Abstract
Stress fibers play a central role in adhesion, motility, and morphogenesis of eukaryotic cells, but the mechanism of how these and other contractile actomyosin structures are generated is not known. By analyzing stress fiber assembly pathways using live cell microscopy, we revealed that these structures are generated by two distinct mechanisms. Dorsal stress fibers, which are connected to the substrate via a focal adhesion at one end, are assembled through formin (mDia1/DRF1)–driven actin polymerization at focal adhesions. In contrast, transverse arcs, which are not directly anchored to substrate, are generated by endwise annealing of myosin bundles and Arp2/3-nucleated actin bundles at the lamella. Remarkably, dorsal stress fibers and transverse arcs can be converted to ventral stress fibers anchored to focal adhesions at both ends. Fluorescence recovery after photobleaching analysis revealed that actin filament cross-linking in stress fibers is highly dynamic, suggesting that the rapid association–dissociation kinetics of cross-linkers may be essential for the formation and contractility of stress fibers. Based on these data, we propose a general model for assembly and maintenance of contractile actin structures in cells.
Introduction
Cell locomotion and adhesion play key roles during embryonic development, tissue regeneration, immune responses, and wound healing in multicellular organisms. Cell migration, changes in cells' shape, and adhesive properties are regulated by continuous remodeling of the actin cytoskeleton. Although multicellular organisms contain a wide array of actin filament assemblies, the actin structures that play fundamental roles in cell migration can be roughly divided into three categories: (1) lamellipodial actin network at the leading edge of the cell, (2) unipolar filopodial bundles beneath the plasma membrane, and (3) contractile actin stress fibers in the cytoplasm (for review see Ridley et al., 2003).
The lamellipodium contains a network of short, branched actin filaments that produce the physical force for protrusion of the leading edge. The formation of new actin filaments at the leading edge is promoted by the Arp2/3 complex, which nucleates new filaments from the sides of preexisting filaments and thus induces the formation of a branched filament network (Mullins et al., 1998; Svitkina and Borisy, 1999). The elongation of newly nucleated filaments is subsequently inhibited by capping proteins to maintain short, stiff filaments as well as to concentrate polymerization to the protruding region close to the plasma membrane (for reviews see Pantaloni et al., 2001; Nicholson-Dykstra et al., 2005). Filopodia are thin cellular processes containing long parallel actin filaments arranged into tight bundles. Recent studies have demonstrated that filopodia are initiated from the dendritic lamellipodial actin network by uncapping and subsequent elongation of subsets of privileged barbed ends (Svitkina et al., 2003). Ena/VASP family proteins and formins appear to play a central role in uncapping and elongation of filopodial actin bundles (Bear et al., 2002; Schirenbeck et al., 2005).
In contrast to relatively well characterized lamellipodia and filopodia, the assembly mechanisms of actin stress fibers are still poorly understood. Stress fibers are contractile actomyosin bundles, which are essential for cell adhesion to the substratum and for changes in cell morphology, specifically the retraction of the trailing edge (“tail”) during migration. Stress fibers are composed of relatively short actin filaments with alternating polarity (Cramer et al., 1997). These filaments are cross-linked by α-actinin and possibly also by other actin-bundling proteins. α-Actinin and myosin display periodic distribution along stress fibers typical also for other types of contractile structures, such as myofibrils of muscle cells. Animal cells contain at least three different categories of stress fibers: ventral stress fibers, transverse arcs, and dorsal stress fibers. Ventral stress fibers are contractile actin filament bundles that are typically associated at both their ends to focal adhesions. These structures are located at the ventral surface of the cell and play an important role in cell adhesion and contraction. Transverse arcs are curved actomyosin bundles that are not directly associated to focal adhesions at their ends. In motile cells, transverse arcs show typical flow from the leading edge toward the cell center. Dorsal stress fibers are actin bundles that insert into focal adhesions at the ventral cell surface, rise toward the dorsal section of the cell, and often terminate to a transverse arc at their proximal ends (Heath, 1983; Heath and Holifield, 1993; Small et al., 1998).
Stress fiber assembly is regulated by a signaling cascade involving the RhoA small GTPase (Ridley and Hall, 1992). The GTP bound form of RhoA activates Rho-associated kinase, which in turn promotes stress fiber formation by inhibiting actin filament depolymerization (through inactivation of actin depolymerizing factor/cofilins via LIM kinase) and by inducing contractility (through phosphorylation of myosin light chains [MLCs]). In addition, RhoA directly activates formins, which have been proposed to induce actin assembly during stress fiber formation (for review see Jaffe and Hall, 2005). However, the actin filament assembly pathways promoting the formation of three types of stress fibers and the mechanism of myosin incorporation into these structures has not been determined. We applied multicolor live cell microscopy and FRAP methods to explore how actin stress fibers are assembled in cells and show that actin filaments for stress fibers are derived from two different sources.
