Abstract
The nuclear pore complex (NPC) is a large channel that spans the two lipid bilayers of the nuclear envelope and mediates transport events between the cytoplasm and the nucleus. Only a few NPC components are transmembrane proteins, and the role of these proteins in NPC function and assembly remains poorly understood. We investigate the function of the three integral membrane nucleoporins, which are Ndc1p, Pom152p, and Pom34p, in NPC assembly and transport in Saccharomyces cerevisiae. We find that Ndc1p is important for the correct localization of nuclear transport cargoes and of components of the NPC. However, the role of Ndc1p in NPC assembly is partially redundant with Pom152p, as cells lacking both of these proteins show enhanced NPC disruption. Electron microscopy studies reveal that the absence of Ndc1p and Pom152p results in aberrant pores that have enlarged diameters and lack proteinaceous material, leading to an increased diffusion between the cytoplasm and the nucleus.
Introduction
All transport events between the cytoplasm and the nucleus occur through a large channel in the nuclear envelope called the nuclear pore complex (NPC). Very little is known about the mechanism of NPC biosynthesis, particularly how the proteins composing this complex are assembled, inserted, or anchored into the nuclear envelope. In metazoans, there are at least two NPC assembly pathways. The first assembly pathway occurs upon the completion of mitosis, when membrane vesicles and nucleoporins (Nups) are recruited to chromatin during nuclear envelope reformation (Rabut et al., 2004). The second pathway occurs in interphase, during which time NPCs are continually synthesized and inserted into intact nuclear envelopes (Maul et al., 1972; Winey et al., 1997). Although the targeting and assembly of membranes and Nups to postmitotic nuclei is somewhat understood, little is known about NPC biogenesis in interphase cells. Many unicellular eukaryotes, such as the yeast Saccharomyces cerevisiae, rely exclusively on NPC insertion into intact double membranes because they undergo a closed mitosis with no nuclear envelope breakdown.
Of the 33 Nups in the yeast S. cerevisiae, three are pore membrane proteins (POMs), and two integral membrane Nups have been described in metazoans. Because of their membrane association, the POMs are attractive candidates for performing functions such as targeting soluble Nup subcomplexes to the nuclear envelope or inserting the NPC into the membrane.
Several studies have been performed on the metazoan transmembrane Nups GP210 and POM121, although results pertaining to their function in NPC assembly are somewhat conflicting. For example, one study in Xenopus laevis extracts found that the inhibition of GP210 by antibody addition or the overexpression of its cytoplasmic tail domain resulted in distorted nuclear envelopes lacking NPCs (Drummond and Wilson, 2002), and another study in the nematode Caenorhabditis elegans found a similar nuclear envelope defect when GP210 was depleted by RNAi (Cohen et al., 2003). More recently (Antonin et al., 2005), both GP210 and POM121 were depleted from HeLa cells and X. laevis extracts by siRNA and immunodepletion, respectively. It was found that when POM121 was depleted postmitotic nuclear envelope reformation was inhibited, and the localization of other Nups in the nuclear envelope during interphase was reduced (Antonin et al., 2005). However, no defects were seen after the depletion of GP210 from cells (Eriksson et al., 2004; Antonin et al., 2005).
Of the three POMs in budding yeast, POM34 and POM152 are not essential for cell viability, whereas NDC1 is an essential gene. Interestingly, Ndc1p localizes to both the NPC and the spindle pole body (SPB), which is comprised of two large protein complexes that span both bilayers of the nuclear envelope (Chial et al., 1998). Most of the work on Ndc1p has focused on its role at the SPB, where it has been shown to be required for duplication of the inner plaque of that structure (Winey et al., 1993). Recently, a conditional allele of NDC1 was shown to prevent the incorporation of newly synthesized Nup49p into the NPC (Lau et al., 2004). However, this mutant did not affect the steady-state localization of Nup49p or nuclear transport.
To examine the role of transmembrane pore proteins in interphase NPC assembly and function in yeast, we depleted Ndc1p, Pom34p, and Pom152p, either alone or in combination. We found that the depletion of Ndc1p led to a partial mislocalization of NPC components and that this phenotype was exacerbated when Pom152p was also absent. However, lack of Ndc1p alone was sufficient to perturb NPC function and to cause mislocalization of nuclear import reporters. This suggests that Ndc1p is required both for efficient nuclear transport and for NPC assembly, although Pom152p can partially compensate for the loss of Ndc1p in NPC assembly. Interestingly, transmission electron microscopy (TEM) images demonstrate that the nuclear envelopes of cells lacking both Ndc1p and Pom152p contain pores with larger diameters than wild-type NPCs and without apparent protein material. In accordance with this, these POM-deficient cells also allowed passive diffusion of proteins that are normally above the exclusion limit of the NPC. These results suggest that the POMs Ndc1p and Pom152p are important for targeting soluble Nups to the nuclear envelope.
Results
Depletion of Ndc1p leads to a cell division arrest
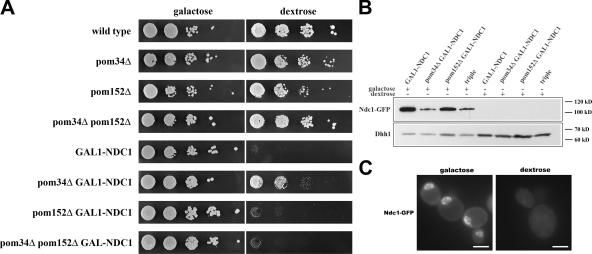
To examine the functions of the three yeast POMs, Ndc1p, Pom34p, and Pom152p, we created strains in which one, two, or all three of the POMs were depleted. Cells with POM34, POM152, or both genes deleted at their genomic loci grew indistinguishably from wild type on either dextrose or galactose plates (Fig. 1 A, top four rows). Because NDC1 is essential for cell viability (Winey et al., 1993), it was depleted from cells using a transcriptional shutoff method. The NDC1 gene was placed under the control of the GAL1 promoter at its endogenous genomic locus (GAL1-NDC1). Thus, NDC1 expression can be induced in the presence of galactose and repressed in the presence of dextrose. To monitor the expression and localization of Ndc1p, GAL1-NDC1 was also tagged with GFP at the COOH terminus. As expected, strains containing GAL1-NDC1 alone or in combination with pom34Δ, pom152Δ, or pom34Δ pom152Δ were inviable when grown on the repressive dextrose plates, but grew like wild type on galactose plates (Fig. 1 A, bottom four rows). Curiously, the pom34Δ GAL1-NDC1 strain consistently displayed slightly increased growth on dextrose plates compared with the other GAL1-NDC1 strains (Fig. 1 A, pom34Δ GAL1-NDC1).
