Abstract
c-Jun NH2-terminal kinases (JNKs) are essential during brain development, when they regulate morphogenic changes involving cell movement and migration. In the adult, JNK determines neuronal cytoarchitecture. To help uncover the molecular effectors for JNKs in these events, we affinity purified JNK-interacting proteins from brain. This revealed that the stathmin family microtubule-destabilizing proteins SCG10, SCLIP, RB3, and RB3′ interact tightly with JNK. Furthermore, SCG10 is also phosphorylated by JNK in vivo on sites that regulate its microtubule depolymerizing activity, serines 62 and 73. SCG10-S73 phosphorylation is significantly decreased in JNK1−/− cortex, indicating that JNK1 phosphorylates SCG10 in developing forebrain. JNK phosphorylation of SCG10 determines axodendritic length in cerebrocortical cultures, and JNK site–phosphorylated SCG10 colocalizes with active JNK in embryonic brain regions undergoing neurite elongation and migration. We demonstrate that inhibition of cytoplasmic JNK and expression of SCG10-62A/73A both inhibited fluorescent tubulin recovery after photobleaching. These data suggest that JNK1 is responsible for regulation of SCG10 depolymerizing activity and neurite elongation during brain development.
Introduction
MAPK signal transduction pathways are among the most ubiquitous mechanisms of cellular regulation. The JNK family of MAPKs is classically activated by stresses leading to transcriptional regulation and apoptotic cell death (Kyriakis and Avruch, 2001). JNKs are highly enriched in the nervous system, where they play additional roles as essential regulators of morphogenesis during early development (Kuan et al., 1999; Weston et al., 2003; Xia and Karin, 2004). Interestingly, the identity of JNK effectors regulating brain development is not clear. The best-characterized target, c-Jun, appears not to be responsible, as c-Jun–deficient mice show no overt brain abnormality (Hilberg et al., 1993).
In addition to the well-documented proapoptotic roles of JNKs (Kyriakis and Avruch, 2001), JNK1-deficient mice display disrupted anterior commissure formation and loss of axonal microtubule integrity (Chang et al., 2003). Microtubules are highly enriched in the brain, where they are major determinants of cell shape, intracellular transport, and cell polarity. Consistent with this, the list of proposed JNK functions that depend on microtubule structure is growing. We have previously shown that JNK activity regulates neuronal architecture (Coffey et al., 2000; Björkblom et al., 2005), and studies in Caenorhabditis elegans suggest that the JNK pathway may regulate protein targeting (Byrd et al., 2001). Nonetheless, the molecular mechanisms of JNK action on the microtubule cytoskeleton remain elusive and the JNK substrates mediating such functions are not clear. Recently identified candidates include MAP2 and doublecortin (DCX). These proteins are phosphorylated by JNK; however, it has not been established whether they mediate JNK action on microtubules (Chang et al., 2003; Gdalyahu et al., 2004; Björkblom et al., 2005). We therefore took a proteomics approach to identify effector molecules that may explain JNK action on neuronal shape.
Affinity purification identified SCG10 as a major, brain-derived, JNK-interacting protein (JIP). SCG10 belongs to a family of proteins known as stathmins, which includes stathmin, SCG10, SCLIP, RB3, RB3′, and RB3″. Biochemical and structural studies show that stathmin proteins each bind two tubulin heterodimers and negatively regulate microtubule stability in vitro (Horwitz et al., 1997; Charbaut et al., 2001; Ravelli et al., 2004). This function is inhibited upon phosphorylation of up to four residues or by mutations that simulate phosphorylation (Mori and Morii, 2002). Although stathmin is ubiquitously expressed, SCG10, SCLIP, RB3, and RB3′ are neuron-specific proteins. Nonetheless, SCLIP, which displays 70% identity to SCG10, is anomalously up-regulated in a variety of human cancers (Walter-Yohrling et al., 2003). Interestingly, stathmin- deficient mice display axonopathy in the nervous system (Liedtke et al., 2002), as do JNK1 knockouts (Chang et al., 2003). The absence of a more major phenotype in these mice likely reflects functional redundancy among other stathmin family proteins in the nervous system. Drosophila melanogaster, on the other hand, possesses only a single gene encoding phosphoproteins bearing homology to stathmins. Disruption of this gene using RNAi produces serious anomalies in nervous system development, with migration and commissure defects in particular (Ozon et al., 2002).
We identify SCG10 as a neuron-specific in vivo substrate for JNK and demonstrate that S73 phosphorylation of SCG10 is reduced in brains from mice lacking JNK1. Phosphorylation of SCG10 by the cytoplasmic pool of JNK controls its ability to influence axodendritic architecture of cortical neurons. Moreover, by monitoring fluorescent tubulin recovery after photobleaching, we establish that inhibition of JNK significantly reduces microtubule polymerization in cultured neurons. Similarly, expression of the SCG10-AA mutant, which cannot be phosphorylated by JNK, disturbs microtubule dynamics with identical kinetics to JNK inhibition, suggesting that JNK1 activity on SCG10 regulates microtubule plasticity. Consistent with this, in embryonic brain, the spatial distribution of JNK site–phosphorylated SCG10 colocalizes with that of active JNK in regions undergoing intense neuronal differentiation. These results indicate that negative regulation of SCG10 microtubule depolymerizing activity by JNK1 contributes to microtubule homeostasis and axondendritic growth during brain development.
Results
JIPs identified by affinity purification
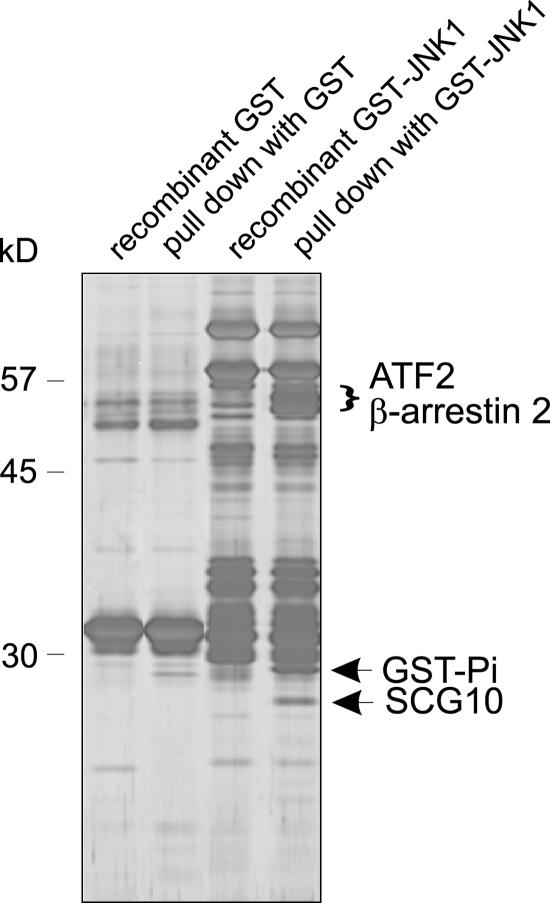
To identify JNK binding partners from brain, we isolated GST-JNK1–interacting proteins from forebrain homogenate. Affinity-purified interacting proteins were separated by SDS-PAGE and visualized by silver staining. Bands that associated with GST-JNK1 and not with GST alone were analyzed by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. The known JIPs GST-Pi, β-arrestin2, and ATF2 (Gupta et al., 1995; Adler et al., 1999; McDonald et al., 2000) were identified as interacting with GST-JNK1 (Fig. 1). The 19-kD JNK1-associating protein was identified as SCG10.
Figure 1.
Novel and known JIPs identified in rat cerebral cortex. To identify novel JIPs, GST-JNK1 or GST bound to Sepharose were incubated with brain extract (pull downs) and associating proteins were visualized by silver staining. Equal proportions of recombinant GST-Sepharose (lane 1), GST-Sepharose pull down (lane 2), GST-JNK1–Sepharose (lane 3), and GST-JNK1–Sepharose pull down (lane 4) were loaded. Lanes 1 and 3 contain fusion proteins that were not incubated with brain extract. Bands from lane 4 that did not correspond with GST-JNK1 or associate with GST were analyzed by mass spectrometry. Identified proteins are indicated.