Results
For multicolor live cell microscopy of stress fiber assembly, we used previously characterized GFP fusion proteins of central stress fiber components: actin (Choidas et al., 1998), α-actinin (Edlund et al., 2001), and myosin II regulatory light chain (Peterson et al., 2004) as well as a focal adhesion marker, zyxin (Rottner et al., 2001; see Materials and methods). These four proteins were also fused with the GFP spectral variants YFP and CFP to simultaneously examine the distribution and dynamics of two different fusion proteins during stress fiber assembly. To avoid possible overexpression artifacts, only cells displaying an intact actin cytoskeleton and expressing the lowest detectable amounts of fusion proteins were chosen for further analysis. It is also important to note that localizations of GFP fusions of actin, α-actinin, zyxin, and MLC were identical as compared with the subcellular localizations of endogenous proteins (unpublished data).
The analysis of stress fiber assembly has been hampered by the fact that most nonmotile cell types contain thick, nondynamic stress fibers, whereas most motile cell types contain very few and thin stress fibers and are thus not suitable for live cell microscopy analysis. Therefore, we first screened several cell lines by phalloidin staining to visualize the actin cytoskeletons in fixed cells as well as by expressing GFP-actin to examine the dynamics of these actin structures. U2OS human osteosarcoma cell line was chosen for further analysis because these cells displayed thick stress fibers that were also relatively dynamic based on GFP-actin distribution in live cells (Fig. 1 and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1).
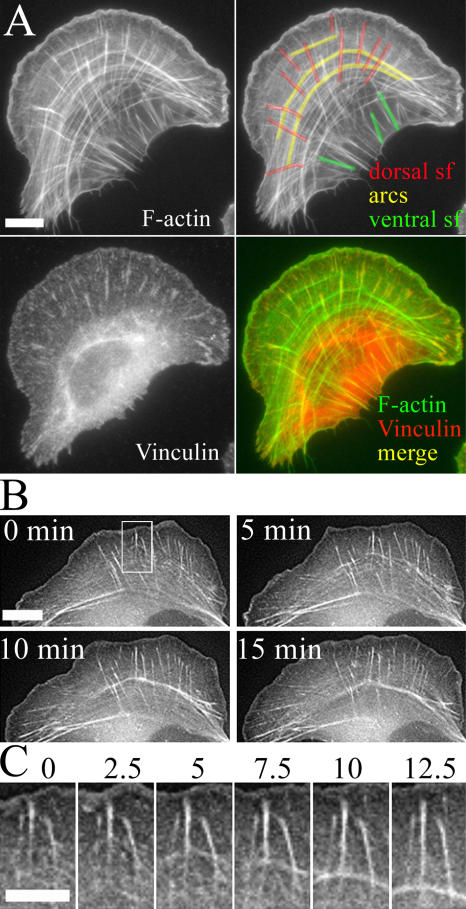
Figure 1.
Contractile actin arrays in U2OS cells. (A) Focal adhesions and F-actin were visualized with anti-vinculin antibodies and phalloidin, respectively. Three categories of contractile actin arrays are highlighted on the F-actin image (top right): dorsal stress fibers (red), transverse arcs (yellow), and ventral stress fibers (green). Bar, 10 μm. (B) Actin dynamics in U2OS cells visualized by GFP-actin. Time-lapse images were taken from Video 1 (available at http://www.jcb.org/cgi/conent/full/jcb.200511093/DC1). Bar, 10 μm. Time-lapse frames of region of interest (indicated with a white rectangle) are displayed in C in higher magnification. (C) Dorsal stress fibers interact with sides of arcs to form a continuous stress fiber network. Time is shown in minutes. Bar, 5 μm.
Ventral stress fibers are generated from arcs and dorsal stress fibers
By analyzing U2OS cells in which F-actin and focal adhesions were visualized by phalloidin and anti-vinculin staining, respectively, we noticed that they contain three different categories of contractile actin arrays that correspond to dorsal stress fibers, transverse arcs, and ventral stress fibers (based on nomenclature described by Small et al., 1998; Fig. 1 A, top right corner). In U2OS cells, dorsal stress fibers typically associate with focal adhesions at one end, ventral stress fibers interact with focal adhesions at both ends, and the ends of transverse arcs do not directly interact with focal adhesions (Fig. 1 A and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1).