Figure 1.
Ndc1p can be effectively depleted from yeast cells. (A) Wild-type cells and strains lacking one, two, or three POMs were grown overnight in galactose, and then plated in 10-fold serial dilutions onto galactose or dextrose plates, starting with OD600 = 0.5. All strains were viable on galactose, whereas all four GAL1-NDC1 strains did not grow on dextrose plates. pom34Δ partially suppressed the GAL1-NDC1 growth defect. (B) The four GAL1-NDC1 strains were grown overnight in galactose media and transferred to dextrose media for 24 h of growth. Ndc1p-GFP protein levels were analyzed by Western blotting against the GFP tag. Western blotting against a cytoplasmic protein, Dhh1p, was used as a loading control. Strains grown in dextrose had >95% of their Ndc1p-GFP depleted. (C) GAL1-NDC1 cells were grown overnight in galactose media, transferred to dextrose media, and grown for 24 h. The cellular distribution of Ndc1p-GFP was visualized under both galactose and dextrose growth conditions. Images were taken with identical exposure times. Bars, 5 μm.
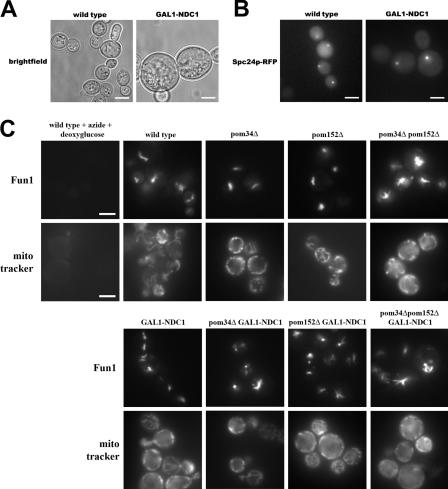
To determine the extent and time course of Ndc1p depletion, we monitored Ndc1p expression in GAL1-NDC1 cells upon the shift to dextrose-containing media. During growth in galactose-containing media, all GAL1-NDC1 strains expressed high levels of Ndc1p-GFP, but >95% of the Ndc1p-GFP had been depleted after 24 h of growth in liquid dextrose media, as determined by Western blotting (Fig. 1 B). We also examined cells from each of the GAL1-NDC1 strains by fluorescence microscopy and observed that most of the cells lacked a detectable Ndc1p-GFP signal after 24 h of growth in dextrose (Fig. 1 C). Depletion of Ndc1p coincided with an increase in cell size (Fig. 2 A), and the GAL1-NDC1 strains arrested as large-budded cells with only one intact SPB, which is illustrated by one bright spot labeling the SPB component Spc24p-RFP (Fig. 2 B). In contrast, wild-type cells with large buds had two spots of Spc24p-RFP signal (Fig. 2 B), confirming that the function of Ndc1p in SPB duplication had been perturbed (Le Masson et al., 2002).
Figure 2.
Ndc1p depletion causes a cell cycle arrest. (A) Wild-type and GAL1-NDC1 cells were grown for 24 h in dextrose media. GAL1-NDC1 cells were enlarged compared with wild-type cells. This phenotype was also seen in the other three GAL1-NDC1 strains (not depicted). (B) Wild-type and GAL1-NDC1 cells expressing the inner SPB component Spc24p-RFP were grown for 24 h in dextrose. In the wild-type strain, large-budded cells had a bright spot of Spc24p-RFP signal in both the mother and the daughter cells, whereas in GAL1-NDC1 only the mother cell had a bright spot. (C) Wild-type cells and strains lacking one, two, or three of the POMs were stained for 30 min with the vital dyes Fun1 (top) or MitoTracker (bottom). Cells were then visualized by fluorescence microscopy. As a control, wild-type cells were treated with NaN3 and 2-deoxyglucose for 30 min to block respiration (top left). Bars, 5 μm.
Because NDC1 is essential, transcriptional shutoff of the gene should inhibit cell division (Fig. 1) and, ultimately, cause cell death. To determine whether the cells in our experiments were still viable after being depleted of Ndc1p for 24 h in dextrose, we stained each strain with two different vital dyes. Both Fun1 and MitoTracker dyes have been shown to stain only cells that are metabolically active (Poot et al., 1996; Millard et al., 1997). Fun1 is actively taken up into live-cell vacuoles and stains intravacuole tubules, whereas MitoTracker is incorporated into actively respiring mitochondria. In each of our strains, both dyes were incorporated into cells, indicating that they remain metabolically active (Fig. 2 C). As a control, neither dye was incorporated into cells treated with sodium azide and 2-deoxyglucose, which stop cellular respiration (Fig. 2 C, left).
Based on these experiments, we conclude that Ndc1p can be effectively depleted from yeast cells either alone or in combination with complete deletions of POM34 and POM152. Furthermore, 24 h in dextrose is sufficient to effectively deplete Ndc1p and induce a cell cycle arrest, although cells remain metabolically active for at least an additional 24 h in the absence of Ndc1p (unpublished data).
Cells lacking both Ndc1p and Pom152p have severe NPC localization defects
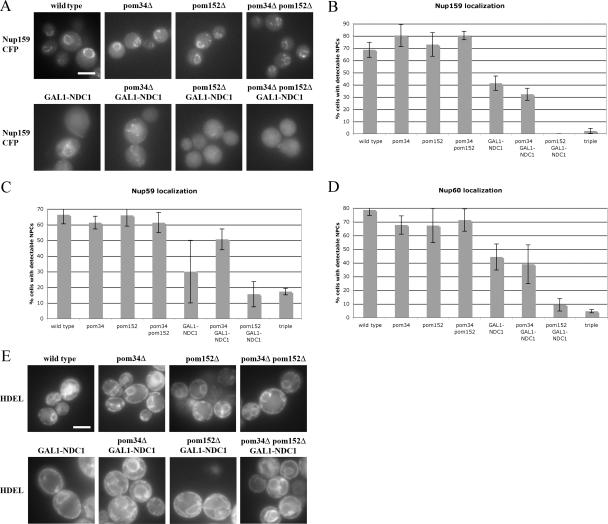
To examine NPC distribution in the absence of transmembrane Nups, each strain was transformed with plasmids expressing Nup159p-CFP, Nup59p-CFP, or Nup60p-CFP under their endogenous promoters. These three Nups were chosen because they each localize to a distinct region of the pore. Nup159p is found on the cytoplasmic filaments, Nup59p is found in the central core, and Nup60p is found on the nuclear side of the NPC (Rout et al., 2000). All strains were grown in dextrose-containing media for 24 h, and live cells were analyzed by fluorescence microscopy (Fig. 3 A). Only cells lacking the Ndc1p-GFP signal were included in the analysis, and cells were scored as having normal NPCs if there was a visible accumulation of Nup-CFP signals around the nucleus.