SCG10, SCLIP, RB3, and RB3′ interact with JNK in intact cells
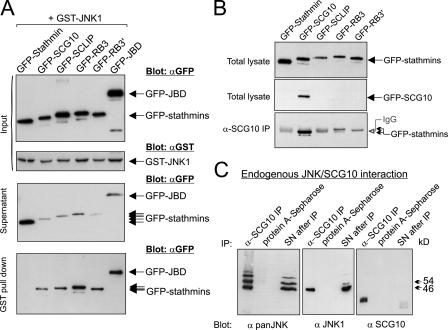
SCG10 belongs to a homologous family of proteins that share a COOH-terminal stathmin domain that is 70–80% conserved and an NH2-terminal sequence that is 45–70% homologous. Given the overall sequence similarity, we anticipated that other stathmin family proteins would interact with JNK. To investigate this, we coexpressed GFP-tagged stathmin, SCG10, SCLIP, RB3, and RB3′ with GST-JNK in COS-7 cells. JIP1 (1–277; JNK binding domain [JBD]), a fragment of JIP1 that shows high-affinity interaction with JNK (Dickens et al., 1997), was used as a positive control. GFP-SCG10, -SCLIP, -RB3, and -RB3′ bound to GST-JNK1 after high salt washing (Fig. 2 A). The extent of interaction with GST-JNK1 was comparable to that of GFP-JBD. Parallel blots of the supernatants after pull down showed a corresponding depletion of JIPs, whereas GFP-stathmin levels remained elevated. Notably, GFP-stathmin, which lacks an NH2-terminal extension, did not bind to GST-JNK1. Given the homology of the stathmin domain in this family, it is likely that the NH2-terminal extension is required for interaction with JNK1.
Figure 2.
Neuron-specific stathmin family proteins interact with JNK in vivo. (A). To examine whether stathmin family proteins interacted with JNK in intact cells, GFP-tagged stathmin, SCG10, SCLIP, RB3, or RB3′ were coexpressed in COS-7 cells with GST-JNK1. GFP-JBD was a positive control for JNK binding. Total lysates (input), supernatants remaining after pull down (supernatant), and GST-JNK1–associating proteins (pull down) were blotted as shown. GFP-SCG10, -SCLIP, -RB3, and -RB3′ interacted with GST-JNK1, whereas GFP-stathmin did not. (B) To test α-SCG10 specificity, COS-7 cell lysates expressing GFP-tagged stathmins were immunoblotted as shown. α-SCG10 only recognized GFP-SCG10 among stathmins. To test α-SCG10 specificity during immunoprecipitation, corresponding fractions were immunoprecipitated with Sepharose bound α-SCG10 and immunoblotted with α-GFP (bottom). (C) To examine whether endogenous SCG10 and JNK interacted in vivo, protein A–Sepharose bound α-SCG10 or protein A–Sepharose alone were incubated with brain extract. Associating proteins or supernatants (SN) after immunoprecipitation (IP) were immunoblotted as shown. JNK1 and additional JNK isoforms copurified with SCG10 from brain.
SCG10 interacts with endogenous JNK in brain
To determine whether SCG10 interacted stably with JNK under physiological conditions, we generated SCG10 antiserum for coimmunoprecipitation assays. Antibody specificity was tested using COS-7 cells expressing GFP-tagged stathmins (Fig. 2 B). Minor cross-reactivity with GFP-RB3 was detectable when used for immunoprecipitation but not for immunoblotting. The diffuse band visible on these immunoblots is antibody heavy chain leached from the Sepharose beads (Fig. 2 B, bottom). Using this serum, we demonstrated coimmunoprecipitation from brain extract of endogenous SCG10 and JNK1, the physiologically active form of JNK in brain (Fig. 2 C; Kuan et al., 2003; Björkblom et al., 2005). Additional JNK isoforms also copurified with SCG10, which is indicated by an extra 54-kD JNK band (Fig. 2 C).
JNK phosphorylates SCG10 with higher specificity than it does stathmin or SCLIP
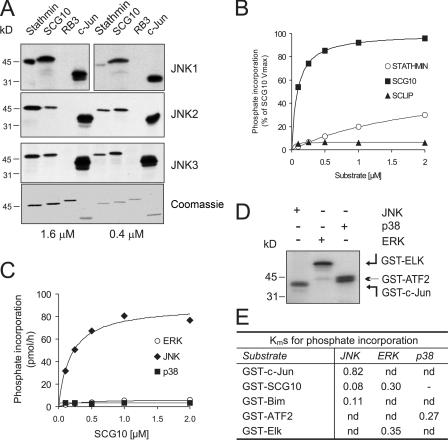
It is notable that potential JNK phosphorylation motifs (SP) exist in stathmin, SCG10, and SCLIP, raising the possibility that these proteins are JNK substrates. It was previously shown that stathmin family proteins are phosphorylated in vitro by MAPKs (Marklund et al., 1993; Parker et al., 1998; Neidhart et al., 2001). However, which MAPK shows the highest activity toward these proteins and whether they phosphorylate SCG10 in vivo was not known. To more closely examine MAPK phosphorylation specificity, we analyzed the kinetics of stathmin family protein phosphorylation (Fig. 3). We first determined JNK isoform–dependent phosphorylation of stathmin family proteins, using the well-characterized JNK substrate c-Jun for comparison. Although GST-stathmin and -SCG10 were similarly phosphorylated by JNKs (1, 2, and 3) at high substrate concentrations (1.6 μM), GST-SCG10 was the preferred JNK1 substrate at lower concentrations (0.4 μM; Fig. 3 A). As expected, GST-RB3 that lacks consensus JNK phosphorylation motifs (SP/TP) was not phosphorylated by JNK. A comparison of JNK phosphorylation kinetics toward GST-stathmin, -SCG10, and -SCLIP revealed that among the stathmin proteins, only GST-SCG10 is a good in vitro substrate for JNK (Fig. 3 B).
Figure 3.
SCG10 is preferentially phosphorylated by JNK, with Km values similar to known JNK substrates. (A) To examine whether stathmins were phosphorylated by JNK, bacterially expressed GST-stathmin, -SCG10, and -RB3 at final concentrations of 1.6 and 0.4 μM were phosphorylated by recombinant active JNKs in vitro. GST–c-Jun(5–89) was used as a control. Representative autoradiographs are shown. The Coomassie-stained gel shows equal protein loading (bottom). (B) A kinetic analysis of phosphate incorporation was performed to examine JNK substrate preference. Quantitative data of JNK-dependent phosphate transfer to GST-tagged SCG10, stathmin, and SCLIP is shown. SCG10 phosphorylation by JNK1 showed substantially higher Vmax and lower Km values than did stathmin and SCLIP. (C) To elucidate which of the MAPKs was the preferred kinase for SCG10, active recombinant JNK1, ERK, and p38 were used for a comparative analysis of SCG10 phosphorylation. (D) An autoradiograph depicting phosphorylation of GST–c-Jun(5–89), -Elk(205–428), and -ATF2(1–109) by preferred kinases JNK, ERK, and p38 is shown. This shows that the activity of kinase preparations used in C were comparable. (E) Km values for MAPK phosphorylation of GST–c-Jun, -SCG10, -Bim, -ATF2, and -Elk were calculated from Lineweaver-Burk plots. The Km value for GST-SCG10 phosphorylation by JNK compares well to those of the better-characterized JNK targets c-Jun and Bim.
The MAPKs ERK and p38 are inefficient kinases for SCG10
Phosphorylation of SCG10 has not been studied as extensively as stathmin, though ERK, p38, and JNK were shown to phosphorylate bacterially expressed SCG10 in vitro (Neidhart et al., 2001). To determine which MAPK showed preferential phosphorylation of SCG10, a kinetic analysis was undertaken (Fig. 3 C). JNK phosphorylated GST-SCG10 with fast kinetics, showing 40-fold higher Vmax than did ERK or p38. Inefficient phosphorylation of GST-SCG10 by ERK and p38 was not due to lower activity of these kinases, as the relative activities of JNK, p38, and ERK toward GST–c-Jun, -ATF2, and -Elk were comparable (Fig. 3 D). We next compared the relative efficiency of GST-SCG10 phosphorylation to known JNK substrates. JNK1 showed higher specificity toward GST-SCG10 than it did toward the known physiological substrates GST–c-Jun and -Bim (Fig. 3 E), indicating that SCG10 is likely to be phosphorylated by JNK in vivo even when concentrations are limiting.