Live cell analysis revealed that dorsal stress fibers and transverse arcs undergo continuous formation and disassembly. Dorsal stress fibers were first detected as small actin-rich spots close to the leading edge of the cell (Fig. 1 B). These actin spots then elongated toward the center of the cell and formed bundles of several micrometers in length. During elongation, the proximal ends of dorsal stress fibers appeared to connect to transverse arcs. In contrast to dorsal stress fibers, transverse arcs appeared to assemble from short actin filament bundles generated at the leading edge, and these structures then condensed as they flowed toward cell center (Fig. 1, B and C; and Video 1).
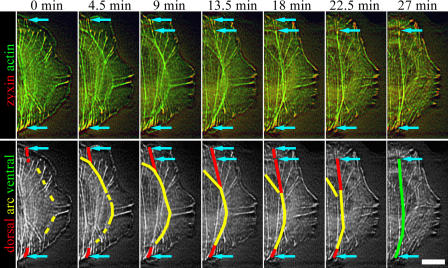
It is important to emphasize that these three distinct categories of contractile actin arrays interact directly with each other and form a continuous and dynamic actin filament network (Fig. 1, B and C; and Video 1). Interestingly, dorsal stress fibers and transverse arcs often converted to ventral stress fibers. During this process, dorsal stress fibers that formed at opposite sides of the cell associated with a transverse arc located between them. During the centripetal flow of actin, the transverse arc (now connected to at least two focal adhesions through dorsal stress fibers) began to contract. During contraction, the transverse arc regions that were not located between the two dorsal stress fibers dissociated, leading to the formation of a ventral stress fiber that associated with focal adhesions at both ends (Fig. 2 and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). Together, these observations indicated that dorsal stress fibers and transverse arcs are generated by the de novo polymerization of actin filaments or condensation of smaller filament fragments into visible structures, whereas ventral stress fibers can be generated through reorganization of the preexisting dorsal stress fiber/transverse arc network. Thus, the assembly of dorsal stress fibers and transverse arcs was studied in more detail.
Figure 2.
Ventral stress fiber assembly. Time-lapse images of U2OS cell expressing YFP-actin (green) and zyxin-CFP (red). The same time-lapse images in grayscale are shown in the bottom panel to highlight the assembly of a single ventral stress fiber. Dorsal stress fibers, transverse arc, and ventral stress fiber are indicated with red, yellow, and green, respectively. The focal adhesions are marked with arrows in both panels. The two arrows at the upper part of the cell (9–18-min frames) indicate two focal adhesions, which both appear to anchor the stress fiber to substrate. The more distal focal adhesion disappears during the maturation of the stress fiber. See Video 3 (available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). Bar, 10 μm.
Mechanism of dorsal stress fiber assembly
To reveal the mechanism of dorsal stress fiber assembly, we first simultaneously visualized focal adhesions (with zyxin-CFP) and actin bundles (with α-actinin–YFP) and observed that after an appearance of a new focal adhesion, an actin filament bundle began to elongate from the focal adhesion (Fig. 3 A and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). During elongation, dorsal stress fibers typically interacted with transverse arcs and thus formed a connection between a transverse arc and a focal adhesion. Occasionally, elongating dorsal stress fibers also interacted with another focal adhesion (Fig. 3 A, cyan arrow).
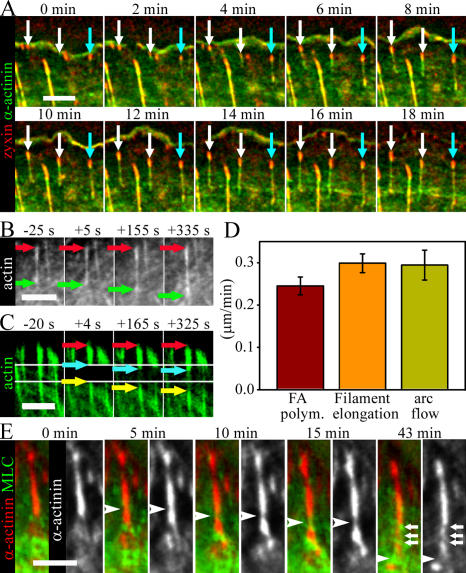
Figure 3.