Figure 3.
Nups are mislocalized in strains lacking both Ndc1p and Pom152p. (A) Wild-type cells and strains lacking one, two, or three of the POMs expressing Nup159p-CFP were grown for 24 h in dextrose media. Nup159p-CFP distribution was visualized in live cells by fluorescence microscopy. (B–D) Cells without detectable Ndc1p-GFP signal were scored for the presence or absence of perinuclear NPC signal. Three independent experiments were performed with each reporter protein, Nup159p-CFP (B), Nup59p-CFP (C), or Nup60p-CFP (D), and the percentage of cells with normal NPCs was quantified. Error bars indicate the SD. (E) Wild-type cells and strains lacking one, two, or three of the POMs expressing dsRED-HDEL. Cells were grown for 24 h in dextrose media. dsRED-HDEL distribution was visualized in live cells by fluorescence microscopy. Error bars indicate the SD. Bars, 5 μm.
Deletion of POM34, POM152, or a combination of both had no significant effect on the localization of Nup159p-CFP (Fig. 3, A and B). Although wild-type yeast had 69 ± 6% of cells with normal Nup159p-CFP localization, this percentage was 80 ± 9% in pom34Δ, 73 ± 10% in pom152Δ, and 80 ± 4% in pom34Δ pom152Δ deletion strains (Fig. 3 B).
In contrast, the strain depleted of Ndc1p displayed a partial reduction in the percentage of cells with properly localized NPCs, with only 41 ± 6% of the cells showing a correct NPC distribution (Fig. 3, A and B). Similar results were obtained in the strain lacking both Ndc1p and Pom34p, in which 32 ± 5% of cells had a normal Nup159p-CFP localization pattern (Fig. 3, A and B). Furthermore, in both GAL1-NDC1 and pom34Δ GAL1-NDC1 cells, the loss of Nup159p NPC staining appeared to coincide with an increased pool of cytoplasmic Nup159p-CFP (Fig. 3 A).
Strikingly, cells that lacked both Ndc1p and Pom152p displayed the most severe mislocalization defects (Fig. 3, A and B). The pom152Δ GAL1-NDC1 strain had no cells with normal Nup159p-CFP, and the triple-mutant strain (pom34Δ pom152Δ GAL1-NDC1) had only 2 ± 2% of cells with proper Nup159p-CFP localization (Fig. 3 B).
Similar results were obtained when the central Nup59p-CFP and the nuclear Nup60p-CFP were visualized and scored as described in the first paragraph of this section (Fig. 3, C and D). In each experiment, Ndc1p depletion led to a partial NPC localization defect, and this effect was exacerbated by the loss of Pom152p. However, the loss of Nup59p from the NPC was not as dramatic as the loss of the peripherally associated Nup159p or Nup60p, suggesting that Nup59p is more stably associated with the core of the NPC.
To rule out the possibility that the observed Nup mislocalizations were caused by a decrease in Nup-CFP protein levels, we performed Western blots on several of these strains after Ndc1p depletion. Although we consistently observed minor variations in Nup-CFP protein expression levels between strains and between experiments, most likely attributable to their plasmid-driven expression, all tested Nups were expressed at comparable levels in dextrose growth conditions (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200506199/DC1, and not depicted). Importantly, there was no correlation between the observed NPC defects and the variations in protein levels.
To determine whether the lack of perinuclear NPC signal in some POM mutant strains was indirectly caused by nuclear envelope defects, we used a reporter protein with a tetrapeptide ER-retention signal (HDEL) fused to dsRED (dsRED-HDEL). In wild-type cells, this reporter localizes to the ER lumen, which in yeast includes the space between the inner and outer nuclear envelopes, as well as the peripheral ER (Fig. 3 E). Although the Ndc1p-depleted cells occasionally had misshapen nuclear envelopes, the dsRED-HDEL reporter was able to properly localize to the lumenal space between the inner and outer nuclear envelopes. This result suggests that the NPC mislocalization is not caused by disruption of the nuclear envelope.
Cells lacking Ndc1p mislocalize nuclear import reporters
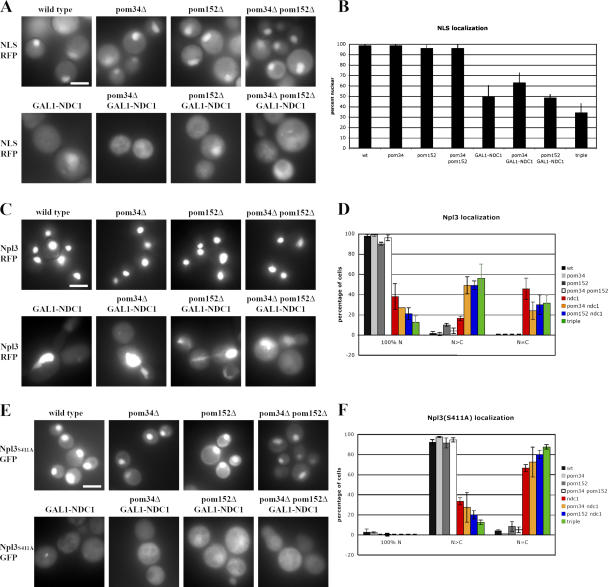
The Nup mislocalization suggested that NPCs were not properly formed in POM-deficient strains. To determine whether the NPCs in these strains were functional for nuclear transport, we tested two different import pathways. First, we monitored the steady-state localization of a classical nuclear import reporter, consisting of the SV40 NLS fused to monomeric red fluorescent protein (mRFP). In wild-type cells, this protein accumulates in the nucleus in 99% of cells (Fig. 4, A and B ). No change in localization of the reporter was seen when either POM34 or POM152 was deleted (Fig. 4, A and B). However, in cells depleted of Ndc1p there was an ∼50% reduction in the number of cells that displayed a concentration of the NLS-mRFP signal in the nucleus (Fig. 4, A and B). In contrast to the Nup localization experiment, no significant additional defect was seen when Ndc1p depletion was combined with pom152Δ.
Figure 4.