SCG10 is phosphorylated by JNK in vivo
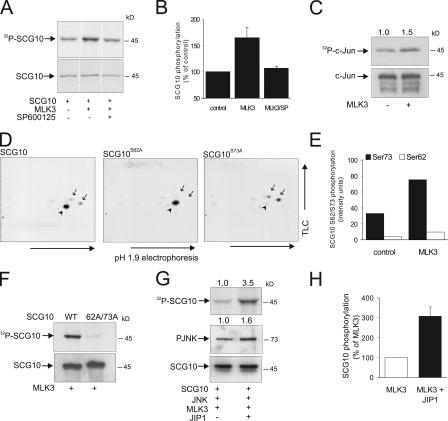
To establish whether SCG10 could be a genuine JNK target in intact cells, COS-7 cells were transfected with GFP-SCG10 in the presence or absence of the JNK cascade activator GFP-MLK3. Activation of JNK resulted in increased phosphorylation of GFP-SCG10. This phosphorylation was prevented by incubation with 3 μM (an effective concentration in intact cells) of the JNK inhibitor SP600125 (Fig. 4, A and B; Cao et al., 2004). The traditional JNK activators UV, anisomycin, and MEKK1 also induced GFP-SCG10 phosphorylation; however, as these stimuli also activated ERK and p38 pathways (not depicted), we did not use them routinely. GFP-SCG10 phosphorylation was moderately induced by coexpression of GFP-MLK3. We postulated that the reason for such a moderate increase resulted from near saturation of GFP-SCG10 phosphorylation because of the presence of GFP-MLK3 before metabolic labeling. We therefore compared the relative phosphorylation of HA–c-Jun in vivo using the same approach (Fig. 4 C). Coexpression of MLK3 resulted in a similar induction of HA–c-Jun phosphorylation (Fig. 4 C). The extent of GFP-SCG10 phosphorylation by JNK in intact cells compared favorably to that of the well-characterized JNK target c-Jun. As kinases and phosphatases are not among the known JNK substrates, these data suggest that SCG10 is a direct target of JNK phosphorylation in vivo.
Figure 4.
SCG10 is phosphorylated by JNK in intact cells. (A) To determine whether SCG10 is a substrate for JNK in vivo, COS-7 cells expressing GFP-SCG10 ± GFP-MLK3 to activate JNK were labeled with 32P ± 3 μM of the JNK inhibitor SP600125. A representative autoradiograph of GFP-SCG10 phosphorylation is shown (top), as is an immunoblot of corresponding samples (bottom). (B) Quantitative data for three sets, mean ± SEM. (C) For comparison, phosphorylation of HA–c-Jun by JNK was examined under identical conditions. An autoradiograph of the resulting gel is shown. (D) COS-7 cells were transfected with GFP-SCG10-WT, GFP-SCG10-S62A, or GFP-SCG10-S73A ± GFP-MLK3. Samples were analyzed by 2D-phosphopeptide mapping. Representative autoradiographs are shown. GFP-SCG10-WT resolved as three phosphopeptide spots (left), GFP-SCG10-S62A as one spot (middle), and GFP-SCG10-S73A as two spots (right; partial cleavage). S62- and S73-phosphorylated peptides are indicated by arrows and arrowheads, respectively. (E) The induction by JNK of SCG10 S62/S73 phosphorylation was measured from two sets of phosphopeptide maps. Means are shown. (F) To evaluate whether JNK phosphorylated GFP-SCG10 on additional sites, GFP-SCG10-WT or a S62A/S73A mutant was coexpressed with GFP-MLK3. SCG10 phosphorylation was measured. A representative autoradiograph (top) and immunoblot (bottom) of immunoprecipitated GFP-SCG10 are shown. (G) To examine whether the JNK scaffold protein JIP1 enhanced SCG10 phosphorylation, COS-7 cells were transfected with GFP-SCG10, -JIP1, -JNK, and -MLK3. A representative autoradiograph is shown (top). Total lysates were immunoblotted with antibodies recognizing PJNK (middle) and GFP (bottom). Reconstitution of the JIP1 scaffolding complex enhanced SCG10 phosphorylation threefold. (H) Means ± range from two datasets are shown.
JNK phosphorylates SCG10 on S62 and S73 in vivo
We identified the JNK phosphorylation sites on SCG10 (S62 and S73) from in vitro–phosphorylated protein using mass spectrometry (unpublished data). To establish whether these sites were phosphorylated by JNK in vivo, GFP-SCG10 wild type (WT), or GFP-SCG10 with candidate phosphorylation sites, S62 and S73 mutated to alanine were expressed in COS-7 cells together with GFP-MLK3. After metabolic labeling, GFP-SCG10 was immunoprecipitated and digested with trypsin. Resulting peptides were separated in two dimensions by thin layer chromatography (Fig. 4 D). GFP-SCG10 phosphopeptides resolved as three spots. By individually mutating S62 and S73, we identified that S73 was more intensely phosphorylated by JNK (Fig. 4, D [middle] and E). The less intensely phosphorylated spots corresponded to a single peptide containing S62 (Fig. 4 D, right). Mutation of both sites resulted in complete loss of GFP-SCG10 phosphorylation, indicating that no additional sites on GFP-SCG10 are phosphorylated by JNK (Fig. 4 F).
Coexpression of JIP1 enhances phosphorylation of SCG10 in vivo
The JIP family of scaffold proteins can facilitate JNK target phosphorylation by recruiting select upstream regulators, including MLK3 (Whitmarsh et al., 2001). To investigate whether SCG10 phosphorylation by JNK was enhanced by reconstitution of the JNK scaffolding machinery, we repeated the earlier experiments with the additional expression of GFP-JNK and -JIP1 (Fig. 4 G). GFP-SCG10 phosphorylation was enhanced threefold by addition of GFP-JIP1 (Fig. 4 H). Together, these results show that GFP-SCG10 phosphorylation by GFP-JNK is amplified but is not dependent on the expression of exogenous GFP-JIP1.
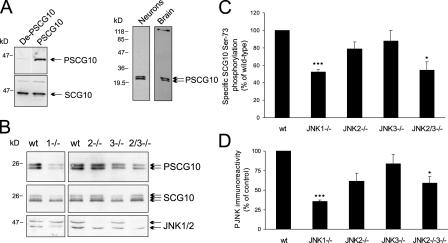
SCG10 S73 phosphorylation is depleted in JNK1−/− cortex
If JNK plays a mandatory role in phosphorylating SCG10 and thereby controls its activity, JNK site phosphorylation of SCG10 should be reduced in JNK-deficient mice. To examine this, we generated antiserum against the major in vivo JNK phosphorylation site on SCG10, S73. After affinity purification, this antibody (PSCG10) recognized only the phosphorylated form of SCG10 (Fig. 5 A [left] and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200511055/DC1) and showed acceptable specificity by immunoblotting (Fig. 5 A, right). To examine the importance of JNK for SCG10 phosphorylation in developing brain, cortex from JNK-deficient mice was immunoblotted with antibodies detecting SCG10 and PSCG10 (Fig. 5 B). JNK site phosphorylation of SCG10 was reduced by ∼50% in brains from JNK1−/− mice (Fig. 5 C) and to a lesser extent in JNK2- and JNK3-deficient mice. A parallel analysis of JNK activity shows that JNK1 is the dominant active form in developing brain (Fig. 5 D).
Figure 5.
SCG10 S73-phosphorylation is decreased in JNK1−/− cortex. (A, left) Antibodies raised against S73-phosphorylated SCG10 (PSCG10) were tested against GFP-SCG10 that was not phosphorylated (DeP-SCG10) or phosphorylated (PSCG10) with active JNK. (right) The specificity of α-PSCG10 toward cortical neurons or brain extract was tested by immunoblotting. (B) Cortex from 7-d JNK-deficient mice (JNK1−/−, JNK2−/−, JNK3−/−, and JNK2−/−3−/−) was immunoblotted with PSCG10, SCG10, and JNK1/2 recognizing antibodies. (C) PSCG10 immunoreactivity was normalized to SCG10 expression and expressed as a percentage of WT tissues. (D) PJNK immunoreactivity was quantified from brain extracts. Means ± SEM are shown for three separate sets of animals (*, P < 0.05; ***, P < 0.005; determined by a t test).
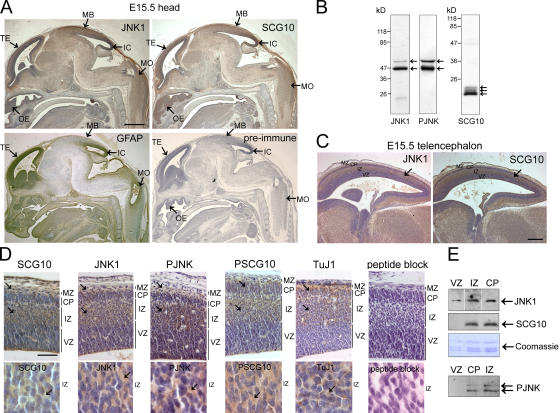
SCG10 and JNK1 site–phosphorylated SCG10 localize to regions of elevated JNK activity in fetal brain
If SCG10 is a genuine JNK target in brain, its regional expression profile should overlap at least partially with that of JNK1, the constitutively active form of JNK in the brain. SCG10 expression is maximal during embryonic development (Stein et al., 1988), although its precise location and function in developing brain was unknown. Examination of SCG10 and JNK1 immunoreactivity in embryonic brain showed a strikingly similar expression pattern that contrasted markedly to that of the glial cell intermediate filament protein (GFAP; Fig. 6 A). JNK1 and SCG10 expression was mid to low throughout the midbrain, with more prominent staining in the telencephalon (developing cortex), the midbrain roof, the olfactory epithelium, the inferior colliculus, and the medulla oblongata (Fig. 6 A; see Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200511055/DC1, for magnified views). Close inspection of the telencephalon revealed concentrated JNK1 and SCG10 staining in the intermediate zone, which has the highest density of postmitotic neurons (Fig. 6, C and D). Similarly, phospho-JNK (PJNK) and JNK1 site–phosphorylated SCG10 localized in the same region as did class III β-tubulin, a marker for early differentiating neurons (Fig. 6 D). In contrast, the ventricular zone, the location of neuronal precursors, showed only an occasional ribbon-like staining pattern for PJNK, JNK1, and SCG10, possibly corresponding to radially migrating early differentiated neurons (Fig. 6 D). These observations were confirmed by microdissection followed by immunoblotting (Fig. 6 E). At the subcellular level, JNK1, PJNK, SCG10, and S73-phosphorylated SCG10 immunoreactivity were cytosolic (Fig. 6 D). This suggests that JNK signaling has functions in the cytoplasm during brain development in addition to the previously described regulation of nuclear events (Weston et al., 2003).