Mechanism of dorsal stress fiber assembly. (A) Time-lapse images of a U2OS cell expressing zyxin-CFP (red) and α-actinin–YFP (green). White arrows indicate elongating dorsal stress fibers, which eventually bind to the sides of the transverse arc. Cyan arrows indicate a growing stress fiber, which binds to another focal adhesion from its proximal end. See Video 4 (available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). (B) The elongation rate of entire dorsal stress fibers was measured from time-lapse images of GFP-actin–expressing U2OS cells. Red and green arrows indicate the positions of distal and proximal ends of the actin bundle, respectively. (C) GFP-actin in a dorsal stress fiber was photobleached from a 2.5-μm-wide region (outlined with white lines) close to the focal adhesion, and elongation of the bright region of the dorsal stress fiber was subsequently measured from time-lapse images acquired after photobleaching. (D) Comparison of elongation rate of entire dorsal stress fibers (filament elongation; mean of 18 stress fibers), growth rate close to focal adhesions (FA polym) as measured by photobleaching experiments (mean of 10 stress fibers), and the rate of centripetal flow of transverse arcs (arc flow; mean of 8 arcs). SEMs are indicated in the graph. (E) Analysis of myosin incorporation into dorsal stress fibers in U2OS cells expressing α-actinin–CFP (red in color images and white in black-and-white images) and YFP-MLC (green). Arrowheads indicate the point of myosin incorporation detected by an appearance of MLC fluorescence and subsequent displacement of α-actinin (black-and-white images). Incorporated myosin bundle moves toward the cell center because of elongation of the dorsal stress fiber. After 43 min, the proximal end of the dorsal stress fiber is decorated by several myosin II dots (indicated by white arrows). In all time-lapse frames, the leading edge of cell is at the top and the cell center is toward the bottom of the panel. Bars, 5 μm.
To determine whether dorsal stress fibers grow by actin polymerization at focal adhesions, along the filament bundles, or at filament ends, we compared the elongation rate of an entire filament bundle with the elongation rate of the filament bundle at the region close to the focal adhesion. Quantification of GFP-actin–labeled dorsal stress fibers yielded a growth rate of 0.3 μm/min (Fig. 3, B and D, filament elongation). The mean growth rates were very similar in dorsal stress fibers that had free proximal ends and in the ones connected to transverse arcs at their proximal end. To determine the rate of stress fiber elongation from focal adhesions, we performed a FRAP analysis in which a 2.5-μm-wide region of a dorsal stress fiber was photobleached near a focal adhesion. In addition to a general recovery of fluorescence at the bleached region of stress fiber (the result of actin turnover within the stress fiber), we observed an elongation of the bright stress fiber region from the focal adhesion (Fig. 3 C, cyan arrows). This elongation was accompanied by a rearward flow of the bleached region (Fig. 3 C, yellow arrows). Quantification of the growth of bright stress fiber regions from focal adhesions yielded an elongation rate of 0.25 μm/min (Fig. 3 D, FA polym). This rate was very similar to the mean value obtained for the elongation rate of entire stress fibers, suggesting that actin polymerization leading to dorsal stress fiber elongation occurs primarily at the focal adhesion. The rearward flow rate of transverse arcs occurred at a speed similar to that of dorsal stress fiber elongation (Fig. 3 D, arc flow).
We next examined the actin filament assembly mechanism at focal adhesions. Three different protein classes have been shown to promote the nucleation of actin filaments in cells: the Arp2/3 complex promotes the formation of a branched actin network at the cell cortex (for review see Welch and Mullins, 2002), formins generate unbranched filament bundles (for review see Zigmond, 2004), and Spire promotes the formation of unbranched filaments in specific cell types (Quinlan et al., 2005). For the following three reasons, formins are the most likely candidates for promoting actin assembly during the formation of dorsal stress fibers: (1) Dorsal stress fibers are composed of unbranched actin bundles (Cramer et al., 1997). Although Spire can also generate unbranched filaments, it is unlikely to play a central role in dorsal stress fiber assembly because mammalian Spire proteins are expressed only in specialized cell types and localize to Rab11-positive recycling endosomes (Kerkhoff et al., 2001; Schumacher et al., 2004). (2) mDia1/DRF1 formin has previously been implicated in regulating the abundance of stress fibers in cells (Watanabe et al., 1999; Tominaga et al., 2000; Higashida et al., 2004). (3) The rate of stress fiber elongation from focal adhesions (0.25 μm/min; Fig. 2 D) is similar to the actin filament polymerization rate of a mammalian formin, mDia1, under physiological actin concentration in vitro (0.4 μm/min; Romero et al., 2004).