Nuclear import reporters mislocalize to the cytoplasm in strains lacking Ndc1p. (A, C, and E) Wild-type cells and strains lacking one, two, or three of the POMs expressing NLS-mRFP (A), Npl3p-GFP (C), or Npl3pS411A-GFP (E). Cells were grown for 24 h in dextrose media. Steady-state distribution of each reporter protein was visualized in live cells by fluorescence microscopy. (B) Cells were scored for nuclear accumulation of the NLS-RFP reporter protein. Three independent experiments were performed, and the percentage of cells with nuclear RFP was quantified. Error bars indicate standard deviation. (D and F) Cells were scored for distribution of the reporter proteins Npl3p-GFP (D) and Npl3pS311A-GFP (F). 100% nuclear indicates no cytoplasmic signal. N > C indicates predominantly nuclear with some cytoplasmic signal. N = C indicates cytoplasmic localization. Two independent experiments were performed for each reporter, and the percentage of cells with nuclear RFP was quantified. Error bars indicate the SD. Bars, 5 μm.
We next examined the localization of the yeast-shutting protein Npl3p, as well as a mutant version, Npl3S411A. Although Npl3p-GFP is exclusively nuclear at steady state, with no detectable cytoplasmic signal in wild-type cells (Fig. 4 C), the Npl3S411A variant partially mislocalizes to the cytoplasm because of the absence of Ser411 phosphorylation (Fig. 4 E; Gilbert et al., 2001). We monitored the localization of these proteins in each strain, and scored individual cells as being exclusively nuclear (100% N), predominantly nuclear with some cytoplasmic signal (N > C), or as having no nuclear accumulation (N = C; Fig. 4, D and F). Although the absence of POM34, POM152, or both had no significant effect on Npl3p localization, each of the four GAL1-NDC1 strains displayed a marked increase in cells with cytoplasmic Npl3p-GFP (both N > C and N = C; Fig. 4, C and D). Similarly, all four GAL1-NDC1 strains displayed a significant increase in the percentage of cells with cytoplasmic (N = C) Npl3pS411A (Fig. 4, E and F).
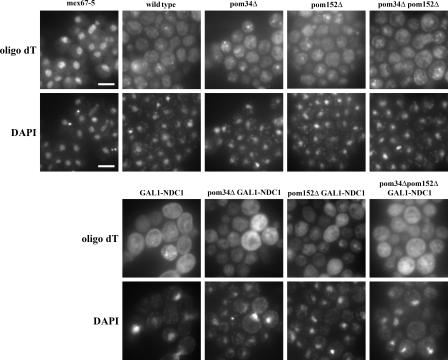
In addition to protein import, we also monitored mRNA export by performing in situ hybridizations against the poly(A)+ tails using an oligo dT probe to monitor nuclear accumulation of poly(A)+ RNA (Fig. 5 ). As a positive control, we shifted mex67-5 cells to the nonpermissive temperature for 30 min, which is known to cause a severe mRNA export defect. Although the probe was strongly concentrated in the nucleus in mex67-5 cells, we did not see nuclear accumulation in any of our POM-deficient strains (Fig. 5).
Figure 5.
POM-deficient strains do not accumulate mRNA in the nucleus. Wild-type cells and strains lacking one, two, or three of the POMs were grown for 24 h in dextrose media. poly(A)+ RNA was detected by in situ hybridization with a dT50 probe, and DNA was stained with DAPI. The known mRNA export mutant mex67-5 was used as a control and showed strong nuclear accumulation of the probe (left). Bars, 5 μm.
Cells lacking Ndc1p and Pom152p have abnormal NPC morphology
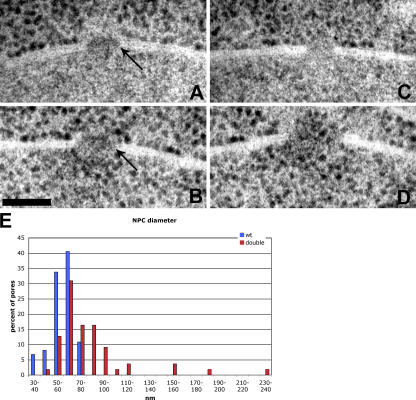
To analyze the ultrastructure of the NPCs, we performed TEM on wild-type and pom152Δ GAL1-NDC1 cells. As we observed by fluorescence microscopy, both the wild-type and mutant cells had intact nuclear envelopes. NPCs in wild-type nuclei were identified by a characteristic gap in the nuclear envelope containing an electron-dense plaque structure (Fig. 6 A, arrow). The pom152Δ GAL1-NDC1 mutant nuclei also contained detectable pores in the nuclear envelope, although there appeared to be fewer pores in each thin section than in the wild-type cells. However, while wild-type NPCs displayed a typical NPC morphology and were of a consistent size, a range of abnormal phenotypes was seen for the pores in the pom152Δ GAL1-NDC1 mutant strain. Only ∼10% of the pores that we observed were comparable to wild type. The majority of the pores fell into the following two phenotypic classes: (a) pores that were roughly the same diameter as wild-type NPCs, but lacked any detectable electron-dense material (Fig. 6 C), and (b) pores that were approximately the same diameter as a wild-type NPC, but contained electron-dense material with an abnormal, less compact morphology (Fig. 6 B, arrow). Another 10% of pores had the most dramatic morphological defect, having a considerably larger diameter than a wild-type NPC and lacking any electron-dense material (Fig. 6 D). We measured the diameters of at least 55 NPCs from each strain, and found that wild-type NPC diameters fell into a narrow range, with a mean diameter of 59 nm and a maximum of 75 nm. However, mutant pore diameters displayed a broader distribution, with a mean diameter of 82 nm and a maximum diameter of 240 nm (Fig. 6 E). Interestingly, in the double-mutant cells we occasionally observed ribosomes in the nuclear compartment (Fig. 6, B and D), whereas ribosomes were strictly cytoplasmic in wild-type nuclei (Fig. 6 A). This suggests that the channel size in these cells has increased enough to allow diffusion of intact ribosomes through the pore.
Figure 6.
pom152ΔGAL1-NDC1 cells have aberrant NPC morphology. Wild-type and pom152Δ GAL1-NDC1 cells were visualized by TEM. In each image, the nuclear compartment is on the bottom and the cytoplasmic compartment is on top. (A) A wild-type NPC. Arrow indicates the electron-dense complex. (B–D) Examples of three types of pom152Δ GAL1-NDC1 pores. (B) A pore containing electron-dense, proteinaceous material, but lacking the typical compact structure in the plane of the membrane. Arrow indicates the indiscreet complex. (C) A pore lacking electron-dense material. (D) An abnormally wide pore lacking electron-dense material. Note the ribosomes in the nucleus in B and D. Bar, 100 nm. (E) Pores were chosen randomly and imaged from wild-type and pom152Δ GAL1-NDC1 cells. Diameters of at least 55 pores per strain were measured, and the distribution is shown. Wild-type NPCs had a mean diameter of 59 nm and a maximum diameter of 75 nm. Mutant pores had a mean diameter of 82 nm and a maximum diameter of 240 nm.