Figure 6.
Expression of JNK1 and SCG10 overlap spatially in developing mouse brain. If SCG10 is a physiological target for JNK, it should localize regionally with active JNK. (A) To test this, sagittal sections of embryonic day (E) 15.5 mouse embryos were immunostained with antibodies recognizing JNK1, SCG10, and GFAP or with preimmune serum for SCG10. Immunoreactivity was detected with 3,3′-diaminobenzedine tetrahydrochloride (brown) and counterstained with hematoxylin (blue). Regions displaying prominent expression of JNK1 and SCG10 are indicated. TE, telencephalon; OE, olfactory epithelium; MB, roof of the midbrain; IC, inferior colliculus; MO, medulla oblongata. Bar, 500 μm. (B) Specificity of the antibodies toward forebrain extract. (C) The telencephalon is divided into the marginal zone (MZ), cortical plate (CP), intermediate zone (IZ), and ventricular zone (VZ) as indicated. Bar, 200 μm. (D) Closer inspection of the telencephalon confirms that SCG10, JNK1, PJNK, PSCG10, and class III β-tubulin (TuJ1) immunoreactivity localizes to the intermediate zone, cortical plate, and marginal zone (arrows) and is predominantly cytosolic (bottom; arrows). Preincubation of α-PSCG10 with phospho-S73 antigenic peptide blocks immunoreactivity. Bar, 50 μm. (E) To verify that the immunoreactivity observed corresponded to the antigens tested, multiple sections of the cortical plate, intermediate zone, and ventricular zone were microdissected and levels of JNK1, PJNK, and SCG10 examined by immunoblotting. Protein loading is indicated by Coomassie staining of histones.
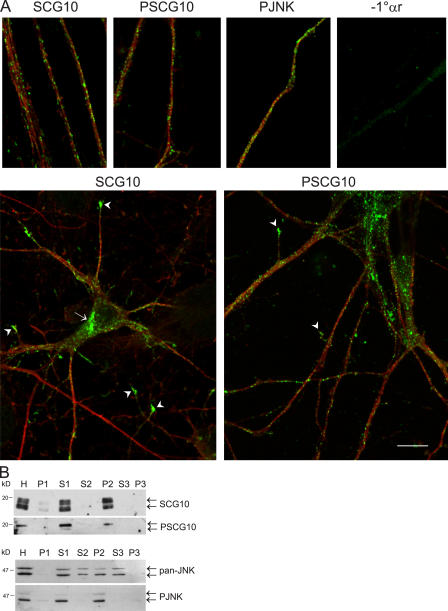
Active JNK and JNK1 site–phosphorylated SCG10 localize to punctate structures on neuronal processes
We previously reported that neuronal JNK1 activity is constitutively elevated and resides in the cytoplasm of neurons differentiating in culture (Coffey et al., 2000; Björkblom et al., 2005). We show that SCG10 and JNK1 associate tightly in vivo and in vitro (Figs. 1 and 2). To examine whether S73-phosphorylated SCG10 localized with active JNK, cortical neurons in culture were stained with PJNK, SCG10, PSCG10, and tubulin antibodies (Fig. 7 A). SCG10 resided in the Golgi apparatus and growth cones but also displayed conspicuous punctate staining in the cytoplasm as previously shown (Lutjens et al., 2000; Gavet et al., 2002). PSCG10 and PJNK showed strikingly similar staining in the neurites and, unlike SCG10, PJNK and PSCG10 immunoreactivity were not elevated at the Golgi compartment. To examine whether S73-phosphorylated SCG10 and PJNK indeed colocalized in vesicles, membrane fractions were prepared from postnatal day 7 brain. Although total brain JNK was present in both soluble and membrane fractions, active JNK, SCG10, and S73-phosphorylated SCG10 concentrated in the P2 fraction, containing predominantly endosomal and Golgi vesicles (Fig. 7 B).
Figure 7.
Active JNK and S73-phosphorylated SCG10 colocalize in vesicular fractions. (A) Confocal sections of cortical neurons at 1 d in vitro immunostained for SCG10, PSCG10, and PJNK (green) and for β-tubulin (red) revealed conspicuously punctate immunoreactivity in the neurites. Corresponding staining in the absence of 1° antibody (−1°αr) is shown. Golgi (arrow) and growth cone–localized SCG10 (arrowheads) are indicated. The specificity of α-PSCG10 for immunostaining is shown in Fig. S1 (available at http://www.jcb.org/cgi/content/full/jcb.200511055/DC1). Bar, 10 μm. (B) To examine whether SCG10 and active JNK colocalized in vesicles, rat forebrain was fractionated. Homogenate (H) was centrifuged at 800 g to isolate intact cells and nuclei (P1). The remaining supernatant (S1) was centrifuged at 16,000 g to yield a pellet containing endosomal and Golgi fractions (P2) and supernatant (S3). The final pellet (P3) contains plasma membranes.
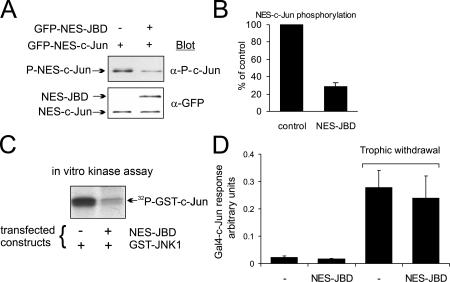
Characterization of a cytosol-specific JNK inhibitor
To test the influence of cytosolic JNK on neuritic architecture without interference from nuclear JNK, we analyzed the efficacy of a cytosol-targeted JNK inhibitor. GFP-NES-JBD expression reduced the phosphorylation of a cytoplasmic JNK target (NES-Jun) in cortical neurons (Fig. 8, A and B) and blocked JNK phosphorylation of recombinant GST–c-Jun (Fig. 8 C). Conversely, GFP-NES-JBD did not inhibit transcriptional activation of GAL4-Jun after withdrawal of trophic support (Fig. 8 D). Together, these data indicate that NES-JBD selectively blocks JNK from phosphorylating cytoplasmic targets.
Figure 8.
NES-JBD selectively blocks cytosolic JNK in cortical neurons. The compartmental specificity of GFP-NES-JBD was tested. (A) 1-d-old cortical neurons were transfected with the JNK inhibitor pEGFP-NES-JBD and pEGFP–NES–c-Jun as shown. Samples were immunoblotted with phospho–S63–c-Jun (P–c-Jun) or GFP antibodies. (B) Quantitative data from A, showing means ± SEM (n = 3). (C) Cortical neurons were transfected with pEBG-JNK1, pEGFP-NES-JBD, and pECFP-SCG10. GST-JNK1 was isolated and kinase activity toward recombinant GST–c-Jun measured. (D) To test whether GFP-NES-JBD interfered with nuclear events, cerebellar granule neurons were transfected with a GAL4-luciferase reporter, GAL4–c-Jun (5–105), Renilla luciferase internal control, and pEGFP-NES-JBD or empty vector (pCMV) as indicated. Trophic withdrawal was for 4 h. Firefly luciferase normalized to Renilla luciferase activity was expressed as arbitrary units. Means ± SEM (n = 3) are shown. GFP-NES-JBD did not inhibit nuclear reporter activity.