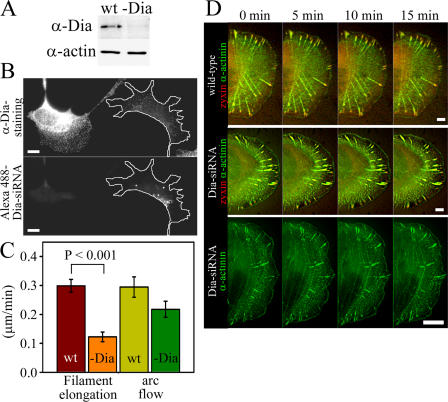
From the Dia family formins, only mDia1/DRF1 has been implicated in the assembly of stress fibers (Watanabe et al., 1999), whereas mDia2 and -3 appear to be involved in other cellular processes (Yasuda et al., 2004; Pellegrin and Mellor, 2005; Schirenbeck et al., 2005). We thus investigated the role of mDia1/DRF1 in dorsal stress fiber elongation by using siRNA gene silencing method. Transfection of U2OS cells with fluorescently labeled siRNA oligonucleotide duplexes targeted to mDia1/DRF1 (Unsworth et al., 2004) resulted in a dramatic decrease in mDia1/DRF1 expression level as compared with nontransfected cells by Western blotting (Fig. 4 A). Correspondingly, a clear reduction in mDia1/DRF1 expression levels was detected by immunofluorescence in cells transfected with fluorescent siRNA oligonucleotides as compared with nontransfected wild-type cells (Fig. 4 B). The elongation rate of dorsal stress fibers and retrograde flow of transverse arcs in mDia1/DRF1 knockdown cells was quantified using the method described for wild-type cells (Fig. 3, B and D). The rate of dorsal stress fiber elongation was reduced from 0.3 (wild type) to 0.1 μm/min in mDia1/DRF1 knockdown cells (Fig. 4 C, filament elongation). Depletion of mDia1/DRF1 also resulted in an abnormal dorsal stress fiber morphology and an accumulation of α-actinin in these structures (Fig. 4 D and Videos 5–7, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). Treatment of cells with transfection reagents or with random RNAi oligonucleotides affected neither the rate of dorsal stress fiber elongation nor accumulation of α-actinin to dorsal stress fibers (see Fig. 6 E; not depicted). Expression of the actin polymerization–defective mDia1/DRF1 mutant in U2OS cells resulted in morphological defects in dorsal stress fibers similar to the ones observed in mDia1/DRF1 knockdown cells (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). These control experiments indicate that the effects were specific for mDia1/DRF siRNA oligonucleotides and that the actin filament nucleation/polymerization activity is crucial for the function of this protein. The transverse arc assembly and morphology were relatively normal in mDia1/DRF1 knockdown cells, although the rate of transverse arc retrograde flow was slightly diminished, as compared with wild-type cells (Fig. 4 C, arc flow; and Videos 5–7).
Figure 4.
mDia1/DRF1 depletion affects elongation rate and morphology of dorsal stress fibers. (A) Western blot analysis demonstrating the mDia1/DRF1 protein levels in wild-type (wt) and mDia1/DRF1 siRNA–transfected (-Dia) U2OS cells. Equal amounts of cell lysates (10 μg) were run on polyacrylamide gel, and mDia1/DRF1 (α-Dia) and actin (α-actin) were visualized by Western blotting. (B) mDia1/DRF1 levels are decreased in mDia1/DRF1 siRNA–transfected cells. U2OS cells were transfected with Alexa 488–labeled mDia1/DRF1 (Dia) siRNA oligonucleotides and replated as a mixture with wild-type cells. mDia1/DRF1 is visualized with anti-mDia1/DRF1 (α-Dia) antibody staining (top). Cells containing Alexa 488–mDia1/DRF1 siRNA oligos (bottom, cell highlighted with a white line) have clearly reduced expression levels of mDia1/DRF1 (top). (C) Comparison of dorsal stress fiber elongation rates in wild-type cells (mean of 18 stress fibers) and in mDia1/DRF1 siRNA–transfected cells (mean of 22 stress fibers). Also, the rate of centripetal flow of transverse arcs (arc flow) was determined from both cell populations (wild type, mean of 8 arcs; -Dia, mean of 9 arcs). SEMs and statistical significance, calculated by Mann-Whitney U test, are indicated in the graph. (D) Live cell analysis of wild-type (top) and mDia1/DRF1 knockdown cells (Dia-siRNA, two examples). Cells were transfected with α-actinin–YFP (green) and with zyxin-CFP (red; at the bottom panel, only α-actinin is shown). In wild-type cells, α-actinin was aligned regularly throughout the dorsal stress fibers, whereas in mDia1/DRF1 knockdown cells irregular accumulation of α-actinin into dorsal stress fibers was detected. See Videos 5–7 (available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). Bars, 10 μm.