Cells lacking Ndc1p and Pom152p have an increased diffusion limit
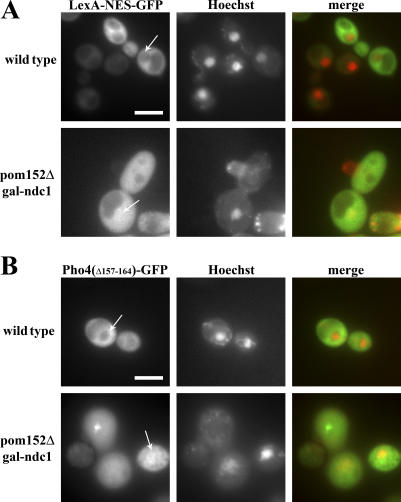
The morphological defects we observed by TEM, together with the Nup-CFP localizations, suggested that the pores in pom152Δ GAL1-NDC1 cells lack many normal nuclear pore components and may no longer provide a fully functional diffusion barrier. To test whether molecules above the size threshold for diffusion through a wild-type NPC are excluded from the nucleus in the mutant cells, we used the following two reporter proteins: (a) the LexA DNA-binding domain fused to a nuclear export signal (NES) and GFP (LexA-NES-GFP), with a molecular mass of 51.5 kD, and (b) a version of Pho4p lacking its NLS (Pho4Δ157–164) fused to GFP (Pho4Δ157–164-GFP), with a molecular mass of 61.8 kD. Previous studies have shown that similar NES-containing constructs with molecular masses as low as 36 kD are unable to passively diffuse through the NPC in wild-type cells (Shulga et al., 2000).
Both of the reporters used were excluded from the nucleus in 80–90% of wild-type cells (Fig. 7, A and B, top). In contrast, the reporters were equilibrated between the nucleus and the cytoplasm in the majority (60–70%) of pom152Δ GAL1-NDC1 cells (Fig. 7, A and B, bottom), indicating that the pores in many POM-depleted cells have an increased limit of passive diffusion. This result confirms our EM observation that pom152Δ GAL1-NDC1 cells have larger pores that are devoid of any detectable proteinaceous material, suggesting that these pores lack a functional central channel. Furthermore, it is consistent with our TEM result that we occasionally observe ribosomes, which have a molecular mass of ∼4 MD, in the nuclear compartment of pom152Δ GAL1-NDC1 cells, suggesting that the diffusion limit in POM-deficient cells is variable and is significantly higher than that in wild-type cells.
Figure 7.
The diffusion limit is increased in pom152ΔGAL1-NDC1 cells. Wild-type and pom152Δ GAL1-NDC1 cells expressing either the 51.5-kD protein LexA-NES-GFP (A) or the 61.8-kD protein Pho4Δ157–164-GFP (B) were grown for 24 h in dextrose media, and then incubated for 30 min with Hoechst to label the nucleus. Steady-state distributions of the reporter proteins were visualized in live cells by fluorescence microscopy. In wild-type cells, the reporters were excluded from the nucleus (top), whereas in the mutant the reporters were equilibrated between the cytoplasm and the nucleus (bottom). Arrows indicate the location of the nucleus. Bars, 5 μm.
Discussion
The assembly of nuclear pore proteins into a functional complex that spans both bilayers of the nuclear envelope is a poorly understood and complicated process. We have characterized the role of the three transmembrane nuclear pore components, Ndc1p, Pom152p, and Pom34p, in interphase NPC assembly and function in yeast. The results presented in this study clearly demonstrate a critical role for Ndc1p in these processes, but also imply a partially redundant function of Pom152p in pore formation, as only the loss of both of these proteins caused complete Nup mislocalization in our assays. In contrast, we were unable to detect any additive effect caused by the loss of Pom34p, suggesting that Pom34p is dispensable for NPC assembly and function.
Although the depletion of Ndc1p and Pom152p had an additive effect with regard to Nup mislocalization, the nuclear import defect that could be observed after Ndc1p depletion was not significantly exacerbated by the absence of POM152. This implies that, although Ndc1p-depleted strains show only a partial Nup localization defect by fluorescence microscopy, the NPCs in these strains are already functionally compromised and, thus, display a phenotype in sensitive transport assays. For example, it is possible that although Ndc1p depletion causes only the partial loss of the three Nups that we monitored, Nup159p, Nup59p, and Nup60p, this could be sufficient to affect the diffusion limit of the central NPC channel.
Because pom152Δ GAL1-NDC1 cells displayed such dramatic Nup and import reporter localization defects, it was originally surprising that we did not observe any mRNA export defects in this strain. However, TEM analysis and nuclear exclusion experiments suggested that POM depletion results in a widening of the central channel and a breakdown of the normal diffusion barrier. Because normal protein import and defective mRNA export both result in an accumulation of substrate inside nuclei, we postulate that NPCs lacking Ndc1p and Pom152p allow the diffusion of molecules that normally rely on facilitated nuclear transport mechanisms. Thus, these molecules are unable to move against their concentration gradients, which is consistent with the observation that neither nuclear import reporters nor mRNA accumulate in the nucleus. In addition, new transcription of mRNA appears to be significantly reduced upon depletion of Ndc1p and Pom152p (unpublished data), further preventing a buildup of nuclear poly(A)+ RNA.
A critical role for transmembrane Nups has now been demonstrated in both yeast NPC assembly (Lau et al., 2004; this study) and postmitotic assembly in metazoans (Drummond and Wilson, 2002; Cohen et al., 2003; Antonin et al., 2005). However, there have been somewhat conflicting results regarding the contributions of the integral membrane proteins GP210 and POM121 in NPC assembly. Interestingly, homologues of yeast Ndc1p have recently been described in humans and other metazoans (Mans et al., 2004) and may be critical for postmitotic NPC assembly in those organisms as well. Based on our data in yeast, which reveal partial redundancy between Ndc1p and Pom152p in NPC assembly, it is conceivable that the conflicting results pertaining to the metazoan POMs may be explained by a similar type of overlapping function between POM121, GP210, or metazoan Ndc1.
Previous studies in X. laevis extracts have also demonstrated important roles for the small GTPase Ran and the Nup107/160 complex in metazoan postmitotic assembly (Hetzer et al., 2000; Zhang and Clarke, 2000; Boehmer et al., 2003; Harel et al., 2003a,b; Walther et al., 2003a,b). However, it has been unclear whether these same proteins are important for NPC assembly into intact nuclear envelopes, which occurs in nonmitotic metazoan cells and throughout the cell cycle in S. cerevisiae. Studies in budding yeast have shown that mutations in the genes encoding Ran and the Nup107/160 orthologue, which is the Nup84/85 complex, disrupt the NPC and, therefore, may be required for yeast NPC biogenesis (Siniossoglou et al., 1996; Ryan et al., 2003). Although NPC assembly has not been tested directly in these mutants, the intriguing similarities between metazoan postmitotic assembly and yeast NPC assembly may indicate that the processes occur by a conserved mechanism.