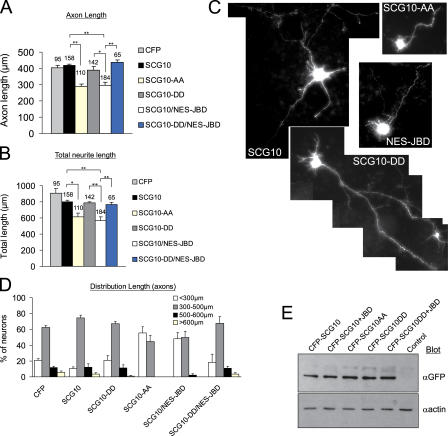
JNK phosphorylation of SCG10 regulates neurite length
Tight regulation of microtubule dynamics is an essential determinant of axonal length. The known function of SCG10 as a microtubule disassembly factor made it a good candidate for mediating JNK regulation of neuronal shape. As active JNK concentrated in cortical regions undergoing differentiation, we examined the effect of JNK on neurite length in cortical neurons expressing CFP-tagged SCG10 variants (Fig. 9). The JNK site–phosphorylation mutants CFP-SCG10-AA (S62A/S73A), which cannot be phosphorylated by JNK, and CFP-SCG10-DD (S62D/S73D), which mimics JNK-phosphorylated SCG10, were used. The expression levels of SCG10 variants did not differ between samples (Fig. 9 E and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200511055/DC1).
Figure 9.
Cytoplasmic JNK controls SCG10 regulation of neurite length. (A) The effect of JNK1 site–phosphorylated SCG10 on neuronal morphology was assessed by transfecting cortical neurons 1 d after plating with pECFP, pECFP-SCG10, pECFP-SCG10-AA, pECFP-SCG10-DD, and pGFP-NES-JBD as indicated. Samples were cotransfected with pEYFP-CAAX as a transfection marker and counterstained with Hoechst 33342. Only viable cells with detectable expression of YFP and CFP were counted. YFP images were analyzed. Axon length refers to the length of the longest cell process. The number of cells analyzed is indicated above the histogram bar. (B) Total neurite length refers to the combined length of processes from a given cell. Means ± SEM from 4–8 experimental sets are shown (*, P < 0.02; **, P < 0.001; determined by paired t test). (C) Representative images of EYFP-CAAX fluorescence. Cells expressing CFP-SCG10-AA or CFP-SCG10 and GFP-NES-JBD show reduced process length, whereas cells expressing CFP-SCG10-DD (mimicking JNK phosphorylation) extend long processes even in the presence of a JNK inhibitor. (D) The distribution of axonal length is shown. (E) To examine the relative expression of CFP-tagged SCG10, SCG10-AA, and SCG10-DD, neurons were transfected as in A–D and lysates were immunoblotted and with GFP antibody.
Mutation of S62/S73 to alanine enhances tubulin depolymerizing activity, whereas mutation to phosphomimicking aspartates inhibits it (Antonsson et al., 1998). Consistent with this, we observed a 25% reduction in axonal length in cortical neurons expressing SCG10-AA for 48 h (Fig. 9, A–D). Under the same conditions, total process length decreased by 30%, indicating that axonal and dendritic compartments were both affected. This is consistent with the proposed axodendritic expression of SCG10 (Gavet et al., 2002). Cells expressing CFP-SCG10-WT grew axons of normal length that did not differ significantly from cells expressing CFP alone. This was as expected because of the elevated JNK activity in cortical neurons (Fig. 5 D; Björkblom et al., 2005), which phosphorylates S62 and S73 (Fig. 5 D), leading to inhibition of its activity (Antonsson et al., 1998). Similarly, cells expressing the functionally inactive, pseudophosphorylated mutant (CFP-SCG10-DD) extended processes of normal length. Notably, inhibition of cytoplasmic JNK using NES-JBD reduced neurite length, similar to CFP-SCG10-AA. These data suggest that endogenous SCG10 may mediate the determination of axodendritic length by JNK. To further test the implication that other JNK effectors were responsible, we expressed CFP-SCG10-DD together with NES-JBD. Inhibition of JNK with NES-JBD failed to influence neurite length in the presence of the pseudophosphorylated CFP-SCG10-DD mutant. This excludes the possibility that other JNK effectors were responsible. The ability of CFP-SCG10-DD to rescue the phenotype induced by NES-JBD suggests that JNK regulation of neurite length is mediated by SCG10.
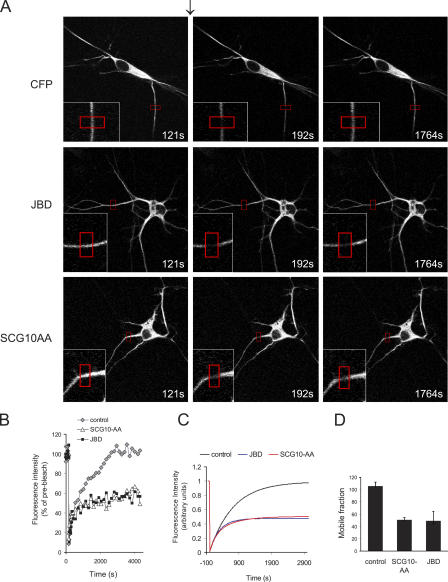
JNK and SCG10 regulate microtubule dynamics in cortical neurons
If JNK negatively regulates SCG10 function, then inhibition of cytosolic JNK or expression of an active SCG10 mutant (CFP-SCG10-AA) should have a similar effect on microtubule dynamics. To test this, we measured microtubule turnover in cortical neurons (Fig. 10). To monitor microtubule dynamics in living neurons, we expressed α-tubulin fused to Venus, a variant of YFP (Nagai et al., 2002). Neurites of cells expressing Venus–α-tubulin were photobleached to the extent that 80% of the initial fluorescence was destroyed. Recovery of Venus–α-tubulin into the bleached region was measured. Coexpression of the JNK inhibitor GFP-NES-JBD reduced Venus–α-tubulin recovery, as did expression of SCG10-AA. Kinetic modeling revealed a 50% decrease in the mobile fraction of Venus–α-tubulin upon JNK inhibition or SCG10-AA expression (Fig. 10, C and D). This is consistent with our proposal that SCG10 mediates the effect of JNK on microtubule dynamics.
Figure 10.
JNK and SCG10-S62/S73 phosphorylation regulate microtubule dynamics. (A) Representative time-lapse images of cortical neurons expressing Venus–α-tubulin, Flag-JBD, and CFP-SCG10-AA as shown before (121 s) directly after (192 s), and 25 min (1764 s) after photobleaching (arrow). The bleached region is highlighted (red). (B) Quantitated fluorescence in the bleached zone, corrected for background and normalized to prebleach intensity, is shown. Means from 3–4 separate experiments are shown. (C) Fitted curves of fluorescence recovery (mean of 3–4 datasets). (D) Mobile fraction of Venus–α-tubulin derived from kinetic plots. Means ± SEM (n = 3–4) are shown.
Discussion
Microtubules are fundamental components of the eukaryotic cytoskeleton. They confer cell shape and polarity and regulate movement, neurite extension, and cell division. They also permit delivery of cargo to and from the cell periphery. Underlying each of these cellular events is the dynamic assembly and disassembly of tubulin (Dent and Gertler, 2003). Microtubule stability is augmented by the binding of microtubule-associated proteins and decreased by destabilizing proteins such as stathmins. Neurite elongation requires a fine balance between microtubule stability and plasticity. Too much or too little stabilization retards neurite extension (Chuckowree and Vickers, 2003). Thus, the activity of destabilizing factors must be tightly regulated to enable extension. The activities of microtubule regulatory proteins are affected by phosphorylation, and inappropriate expression and phosphorylation of these proteins are hallmarks of cancer and neurodegenerative diseases (Walter-Yohrling et al., 2003; Stoothoff and Johnson, 2005). In spite of this knowledge, it remains unclear which are the pivotal protein kinases determining the activities of these microtubule regulators. JNK1 has recently been implicated in regulating microtubule integrity in the nervous system. Thus, mice lacking this kinase display decreased microtubule polymer length, axonal commissure defects (Chang et al., 2003), and disturbed dendritic architecture (Björkblom et al., 2005). These architectural anomalies are believed to be mediated via phosphorylation of the microtubule-stabilizing protein MAP2; however, additional JNK1 effectors have not been excluded.
JNK1 and SCG10 regulate microtubule dynamics in differentiating neurons
We demonstrate that SCG10 is phosphorylated on S62 and S73 by JNK in intact cells. We investigate the functional importance of this phosphorylation on microtubule dynamics in vivo using FRAP. Expression of an SCG10 mutant that cannot be phosphorylated by JNK (CFP-SC10-AA) dramatically inhibits microtubule turnover. Inhibition of the cytoplasmic pool of JNK is as effective in disrupting microtubule dynamics as the phosphorylation site mutant. Both conditions reduce the mobile fraction of fluorescent tubulin by 50%. Interestingly, there is a 50% recovery of Venus–α-tubulin that persists under conditions when JNK is inhibited or SCG10 is active. This recovery, which displays a half-life of 200 s, may be due to slow diffusion of Venus–α-tubulin:β-tubulin dimers. Tubulin has many binding partners within cells, and the soluble pool of fluorescent tubulin may be low, leading to a slower diffusion rate than expected based on size alone.