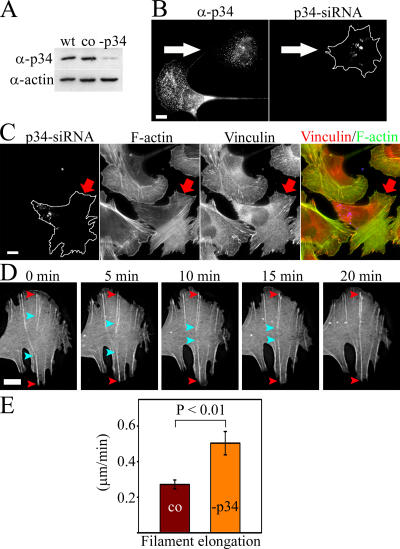
Figure 6.
p34 depletion results in a loss of lamellipodia and transverse arcs. (A) Western blot analysis demonstrating the p34 protein levels in wild-type (wt) cells, in control cells treated with transfection reagents (co), and in p34 siRNA–transfected (-p34) U2OS cells. Equal amounts of cell lysates (15 μg) were run on polyacrylamide gel, and p34 (α-p34) and actin (α-actin) were visualized by Western blotting. (B) U2OS cells were transfected with Alexa 488–labeled p34 siRNA oligonucleotides and replated as a mixture with wild-type cells. p34 was visualized with anti-p34 antibody (left, α-p34). The cell containing Alexa 488–p34 siRNA oligonucleotides (right, arrow) displays reduced p34 levels (left, arrow). (C) Cells containing Alexa 488–p34 siRNA oligonucleotides (left, arrow) displayed a loss of lamellipodal actin network and transverse arcs. F-actin was visualized by phalloidin (F-actin), and focal adhesions were labeled with anti-vinculin antibody (vinculin). (D) Live cell analysis of p34 knockdown cell expressing GFP-actin. Only the lamella of a polarized cell is shown. Dorsal stress fibers elongate from sides of lamella toward the center of lamella. The ends of two dorsal stress fibers growing from the opposite sides of the lamella are highlighted with red and cyan arrowheads. Endwise annealing of the two dorsal stress fibers leads to a formation a stress fiber attached to substrate from both ends. See Video 9 (available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). (E) Comparison of dorsal stress fiber elongation rates in control cells treated with transfection reagents (mean of 25 stress fibers) and in p34 siRNA–transfected cells (mean of 13 stress fibers). Statistical significance, calculated by Mann-Whitney U test, and SEMs are indicated in the graph. Bars, 10 μm.
Collectively, these data show that mDia1/DRF1 formin plays an important role in actin polymerization during dorsal stress fiber elongation. However, it is important to note that although the elongation and morphology of dorsal stress fibers in mDia1/DRF1 knockdown cells were severely abnormal, some actin polymerization still occurred at focal adhesions. This suggests that in addition to mDia1/DRF1-mediated filament elongation, other formin family proteins may also contribute to actin polymerization at focal adhesions.
To reveal the mechanism and timing of myosin incorporation into elongating dorsal stress fibers, we simultaneously visualized α-actinin–CFP and YFP-MLC in live U2OS cells. Dorsal stress fibers first elongated from focal adhesions as α-actinin cross-linked bundles. In general, elongating dorsal stress fibers did not contain detectable amounts of myosin II. Myosin spots only appeared in these bundles after they reached a length of several micrometers and connected to transverse arcs (or converted to ventral stress fibers). Interestingly, myosin bundles were inserted in the middle of the α-actinin cross-linked bundles. Simultaneous with the appearance of a myosin spot in the stress fiber, α-actinin was displaced from this region, demonstrating that myosin and α-actinin binding to actin filaments in stress fibers are mutually exclusive (Fig. 3 E, white arrowheads). As more myosin bundles were integrated into the actin bundle, the dorsal stress fibers gained a periodic α-actinin–myosin pattern (Fig. 3 E, last frame, arrows).