Our results in yeast imply that Ndc1p plays an important structural role at the NPC, although Pom152p may partially compensate for the loss of this transmembrane protein. Although the detailed mechanistic role of transmembrane Nups remains unclear, multiple possible mechanisms by which transmembrane Nups could function in NPC assembly can be envisioned. For example, transmembrane Nups could remodel the two nuclear envelope membranes to induce membrane curvature, thus forming a prepore-type structure that is needed for NPC insertion. Furthermore, they could be required to anchor or target certain Nup subcomplexes to the nuclear envelope. The dramatic mislocalization of three spatially distinct Nups (Nup60p, Nup59p, and Nup159p) is consistent with a model in which Ndc1p and Pom152p act at an early stage of NPC assembly, possibly in the recruitment of soluble Nup subcomplexes to the nuclear envelope. This model is also supported by TEM experiments demonstrating that pom152Δ GAL1-NDC1 cells have pores that appear to be lacking the bulk of their protein content. Nuclear exclusion assays confirmed that the diffusion limit in pom152Δ GAL1-NDC1 cells is increased, further strengthening the conclusion that POM depletion results in pores that lack a functional central NPC channel. Based on the data presented in this study, we suggest that Ndc1p, together with Pom152p, acts as an essential tether between the membrane and soluble components of the NPC.
Materials and methods
Yeast strain construction
Yeast media and strain constructions were performed according to established protocols. All plasmids used in this study are listed in Table I, and all yeast strains are listed in Table II.
Table I.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pKW1368 | pRS306-GAL1-NDC1(N-term) | this study |
| pKW1552 | pRS315-NUP159-CFP | this study |
| pKW1706 | pRS425-NUP59-CFP | this study |
| pKW1752 | pRS425-NUP60-CFP | this study |
| pKW1219 | pRS425-NLS-mRFP1 | this study |
| pKW1783 | pRS425-Spc24-mRFP | this study |
| pKW551 | pRS315-Npl3-GFP | Siebel and Guthrie, 1996 |
| pKW776 | pRS315-Npl3(S411A)-GFP | Duncan et al., 2000 |
| pKW1803 | TIPlac-dsRED-HDEL; NatMX | this study |
| pKW409 | pRS314-LexA-NES-GFP | this study |
| pKW1898 | pRS315-Pho4(Δ157–164)-GFP | this study |
Table II.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| KWY165 | wild type | this study |
| KWY1105 | pom34∷Kan | this study |
| KWY742 | pom152∷Kan | this study |
| KWY1117 | pom34∷Kan pom152∷Kan | this study |
| KWY958 | GAL1(∷URA3)-NDC1-GFP∷HIS3 | this study |
| KWY1118 | GAL1(∷URA3)-NDC1-GFP∷HIS3 pom34∷Kan | this study |
| KWY1120 | GAL1(∷URA3)-NDC1-GFP∷HIS3 pom152∷Kan | this study |
| KWY1127 | GAL1(∷URA3)-NDC1-GFP∷HIS3 pom34∷Kan pom152∷Kan | this study |
| KWY1400 | HDEL-dsRED | this study |
| KWY1484 | HDEL-dsRED pom34∷Kan | this study |
| KWY1483 | HDEL-dsRED pom152∷Kan | this study |
| KWY1401 | HDEL-dsRED pom34∷Kan pom152∷Kan | this study |
| KWY1482 |
HDEL-dsRED GAL1(∷URA3)-NDC1- GFP∷HIS3 |
this study |
| KWY1402 |
HDEL-dsRED GAL1(∷URA3)-NDC1- GFP∷HIS3 pom34∷Kan |
this study |
| KWY1403 |
HDEL-dsRED GAL1(∷URA3)-NDC1- GFP∷HIS3 pom152∷Kan |
this study |
| KWY1404 |
HDEL-dsRED GAL1(∷URA3)-NDC1- GFP∷HIS3 pom34∷Kan pom152∷Kan |
this study |
| KWY812 | mex67∷Kan + pUN100-mex67-5 | this study |
All strains were derived from W303: his3-11,15 ura3-1 leu2-3 trp1-1 ade2-1.
To delete POM34 at its genomic locus in our strain background, PCR was performed using oligos (forward, GATGAACAAAAAGATTTATA; reverse, GCTAATCATATGTAAAATAT) and genomic DNA from the pom34Δ Yeast Deletion Consortium strain (Winzeler et al., 1999) as templates. The PCR product was transformed into KWY165 (W303 MATa) and integrated by homologous recombination. To delete POM152 at its genomic locus, PCR was performed using oligos (forward, GATAAACGGATTATAGATTTATCATACCAGATACGTTTATCAGGGCAGATCCGCTAGGGATAA; reverse, TATATTATACATTACAATTGTACAAAAATATTGCGGGAAAGAATTCGAGCTCGTTTAAAC) and plasmid pAF6-GFP(S65T)-kan (Longtine et al., 1998) as templates. The PCR product was transformed into KWY390 (W303 MATα) and integrated by homologous recombination. GAL1-NDC1-GFP strains were created by first fusing GFP to the COOH terminus of NDC1 using oligos (forward, GTTTCTAGAAGTGTACGCCTCAGGCAACCCTAATGCTACGCGGATCCCCGGGTTAATTAA; reverse, CCGGAAACGATAAAGGTAGCTTTTTGCTCTTTTTGCTCATGAATTCGAGCTCGTTTAAA) and the plasmid pFA6a-GFP(S65T)-hisMX6 (Longtine et al., 1998) as templates. The PCR was transformed into KWY165 (W303 MATa) and integrated by homologous recombination. The GAL1 promoter was integrated 5′ of the NDC1-coding region by linearizing the integration plasmid pKW1368 (GAL1-NDC1Nterm) with NheI. HDEL-dsRED strains were created by linearizing pKW1803 with EcoRV and integrating it at the TRP1 locus.