JNK1 is the predominant active form of JNK in cortical neurons and it resides in the cytoplasm, as we have previously reported in other neuronal types (Coffey et al., 2002; Björkblom et al., 2005). We show that JNK1 phosphorylates S73 of SCG10 in developing brain. However, published data shows that, at high-kinase concentrations, SCG10 is equally phosphorylated by ERK, p38, and JNK3 in vitro (Neidhart et al., 2001). We find that ERK and p38 phosphorylated SCG10 very poorly under conditions that favor kinase substrate specificity and, furthermore, that JNK1 phosphorylates S73 in brain, whereas JNK3 plays only a minor role. Our data suggest that JNK1 and SCG10 cooperate in developing neurons to determine microtubule turnover.
Active JNK stabilizes the neuritic shaft during outgrowth, and SCG10 phosphorylation may be pivotal
Turnover of microtubules occurs along the length of the neurite (Lim et al., 1990). We show that overexpression of a functionally active SCG10 mutant (GFP-SCG10-AA) hinders microtubule recovery in this region. Endogenous SCG10, on the other hand, is abundantly distributed in punctae throughout the neurites in resting neurons, yet microtubule homeostasis is not disturbed. Therefore, SCG10 activity in neurites must be tightly controlled. Consistent with this, we find that in this region, SCG10 is phosphorylated on S73, which is sufficient to inactivate the protein (Antonsson et al., 1998). To extend long axons and dendrites, neurons must exert precise control over the dynamics of microtubules, and inadvertent disassembly along the neurite would impede outgrowth (Dent and Gertler, 2003). We show that JNK can confer this essential stabilizing influence to growing neurites. We propose that JNK1, by phosphorylating SCG10 locally within the neurite, represses inappropriate destabilizing events. Such a role for JNK1 in fine-tuning SCG10 activity in the developing nervous system may be widespread, as active JNK and JNK site–phosphorylated SCG10 localize regionally in embryonic brain.
JNK regulation of SCG10 phosphorylation appears to be confined to distinct subcellular compartments. For instance, there is no prominent PJNK immunoreactivity at the Golgi complex (unpublished data). Similarly, there is a striking absence of concentrated S73-phosphorylated SCG10 immunoreactivity at the Golgi, despite the clear presence of SCG10. This suggests that SCG10 may be active in this region. However, it is also possible that Golgi-localized SCG10 is phosphorylated on a separate site, leading to inactivation. The function of SCG10 at the Golgi is not reported and may require some effort to elucidate. On the contrary, in growth cones, SCG10 is believed to be active, and this activity is required for neurite extension (Suh et al., 2004). In spite of this, we detect moderate levels of inactive S73-phosphorylated SCG10 in growth cones. However, the proportion of phosphorylated SCG10 at the growing tips is low relative to total SCG10, suggesting that the dephosphorylated active form may be enriched here. Consistent with this, PJNK does not concentrate in growth cones (unpublished data). Together, these findings suggest that in contrast to neurites, where we show that JNK maintains microtubule stability, JNK is inactive in areas undergoing dynamic growth. The characteristic triangular shape of growth cone–localized active SCG10 shown in young neurons suggests that it extends into the lamellipodia, where it could contribute to motility by repressing rigid microtubule formation. Consistent with this idea, active stathmin has been shown to concentrate in the highly motile lamellipodia of migrating cells (Niethammer et al., 2004).
JNK1 and SCG10 are likely partners during brain development
Normal brain morphogenesis requires functional JNK at critical events during development, including closure of the neural tube (Kuan et al., 1999), migration of telencephalic neurons (Hirai et al., 2002), and development of the optic fissure and eye lens (Weston et al., 2003). The consequences of interfering with the cytosolic pool of physiologically active JNK are disturbed stability of the microtubule cytoskeleton and stunted neurite growth (Figs. 9 and 10). Yet, the JNK targets mediating these events are unclear. SCG10 is a good candidate for mediating JNK regulation of microtubule homeostasis in embryonic brain. Its expression peaks during embryogenesis (Stein et al., 1988), when we show that JNK1 site–phosphorylated SCG10 colocalizes with active JNK in the intermediate zone of the developing cortex. Neurons in the intermediate zone migrate radially from their place of origin in the ventricular zone, through the layers generated earlier, to form the cortical layers that finally make up the mature cortex. To accomplish this, neurons extend elongated processes that are rich in class III β-tubulin. JNK is implicated in regulating microtubules in the intermediate zone at this developmental stage (Hirai et al., 2002). Similarly, the SCG10 homologue in D. melanogaster (D-stathmin) is associated with neurite elongation during embryogenesis (Ozon et al., 2002). Thus, it is likely that the colocalization of JNK1 with JNK1 site–phosphorylated SCG10 to this region is functionally important.
Alternative mediators of JNK action
Alternative JNK substrates that could contribute to the neuronal shape changes described are MAP2 and DCX. Both proteins function to stabilize microtubules. High molecular weight MAP2 is a likely JNK effector maintaining dendritic architecture in the brain (Björkblom et al., 2005), although it is unlikely to mediate JNK regulation of axonal length in developing cortex, as it is dendrite specific and expressed in mature neurons. DCX, on the other hand, is expressed in embryonic brain and resides, like SCG10, in the intermediate zone of the developing telencephalon (Gdalyahu et al., 2004). However, the impact of JNK on DCX regulation of microtubules is not known. Our model proposes that JNK phosphorylation of SCG10 reduces microtubule depolymerizing activity in the neuritic shaft, where microtubules are more stable (Li and Black, 1996), and is required for neurite elongation. The functional significance of JNK interaction with the neural-enriched stathmin family members SCG10, SCLIP, RB3, and RB3′ is not yet clear. It is unlikely to be significant during early brain development, as expression of SCLIP and RB3 increase postnatally (Mori and Morii, 2002). However, in adult RB3, expression like JNK activity is induced after seizures in the hippocampus (Beilharz et al., 1998; Mielke et al., 1999), where JNK activation precedes RB3 induction. Whether these proteins cooperate in functional plasticity changes during learning and memory remains to be seen.
The JNK family of protein kinases is a central control point of cell fate in disease and physiological situations. It is only relatively recently that researchers have realized that they may also have additional physiological functions in the central nervous system (Coffey et al., 2000; Byrd et al., 2001; Chang et al., 2003; Weston et al., 2003; Björkblom et al., 2005), indicating that we must reach an understanding of precisely how JNKs participate in disease mechanisms on the one hand and homeostatic, developmental, and regenerative mechanisms on the other. Key to this understanding is a detailed knowledge of JNK effectors mediating physiological responses in the brain. We identify SCG10, a microtubule depolymerizing factor, as an in vivo substrate for JNK on sites that regulate its destabilizing activity. Our data indicate that phosphorylation of SCG10 by JNK can play an unequivocal role in maintaining neuronal microtubule stability and efficient neurite elongation.
Materials and methods
Plasmid construction
Rat stathmins and α-tubulin were isolated by PCR from rat cerebellar granule neuron cDNA and inserted into pEGFP-C1, pECFP-C1, pEYFP-C1 (CLONTECH Laboratories, Inc.), Venus-encoding vector (a gift from A. Miyawaki, Institute of Physical and Chemical Research, Saitama, Japan), pEBG (a gift from B. Mayer, Children's Hospital, Boston, MA), and pGEX-KG (a gift from J. Kyriakis, Massachusetts General Hospital, Boston, MA) vectors as described previously (Coffey et al., 2002). ECFP-tagged SCG10 phosphorylation site mutants S73A, S62A, S62A/S73A, S62D, S73D, and S62D/S73D were prepared by insertional overlapping PCR using mutagenic and flanking primers. The pEBG-JNK1, JNK2, JNK3, pEGFP-MEKK1Δ, pcDNA3-GAL4-Jun(5–105), and pGL3-G5E4▵38 constructs were described previously (Coffey et al., 2000, 2002; Hongisto et al., 2003). pEGFP-JIP-JBD, NES–c-Jun(1–146), and MLK3 were constructed by PCR-based methods from pcDNA3-mJIP1a, pMT108, and HeLa cDNA, respectively. c-Jun(5–89), JNK1, ATF2(1–109), and Elk1(205–428) in pGEX, ERK, and p38 in pEBG, pEF-EE-Bim-EL, and pCMV were gifts from J. Woodgett (Ontario Cancer Institute, Toronto, Canada), R. Davis (Howard Hughes Medical Institute, Worcester, MA), A. Sharrocks (University of Manchester, Manchester, UK), B. Mayer, D. Huang (The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia), and S. van den Heuvel (Massachusetts General Hospital, Boston, MA), respectively. The coding sequence for Bim was cut from pEF-EE-Bim-EL and inserted into pGEX-KG.