Mechanism of transverse arc assembly
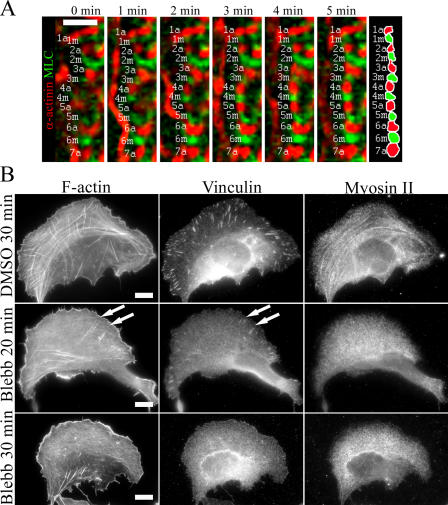
We next examined the assembly mechanism of the transverse arcs. As described above (Fig. 1 A), transverse arcs do not directly associate from their ends to focal adhesions and thus are likely to be assembled through a mechanism distinct from the one described in Fig. 3 for dorsal stress fibers. Live cell analysis of GFP-actin– and α-actinin–YFP–expressing cells (Fig. 1 B and Videos 1 and 8, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1) suggested that transverse arcs assemble from short actin bundles generated at the leading lamellipodium. These structures then flow toward the cell center and condense to form contractile transverse arcs. To reveal how myosin is incorporated into these structures, we followed the formation of transverse arcs in U2OS cells expressing α-actinin–CFP and YFP-MLC. In contrast to dorsal stress fibers, where myosin was incorporated into α-actinin cross-linked actin bundles, the periodic α-actinin–myosin pattern in transverse arcs arose from end-to-end annealing of α-actinin– and myosin-containing bundles (Fig. 5 A, left frames). Myosin II and α-actinin cross-linked actin filaments subsequently formed more regular structures through an equalization of the widths of the α-actinin and myosin bands and by straightening the entire bundle (Fig. 5 A, right frames). The intensity of some myosin II bands appeared to increase during the transverse arc formation, suggesting that additional myosin II molecules can be added to myosin II bundles during the maturation and centripetal flow of the transverse arc.
Figure 5.
Mechanism of transverse arc assembly. (A) A U2OS cell expressing α-actinin–CFP (red) and YFP-MLC (green) was monitored by time-lapse imaging. Individual α-actinin (a) and myosin (m) bands are indicated by numbers (see diagram on the right). Cell edge is located on the left side and the center of the cell on the right side. Transverse arcs are generated by endwise annealing of short α-actinin cross-linked actin and myosin bundles. Bar, 5 μm. (B) U2OS cells were treated with 90% DMSO (control) for 30 min (top) or with 50 μM blebbistatin for 20 min (middle) or 30 min (bottom). Cells were fixed, and F-actin, vinculin (focal adhesions), and myosin II were visualized by phalloidin and anti-vinculin and anti–myosin II antibodies, respectively. White arrows indicate remaining focal adhesions and dorsal stress fibers in blebbistatin-treated cells, whereas transverse arcs were no longer detected in these cells after 20 min of blebbistatin treatment. Bars, 10 μm.
Because transverse arc formation was not severely affected by mDia1/DRF1 depletion (Fig. 4) and the filaments for transverse arcs appeared to originate from the lamellipodial network, we examined the role of the Arp2/3 complex in transverse arc assembly. Transfection of U2OS cells with fluorescently labeled siRNA oligonucleotide duplexes targeted to the p34 subunit of the Arp2/3 complex substantially depleted p34 expression as determined by Western blotting. Treatment of cells with transfection reagents did not alter p34 expression level in comparison to wild-type cells (Fig. 6 A). Correspondingly, a clear reduction in p34 levels was detected by immunofluorescence in cells transfected with fluorescent p34 siRNA oligonucleotides as compared with nontransfected wild-type cells (Fig. 6 B). Depletion of the p34 led to nearly total disappearance of lamellipodial actin structures from U2OS cells, supporting the central role of this actin-nucleating complex at the leading edge of motile cells (Fig. 6 C). Interestingly, p34 depletion also led to an almost total disappearance of transverse arcs, whereas the knockdown cells still displayed dorsal stress fibers. Focal adhesions in p34 knockdown cells were located very close to the cell edge, at the sides of the lamellae (Fig. 6 C). Control cells treated with transfection reagents or random oligonucleotide duplexes did not show reduction in lamellipodial actin or absence of transverse arcs (unpublished data). The disruption of the Arp2/3 localization in cells by expressing Scar1-WA (Machesky and Insall, 1998) led to a phenotype similar to that of p34 siRNA treatment (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). These findings suggest that the p34 knockdown phenotype described was specific for depletion of the Arp2/3 complex and did not result from the nonspecific effects of transfection reagents or siRNA.