Plasmid construction
To generate plasmid pKW1368 (pRS306-GAL1-NDC1Nterm), the GAL1 promoter was cloned into pRS306 as a NotI–EcoRI fragment, and the NH2 terminus of NDC1 was PCR amplified from genomic DNA using oligos (forward, AAAGAATTCATGATACAGACGCCA; reverse, AAACTCGAGCGAAAGTGATGAAGA) and inserted as an EcoRI–XhoI fragment. To generate the plasmid pKW1552 (pRS315-NUP159-CFP), CFP was PCR amplified as an EcoRI–NotI fragment to replace the triple GFP from pKW1220 (pRS313-NUP159-GFP3) to get pRS313-NUP159-CFP. This was subsequently subcloned into pRS315 (Sikorski and Hieter, 1989) to change the marker. To generate plasmid pKW1706 (pRS425-NUP59-CFP), NUP59, along with its promoter, was PCR amplified from yeast genomic DNA and subcloned into pRS425 using XhoI and NaeI sites. CFP was PCR amplified as a NaeI–DraIII fragment and subcloned at the 3′ end of NUP59. pKW1752 (pRS425-NUP60-CFP) was generated by PCR amplifying NUP60 and its promoter from yeast genomic DNA and subcloning it into pKW1706 using NaeI–DraIII, thus, replacing NUP59 with NUP60. The plasmid pKW1219 (pRS425-NLS-mRFP) was created by replacing the GFP from pKW674 (pRS426-PADH1-NLS-NES[P12]-GFP) with mRFP. This construct was then subcloned into pRS425 using XhoI–SacI. To generate pKW1783 (pRS425-Spc24-mRFP), SPC24, along with its promoter, was amplified from genomic DNA as a SacI–BamHI fragment and cloned into pKW1219, replacing the ADH1 promoter and NLS. pKW1803 (YIPlac-dsRED-HDEL-NatMX) was generated from YIPlac-dsRED-HDEL (a gift from C. Reinke and B. Glick, University of Chicago, Chicago, IL) by cloning in the NatMX cassette as a HindII–SapI fragment. To generate pKW409 (pRS314-LexA-NES-GFP), a SacI–KpnI LexA PCR product was amplified from pEG202 (Gyuris et al., 1993) and cloned into pRS314. The NES from PKI was inserted an EcoRI–BamHI fragment by annealing the oligos (forward, AATTCAATGAATTAGCCTTGAAATTAGCAGGTCTTGATATCAACAAGACAGG; reverse, GATCCCTGTCTTGTTGATATCAAGACCTGCTAATTTCAAGGCTAATTCATTG). Finally, GFP(S65T) was inserted as a BamHI–XhoI fragment. Plasmid pKW1898 (pRS315-Pho4 Δ157-164-GFP) was constructed from EBO383 (a gift from E. O'Shea, Harvard University, Cambridge, MA; Kaffman et al., 1998) to change the marker from URA3 to LEU2. A SalI–BamHI fragment was cut out of EBO383 and ligated into pRS315. Plasmids pKW551 and pKW776 were a gift from C. Guthrie, University of California, San Francisco, San Francisco, CA.
Ndc1p depletion
Cells were grown at 30°C in galactose media overnight, washed, and diluted to OD600 = 0.1 in dextrose media. Cells were then grown at 30°C in dextrose for 24 h to deplete GAL1-NDC1 cells of Ndc1p. The efficiency of Ndc1p depletion was verified by the visualization of Ndc1p-GFP by fluorescence microscopy.
In vivo protein localization
Cells were grown under the conditions indicated in the text and figure legends, and embedded in 0.5% agarose for visualization. Images were acquired with a digital camera (model CA742-98; Hamamatsu) controlled by the Metamorph software program (Universal Imaging Corp.). Images were processed using Photoshop CS (Adobe), and figures were assembled using Photoshop CS and Illustrator CS (Adobe). DNA was stained in live cells by adding either 2.5 μg/ml of DAPI (Sigma-Aldrich) or 5 μg/ml Hoechst dye (Sigma-Aldrich) directly to cell culture and growing for 30 min before visualization.
Vital staining
The vital dyes Fun1 (Invitrogen) or MitoTracker red (Invitrogen) were added directly to cells in growth media at final concentrations of 4 μM and 100 ng/ml, respectively. Cells were grown for 30 min and then visualized by fluorescence microscopy. As a negative control, wild-type cells were treated with 10 mM sodium azide and 10 mM 2-deoxyglucose for 30 min to stop respiration, and were then dyed and visualized the same as the cells in growth media.
In situ hybridization
Cells were grown for 24 h in dextrose before being fixed in paraformaldehyde and permeabilized with zymolyase T100 (MP Biomedicals). poly(A)+ RNA was detected by hybridization of a digoxigenin-labeled oligo (dT)50 probe and a rhodamine-labeled anti–mouse antibody, as described previously (de Bruyn Kops and Guthrie, 2001). DNA was stained with DAPI (Sigma-Aldrich). The known mRNA export mutant mex67-5 was used as a positive control, and was shifted to 37°C for 30 min before fixation.
Western blotting
Yeast cultures were normalized by OD600, and protein samples were prepared by spinning down the cells and resuspending them in 0.1 M NaOH for 5 min before spinning down and resuspending them in 1× SDS loading buffer. Protein samples were loaded onto a 10% polyacrylamide gel and separated by SDS-PAGE. Proteins were transferred to nitrocellulose membranes, which were cut in half, and the top half was blotted with a mouse monoclonal anti-GFP antibody (Roche) to visualize Ndc1p-GFP and either Nup159p-CFP, Nup59p-CFP, or Nup60p-CFP. The bottom half was blotted with a rabbit polyclonal anti-Dhh1 antibody (Fischer and Weis, 2002) as a loading control. For Fig. S1, protein levels were quantified using ImageJ software (National Institutes of Health), and a ratio of GFP/Dhh1 signals was calculated. This ratio was normalized to the ratio in the wild-type strain.
TEM analysis
High pressure freezing of cells followed a protocol similar to that explained by McDonald (1999). In brief, cells were harvested from liquid cultures by vacuum filtration onto filters (Millipore), transferred to sample chambers, and frozen with a high pressure freezer (model EM PACT2; Leica). Frozen samples were stored in liquid N2 and transferred to a freeze substitution apparatus (model AFS2; Leica) and fixed with 0.2% OsO4 and 0.05% uranyl acetate. Cells were then infiltrated and embedded with Epon resin. Serial sections were cut at 60-nm thick using an Ultracut E (Reichert). Sections were then picked up on formvar-coated grids and stained with 2% uranyl acetate in 70% methanol and Reynolds lead citrate. Cells were then observed and imaged on an electron microscope (model 1200 CX; JEOL) operating at 80 kV.