Recombinant protein preparation
Recombinant c-Jun, ATF2, Elk, Bim, and stathmins were prepared from bacterial extracts as described previously (Courtney and Coffey, 1999; Coffey et al., 2000). Recombinant active JNK1, JNK2, JNK3, p38, and ERK were prepared as described previously (Hongisto et al., 2003).
Affinity purification and mass spectrometry
For affinity purification of JIPs, postnatal day 7 rat cortex was homogenized in lysis buffer (20 mM Hepes, pH 7.4; 2 mM EGTA; 50 mM β-glycerophosphate; 1 mM DTT; 1 mM Na3VO4; 1% Triton X-100; 10% glycerol; 50 mM NaF; 1 mM benzamidine; 1 μg/ml aprotinin, leupeptin, and pepstatin; and 100 μg/ml PMSF) with an ultra-turrax (IKA) and a 27-gauge needle and centrifuged for 15 min at 10,000 g at 4°C to remove insoluble material. 10 mg/ml of extract was incubated with 100 μg of GST coupled to glutathione–Sepharose for 1 h at 4°C. Precleared extract was rotated overnight at 4°C with 100 μg of GST-JNK1 conjugated to glutathione–Sepharose (Sigma-Aldrich). Immobilized proteins were washed in 2× lysis buffer, 3× LiCl buffer (100 mM Tris, pH 7.6, 0.5 M LiCl, 0.1% Triton X-100, and 1 mM DTT), and 2× lysis buffer. Equal proportions were separated by SDS-PAGE and visualized by silver staining. For identification of JNK1-interacting proteins, in-gel digestion was performed as described previously (Björkblom et al., 2005) and analyzed by matrix-assisted laser-desorption/ionization time-of-flight using a Voyager DE PRO. Peptide mass fingerprints were analyzed with Mascot (Matrix Science).
Antibody preparation
Polyclonal antibodies were raised against bacterially expressed GST-SCG10 and against the phosphopeptide EAPRTLAS(PO3)PKKKDLSLEE. Phosphospecific antibodies were affinity purified sequentially on phospho- and dephosphopeptide columns.
Protein phosphorylation analysis
Transfected COS-7 cells were serum starved overnight (0.1% FCS) and incubated in phosphate-free Eagle's minimum essential medium (Sigma-Aldrich) for 2 h. ATP pools were isotopically labeled by incubating cells with 0.5 mCi/ml [32P]phosphate for 2 h at 37°C and 5% CO2. Cells were washed with PBS, lysed in lysis buffer, and centrifuged at 13,000 rpm and 4°C for 10 min. Supernatants were immunoprecipitated using cross-linked SCG10 antiserum (5 μl) or with anti-HA (5 μl; Santa Cruz Biotechnology, Inc.). Phosphorylated proteins were visualized by autoradiography. Phospho- SCG10 was excised from the gel and trypsinized overnight at 37°C. Tryptic digests were separated by 2D-TLC with electrophoresis, pH 1.9, followed by ascending chromatography, pH 3.5. TLC-separated phosphopeptides were quantified by phosphorimaging. For in vitro analysis, active recombinant JNKs were used to phosphorylate bacterially purified GST fusion proteins using γ-[32P]ATP as described previously (Coffey et al., 2000; Hongisto et al., 2003). Kinetic analyses were performed as described previously (Björkblom et al., 2005).
Cell culture, transfection, and morphological analysis
Cortical neuron cultures were prepared as described previously (Björkblom et al., 2005). Cells for morphological analysis were plated at a density of 0.5 × 106. 24 h after plating, cells were transfected using Lipofectamine 2000 (Invitrogen) with 25% of total DNA as pEYFP-CAAX, 25% pECFP-C1, or ECFP-tagged SCG10 WT, S62A/S73A, or 62D/S73D and 50% pEGFP-NES-JBD as indicated. Cells were fixed 24 h later, and fluorescence was examined with a microscope (Axiovert 200; Carl Zeiss MicroImaging, Inc.) using a 20× objective. Digitized images (∼15 cells per coverslip) of EYFP fluorescence were acquired using a camera (ORCA II ERG; Hamamatsu) and Wasabi software (Hamamatsu). Overlapping images were taken where necessary to encompass the entire process length. Neurite length was measured from size-calibrated images using MetaMorph 6.1 imaging software (Universal Imaging Corp.).
COS-7 cells were transfected using Lipofectamine. For analysis of protein–protein interactions, COS-7 cells were transfected as follows: 50% of total DNA as EGFP-tagged stathmin, SCG10, SCLIP, RB3, or RB3′ together with 50% of pEBG-JNK. Cells were lysed 48 h after transfection in lysis buffer. Precleared supernatants were incubated with S-hexylglutathione–Sepharose at 4°C. Pull downs were washed (2× lysis buffer, 3 × LiCl wash, and 1× lysis buffer), suspended in laemmli buffer, and immunoblotted.
Online supplemental material
Fig. S1 shows characterization of PSCG10 antibody specificity for immunostaining. Fig. S2 shows magnified views of JNK1, SCG10, and GFAP staining in E15 embryo. Fig. S3 shows composite images of YFP-CAAX/CFP-SCG10 mutant expression in cortical neurons. The supplementary text describes reporter gene expression analysis, immunohistochemistry, immunofluorescence, subcellular fractionation, and photobleaching procedures. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200511055/DC1.
Supplementary Material
Acknowledgments
We thank the Turku Centre for Biotechnology Cell Imaging Core and Proteomics Unit for use of facilities.
This work was funded by the Academy of Finland (grants 206497, 49949, and 47536 to E.T. Coffey and 203520 to M.J. Courtney), Åbo Akademi University, the Turku Graduate School of Biomedical Sciences, the Finnish Graduate School of Neuroscience, the National Graduate School in Information and Structural Biology, the Drug Discovery Graduate School, and the Centre for International Mobility (grant TM-02-886).
N. Östman and W. Li contributed equally to this paper.
Abbreviations used in this paper: DCX, doublecortin; GFAP, glial cell intermediate filament protein; JBD, JNK binding domain; JIP, JNK-interacting protein; PJNK, phospho-JNK; WT, wild type.
