Terns, and M.P. Terns. 2001. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 276:38645–38651. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. 2004. Biogenesis of small nuclear RNPs. J. Cell Sci. 117:5949–5951. [DOI] [PubMed] [Google Scholar]
- Kiss, T., and W. Filipowicz. 1995. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 9:1411–1424. [DOI] [PubMed] [Google Scholar]
- Kiss, T., M.L. Bortolin, and W. Filipowicz. 1996. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol. Cell. Biol. 16:1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D.L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D.L.J., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade, L., and M.J. Weber. 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, U.T. 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma. 114:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, U.T., and G. Blobel. 1992. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 70:127–138. [DOI] [PubMed] [Google Scholar]
- Meier, U.T., and G. Blobel. 1994. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 127:1505–1514. (published erratum appears in. J. Cell Biol. 1998. 140:447) [DOI] [PMC free article] [PubMed]
- Pellizzoni, L., J. Baccon, B. Charroux, and G. Dreyfuss. 2001. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 11:1079–1088. [DOI] [PubMed] [Google Scholar]
- Platani, M., I. Goldberg, J.R. Swedlow, and A.I. Lamond. 2000. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 151:1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacic, V., F. Dragon, and W. Filipowicz. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20:9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R., A.E.M. Adams, D.G. Drubin, and B.K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565–602. [DOI] [PubMed] [Google Scholar]
- Robinett, C.C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A.S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y., X. Darzacq, S.M. Shenoy, D. Fusco, S.M. Janicki, D.L. Spector, and R.H. Singer. 2004. a. Dynamics of single mRNPs in nuclei of living cells. Science. 304:1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y., R.H. Singer, and X. Darzacq. 2004. b. Imaging gene expression in single living cells. Nat. Rev. Mol. Cell Biol. 5:855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.M., and J.A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 89:669–672. [DOI] [PubMed] [Google Scholar]
- Smith, K.P., and J.B. Lawrence. 2000. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol. Biol. Cell. 11:2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp, E.L., R.S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub, K., J. Moss, and P. Walter. 1991. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol. Cell. Biol. 11:3949–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto, T., N. Hashiguchi, S.M. Janicki, T. Tumbar, A.S. Belmont, and D.L. Spector. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871–878. [DOI] [PubMed] [Google Scholar]
- Verheggen, C., D.L. Lafontaine, D. Samarsky, J. Mouaikel, J.M. Blanchard, R. Bordonne, and E. Bertrand. 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 21:2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., and U.T. Meier. 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23:1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., C.C. Query, and U.T. Meier. 2002. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 22:8457–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N.J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Lührmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all box H/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 4:1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N.J., A. Dickmanns, and R. Luhrmann. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22:8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann, A.S., S.M. Bartley, T. Zhang, M.Q. Zhang, and P.J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, S.E., K.W. Jones, X. Zhang, X. Cheng, R.M. Terns, and M.P. Terns. 2002. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 277:48087–48093. [DOI] [PubMed] [Google Scholar]
- Yang, P.K., G. Rotondo, T. Porras, P. Legrain, and G. Chanfreau. 2002. The Shq1p·Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 277:45235–45242. [DOI] [PubMed] [Google Scholar]
- Yang, P.K., C. Hoareau, C. Froment, B. Monsarrat, Y. Henry, and G. Chanfreau. 2005. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 25:3295–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., C. Isaac, C. Wang, F. Dragon, V. Pogacic, and U.T. Meier. 2000. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell. 11:567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]