Online supplemental material
Fig. S1 shows that Nup159p and Nup60p proteins are expressed in all POM-depleted strains, as determined by Western blotting. Online supplemental material is available at http://www.jcb.org/cgi/content/full/ jcb.200506199/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Mark Winey for reagents and for communicating results before publication. We would like to thank Ben Glick, Catherine Reinke, Christine Guthrie, and Erin O'Shea for their generous gifts of plasmids. We also thank Kent McDonald for use of the electron microscopy facility, Matthew Welch, Mike Blower, and Zain Dossani for critically reading the manuscript, and all the members of the Weis laboratory for helpful discussions.
This work was supported by grants from the Searle Scholars program and the National Institutes of Health to K. Weis (GM58065) and W.Z. Cande (GM067992).
Note added in proof. A role for a homologue of Ndc1p in NPC assembly in vertebrate cells has now been reported (Mansfeld, J., S. Güttinger, L.A. Hawryluk-Gara, N. Panté, M. Mall, V. Galy, U. Haselmann, P. Mühlhäusser, R.W. Wozniak, I.W. Mattaj, U. Kutay, W. Antonin. 2006. Mol. Cell. 22:93–103).
Abbreviations used in this paper: mRFP, monomeric red fluorescent protein; NES, nuclear export signal; NPC, nuclear pore complex; Nup, nucleoporin; POM, pore membrane protein; SPB, spindle pole body; TEM, transmission electron microscopy.
References
- Antonin, W., C. Franz, U. Haselmann, C. Antony, and I.W. Mattaj. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell. 17:83–92. [DOI] [PubMed] [Google Scholar]
- Boehmer, T., J. Enninga, S. Dales, G. Blobel, and H. Zhong. 2003. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 100:981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial, H.J., M.P. Rout, T.H. Giddings, and M. Winey. 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143:1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, M., N. Feinstein, K.L. Wilson, and Y. Gruenbaum. 2003. Nuclear pore protein gp210 is essential for viability in HeLa cells and Caenorhabditis elegans. Mol. Biol. Cell. 14:4230–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyn Kops, A., and C. Guthrie. 2001. An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J. 20:4183–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, S.P., and K.L. Wilson. 2002. Interference with the cytoplasmic tail of gp210 disrupts “close apposition” of nuclear membranes and blocks nuclear pore dilation. J. Cell Biol. 158:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, K., J.G. Umen, and C. Guthrie. 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 10:687–696. [DOI] [PubMed] [Google Scholar]
- Eriksson, C., C. Rustum, E. Hallberg, M. Cohen, N. Feinstein, K.L. Wilson, and Y. Gruenbaum. 2004. Dynamic properties of nuclear pore complex proteins in gp210 deficient cells. FEBS Lett. 572:261–265. [DOI] [PubMed] [Google Scholar]
- Fischer, N., and K. Weis. 2002. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 21:2788–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W., C.W. Siebel, and C. Guthrie. 2001. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA. 7:302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 75:791–803. [DOI] [PubMed] [Google Scholar]
- Harel, A., R.C. Chan, A. Lachish-Zalait, E. Zimmerman, M. Elbaum, and D.J. Forbes. 2003. a. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell. 14:4387–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel, A., A.V. Orjalo, T. Vincent, A. Lachish-Zalait, S. Vasu, S. Shah, E. Zimmerman, M. Elbaum, and D.J. Forbes. 2003. b. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 11:853–864. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., D. Bilbao-Cortes, T.C. Walther, O.J. Gruss, and I.W. Mattaj. 2000. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell. 5:1013–1024. [DOI] [PubMed] [Google Scholar]
- Kaffman, A., N.M. Rank, and E.K. O'Shea. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.K., T.H. Giddings Jr., and M. Winey. 2004. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot. Cell. 3:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Masson, I., C. Saveanu, A. Chevalier, A. Namane, R. Gobin, M. Fromont-Racine, A. Jacquier, and C. Mann. 2002. Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol. Microbiol. 43:1431–1443. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mans, B.J., Anantharaman, V., Aravind, L., Koonin, E.V. 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 3:1612–1637. [DOI] [PubMed] [Google Scholar]
- Mansfeld, J., S. Güttinger, L.A. Hawryluk-Gara, N. Panté, M. Mall, V. Galy, U. Haselmann, P. Mühlhäusser, R.W. Wozniak, I.W. Mattaj, U. Kutay, W. Antonin. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 22:93–103. [DOI] [PubMed] [Google Scholar]
- Maul, G.G., H.M. Maul, J.E. Scogna, M.W. Lieberman, G.S. Stein, B.Y. Hsu, and T.W. Borun. 1972. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 55:433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, K. 1999. High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 117:77–97. [DOI] [PubMed] [Google Scholar]
- Millard, P.J., B.L. Roth, H.P. Thi, S.T. Yue, and R.P. Haugland. 1997. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 63:2897–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot, M., Y.Z. Zhang, J.A. Kramer, K.S. Wells, L.J. Jones, D.K. Hanzel, A.G. Lugade, V.L. Singer, and R.P. Haugland. 1996. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 44:1363–1372. [DOI] [PubMed] [Google Scholar]
- Rabut, G., P. Lenart, and J. Ellenberg. 2004. Dynamics of nuclear pore complex organization through the cell cycle. Curr. Opin. Cell Biol. 16:314–321. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., J.M. McCaffery, and S.R. Wente. 2003. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160:1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga, N., N. Mosammaparast, R. Wozniak, and D.S. Goldfarb. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel, C.W., and C. Guthrie. 1996. The essential yeast RNA binding protein Np13p is methylated. Proc. Natl. Acad. Sci. USA. 93:13641–13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., C. Wimmer, M. Rieger, V. Doye, H. Tekotte, C. Weise, S. Emig, A. Segref, and E.C. Hurt. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 84:265–275. [DOI] [PubMed] [Google Scholar]
- Thomas, J.H., and D. Botstein. 1986. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 44:65–76. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., A. Alves, H. Pickersgill, I. Loiodice, M. Hetzer, V. Galy, B.B. Hulsmann, T. Kocher, M. Wilm, T. Allen, et al. 2003. a. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 113:195–206. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., P. Askjaer, M. Gentzel, A. Habermann, G. Griffiths, M. Wilm, I.W. Mattaj, and M. Hetzer. 2003. b. RanGTP mediates nuclear pore complex assembly. Nature. 424:689–694. [DOI] [PubMed] [Google Scholar]
- Winey, M., M.A. Hoyt, C. Chan, L. Goetsch, D. Botstein, B. Byers, and J.H. Thomas. 1993. NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 122:743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., D. Yarar, T.H. Giddings Jr., and D.N. Mastronarde. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 8:2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A., D.D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J.D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. [DOI] [PubMed] [Google Scholar]
- Zhang, C., and P.R. Clarke. 2000. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 288:1429–1432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.