References
- Adler, V., Z. Yin, S.Y. Fuchs, M. Benezra, L. Rosario, K.D. Tew, M.R. Pincus, M. Sardana, C.J. Henderson, C.R. Wolf, et al. 1999. Regulation of JNK signaling by GSTp. EMBO J. 18:1321–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson, B., D.B. Kassel, G. Di Paolo, R. Lutjens, B.M. Riederer, and G. Grenningloh. 1998. Identification of in vitro phosphorylation sites in the growth cone protein SCG10. Effect of phosphorylation site mutants on microtubule-destabilizing activity. J. Biol. Chem. 273:8439–8446. [DOI] [PubMed] [Google Scholar]
- Beilharz, E.J., E. Zhukovsky, A.A. Lanahan, P.F. Worley, K. Nikolich, and L.J. Goodman. 1998. Neuronal activity induction of the stathmin-like gene RB3 in the rat hippocampus: possible role in neuronal plasticity. J. Neurosci. 18:9780–9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkblom, B., N. Östman, V. Hongisto, V. Komarovski, J. Filen, T. Nyman, T. Kallunki, M. Courtney, and E.T. Coffey. 2005. Constitutively active cytoplasmic c-Jun N-terminal kinase 1 is a dominant regulator of dendritic architecture: role of microtubule-associated protein 2 as an effector. J. Neurosci. 25:6350–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, D.T., M. Kawasaki, M. Walcoff, N. Hisamoto, K. Matsumoto, and Y. Jin. 2001. UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron. 32:787–800. [DOI] [PubMed] [Google Scholar]
- Cao, J., M.M. Semenova, V.T. Solovyan, J. Han, E.T. Coffey, and M.J. Courtney. 2004. Distinct requirements for p38-alpha and JNK stress-activated protein kinases in different forms of apoptotic neuronal death. J. Biol. Chem. 279:35903–35913. [DOI] [PubMed] [Google Scholar]
- Chang, L., Y. Jones, M.H. Ellisman, L.S. Goldstein, and M. Karin. 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell. 4:521–533. [DOI] [PubMed] [Google Scholar]
- Charbaut, E., P.A. Curmi, S. Ozon, S. Lachkar, V. Redeker, and A. Sobel. 2001. Stathmin family proteins display specific molecular and tubulin binding properties. J. Biol. Chem. 276:16146–16154. [DOI] [PubMed] [Google Scholar]
- Chuckowree, J.A., and J.C. Vickers. 2003. Cytoskeletal and morphological alterations underlying axonal sprouting after localized transection of cortical neuron axons in vitro. J. Neurosci. 23:3715–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey, E.T., V. Hongisto, M. Dickens, R.J. Davis, and M.J. Courtney. 2000. Dual roles for c-Jun N-terminal kinase in developmental and stress responses in cerebellar granule neurons. J. Neurosci. 20:7602–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey, E.T., G. Smiciene, V. Hongisto, J. Cao, S. Brecht, T. Herdegen, and M.J. Courtney. 2002. c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J. Neurosci. 22:4335–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney, M.J., and E.T. Coffey. 1999. The mechanism of Ara-C-induced apoptosis of differentiating cerebellar granule neurons. Eur. J. Neurosci. 11:1073–1084. [DOI] [PubMed] [Google Scholar]
- Dent, E.W., and F.B. Gertler. 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 40:209–227. [DOI] [PubMed] [Google Scholar]
- Dickens, M., J.S. Rogers, J. Cavanagh, A. Raitano, Z. Xia, J.R. Halpern, M.E. Greenberg, C.L. Sawyers, and R.J. Davis. 1997. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 277:693–696. [DOI] [PubMed] [Google Scholar]
- Gavet, O., S. El Messari, S. Ozon, and A. Sobel. 2002. Regulation and subcellular localization of the microtubule-destabilizing stathmin family phosphoproteins in cortical neurons. J. Neurosci. Res. 68:535–550. [DOI] [PubMed] [Google Scholar]
- Gdalyahu, A., I. Ghosh, T. Levy, T. Sapir, S. Sapoznik, Y. Fishler, D. Azoulai, and O. Reiner. 2004. DCX, a new mediator of the JNK pathway. EMBO J. 23:823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., D. Campbell, B. Derijard, and R.J. Davis. 1995. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 267:389–393. [DOI] [PubMed] [Google Scholar]
- Hilberg, F., A. Aguzzi, N. Howells, and E.F. Wagner. 1993. c-Jun is essential for normal mouse development and hepatogenesis. Nature. 365:179–181. [DOI] [PubMed] [Google Scholar]
- Hirai, S., A. Kawaguchi, R. Hirasawa, M. Baba, T. Ohnishi, and S. Ohno. 2002. MAPK-upstream protein kinase (MUK) regulates the radial migration of immature neurons in telencephalon of mouse embryo. Development. 129:4483–4495. [DOI] [PubMed] [Google Scholar]
- Hongisto, V., N. Smeds, S. Brecht, T. Herdegen, M.J. Courtney, and E.T. Coffey. 2003. Lithium blocks the c-Jun stress response and protects neurons via its action on glycogen synthase kinase 3. Mol. Cell. Biol. 23:6027–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz, S.B., H.J. Shen, L. He, P. Dittmar, R. Neef, J. Chen, and U.K. Schubart. 1997. The microtubule-destabilizing activity of metablastin (p19) is controlled by phosphorylation. J. Biol. Chem. 272:8129–8132. [DOI] [PubMed] [Google Scholar]
- Kuan, C.Y., D.D. Yang, D.R. Samanta Roy, R.J. Davis, P. Rakic, and R.A. Flavell. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 22:667–676. [DOI] [PubMed] [Google Scholar]
- Kuan, C.Y., A.J. Whitmarsh, D.D. Yang, G. Liao, A.J. Schloemer, C. Dong, J. Bao, K.J. Banasiak, G.G. Haddad, R.A. Flavell, et al. 2003. A critical role of neural-specific JNK3 for ischemic apoptosis. Proc. Natl. Acad. Sci. USA. 100:15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis, J.M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807–869. [DOI] [PubMed] [Google Scholar]
- Li, Y., and M.M. Black. 1996. Microtubule assembly and turnover in growing axons. J. Neurosci. 16:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke, W., E.E. Leman, R.E. Fyffe, C.S. Raine, and U.K. Schubart. 2002. Stathmin-deficient mice develop an age-dependent axonopathy of the central and peripheral nervous systems. Am. J. Pathol. 160:469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S., K.J. Edson, P.C. Letourneau, and G.G. Borisy. 1990. A test of microtubule translocation during neurite elongation. J. Cell Biol. 111:123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutjens, R., M. Igarashi, V. Pellier, H. Blasey, G. Di Paolo, E. Ruchti, C. Pfulg, J.K. Staple, S. Catsicas, and G. Grenningloh. 2000. Localization and targeting of SCG10 to the trans-Golgi apparatus and growth cone vesicles. Eur. J. Neurosci. 12:2224–2234. [DOI] [PubMed] [Google Scholar]
- Marklund, U., G. Brattsand, V. Shingler, and M. Gullberg. 1993. Serine 25 of oncoprotein 18 is a major cytosolic target for the mitogen-activated protein kinase. J. Biol. Chem. 268:15039–15047. [PubMed] [Google Scholar]
- McDonald, P.H., C.W. Chow, W.E. Miller, S.A. Laporte, M.E. Field, F.T. Lin, R.J. Davis, and R.J. Lefkowitz. 2000. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 290:1574–1577. [DOI] [PubMed] [Google Scholar]
- Mielke, K., S. Brecht, A. Dorst, and T. Herdegen. 1999. Activity and expression of JNK1, p38 and ERK kinases, c-Jun N-terminal phosphorylation, and c-jun promoter binding in the adult rat brain following kainate-induced seizures. Neuroscience. 91:471–483. [DOI] [PubMed] [Google Scholar]
- Mori, N., and H. Morii. 2002. SCG10-related neuronal growth-associated proteins in neural development, plasticity, degeneration, and aging. J. Neurosci. Res. 70:264–273. [DOI] [PubMed] [Google Scholar]
- Nagai, T., K. Ibata, E.S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90. [DOI] [PubMed] [Google Scholar]
- Neidhart, S., B. Antonsson, C. Gillieron, F. Vilbois, G. Grenningloh, and S. Arkinstall. 2001. c-Jun N-terminal kinase-3 (JNK3)/stress-activated protein kinase-beta (SAPKbeta) binds and phosphorylates the neuronal microtubule regulator SCG10. FEBS Lett. 508:259–264. [DOI] [PubMed] [Google Scholar]
- Niethammer, P., P. Bastiaens, and E. Karsenti. 2004. Stathmin-tubulin interaction gradients in motile and mitotic cells. Science. 303:1862–1866. [DOI] [PubMed] [Google Scholar]
- Ozon, S., A. Guichet, O. Gavet, S. Roth, and A. Sobel. 2002. Drosophila stathmin: a microtubule-destabilizing factor involved in nervous system formation. Mol. Biol. Cell. 13:698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, C.G., J. Hunt, K. Diener, M. McGinley, B. Soriano, G.A. Keesler, J. Bray, Z. Yao, X.S. Wang, T. Kohno, and H.S. Lichenstein. 1998. Identification of stathmin as a novel substrate for p38 delta. Biochem. Biophys. Res. Commun. 249:791–796. [DOI] [PubMed] [Google Scholar]
- Ravelli, R.B., B. Gigant, P.A. Curmi, I. Jourdain, S. Lachkar, A. Sobel, and M. Knossow. 2004. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 428:198–202. [DOI] [PubMed] [Google Scholar]
- Stein, R., N. Mori, K. Matthews, L.-C. Lo, and D.J. Anderson. 1988. The NGF-inducible SCG10 mRNA encodes a novel membrane-bound protein present in growth cones and abundant in developing neurons. Neuron. 1:463–476. [DOI] [PubMed] [Google Scholar]
- Stoothoff, W.H., and G.V. Johnson. 2005. Tau phosphorylation: physiological and pathological consequences. Biochim. Biophys. Acta. 1739:280–297. [DOI] [PubMed] [Google Scholar]
- Suh, L.H., S.F. Oster, S.S. Soehrman, G. Grenningloh, and D.W. Sretavan. 2004. L1/Laminin modulation of growth cone response to EphB triggers growth pauses and regulates the microtubule destabilizing protein SCG10. J. Neurosci. 24:1976–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter-Yohrling, J., X. Cao, M. Callahan, W. Weber, S. Morgenbesser, S.L. Madden, C. Wang, and B.A. Teicher. 2003. Identification of genes expressed in malignant cells that promote invasion. Cancer Res. 63:8939–8947. [PubMed] [Google Scholar]
- Weston, C.R., A. Wong, J.P. Hall, M.E. Goad, R.A. Flavell, and R.J. Davis. 2003. JNK initiates a cytokine cascade that causes Pax2 expression and closure of the optic fissure. Genes Dev. 17:1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh, A.J., C.Y. Kuan, N.J. Kennedy, N. Kelkar, T.F. Haydar, J.P. Mordes, M. Appel, A.A. Rossini, S.N. Jones, R.A. Flavell, et al. 2001. Requirement of the JIP1 scaffold protein for stress-induced JNK activation. Genes Dev. 15:2421–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., and M. Karin. 2004. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 14:94–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.