Live cell analysis of the p34 knockdown cells revealed that dorsal stress fibers grew from focal adhesions toward the cell center (Fig. 6 D and Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). Interestingly, the growth rate was faster (0.50 μm/min) than in wild-type cells and in control cells treated with transfection reagents (0.27 μm/min; Fig. 6 E), indicating that the Arp2/3 complex does not promote elongation of dorsal stress fibers. We propose that the enhanced growth rate of dorsal stress fibers in p34 knockdown cells may result from an increase in the cytoplasmic G-actin concentration in the absence of Arp2/3-nucleated barbed ends. This is supported by the results demonstrating that the elongation rate of Mg-ATP-actin in the presence of formins is directly proportional to the monomer concentration (Kovar et al., 2006). By following the growth of dorsal stress fibers in p34 knockdown cells, we also detected that dorsal stress fibers growing from the opposite sides of lamellae could anneal together in an endwise manner to generate a ventral stress fiber attached to the substrate from both its ends (Fig. 6 D, right frames; and Video 9).
To investigate the role of myosin II activity in transverse arc formation, we treated cells with blebbistatin, an inhibitor of myosin ATPase activity (Straight et al., 2003). Treatment of cells with 50 μM blebbistatin for 20 min led to a complete loss of transverse arcs and myosin II localization in most cells. In contrast, a fraction of focal adhesions and dorsal stress fibers were still visible after 20 min of blebbistatin treatment. Extension of the incubation time to 30 min resulted in a disappearance of focal adhesions and thus probably disappearance of dorsal stress fibers (Fig. 5 B). The chronology of the disappearance of different structures was further examined by video microscopy using GFP-actin–transfected U2OS cells (Video 10, available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1). Acquisition of time-lapse images every 3 min confirmed that transverse arcs disappeared first after myosin inhibition, followed by the disappearance of dorsal stress fibers. Sensitivity of blebbistatin to blue light (Sakamoto et al., 2005) may explain the less extensive disappearance of stress fibers in time-lapse images as compared with fixed cells. Together, these data suggest that transverse arc formation is strictly dependent on myosin activity, whereas effects of myosin inhibition to dorsal stress fibers may be more indirect and mediated by the disappearance of focal adhesions. It is also possible that the delayed dorsal stress fiber disappearance after blebbistatin treatment results from lower sensitivity of dorsal stress fiber myosin to blebbistatin or from the fact that dorsal stress fibers typically contain fewer myosin bundles than transverse arcs.
Dynamics of stress fiber components
To obtain a more accurate picture of the dynamics of contractile actin arrays, we determined the association/dissociation rates of the three main stress fiber components—actin, α-actinin, and myosin—by FRAP analysis. All three types of contractile actin arrays—dorsal stress fibers, transverse arcs, and ventral stress fibers—were analyzed. In contrast to the FRAP analysis shown in Fig. 3 C, we examined the general recovery rates of the bleached regions and, in the case of elongating dorsal stress fibers, excluded the “growth region” from the analyses. Thus, these analyses were expected to reflect the association–dissociation dynamics of proteins to stress fibers.
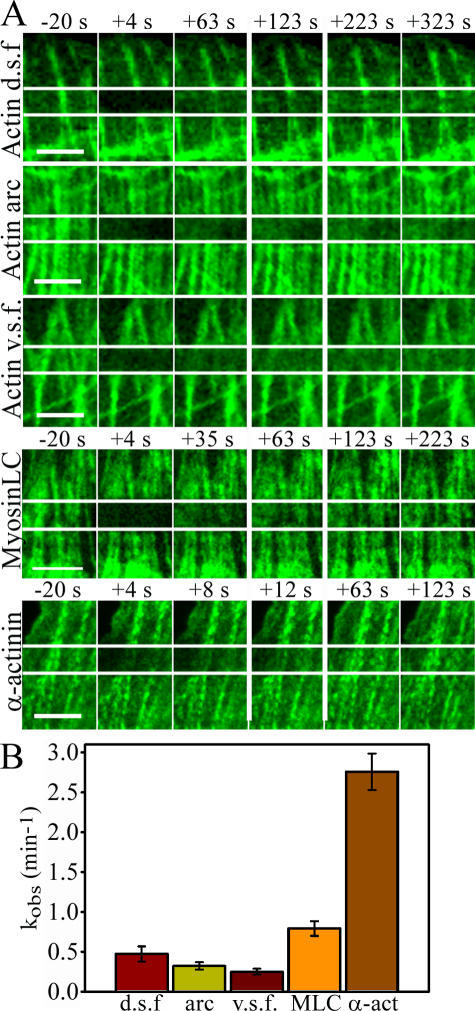
Relative to α-actinin and MLC recovery rates, the recovery of GFP-actin in the bleached regions of all stress fiber types was slow, indicating that the actin filaments in stress fibers are relatively stable (Fig. 7). Also, dynamics of MLC (Fig. 7) and myosin heavy chain (not depicted) in stress fibers were relatively slow in U2OS cells. Interestingly, our FRAP analyses revealed that α-actinin associated with all three types of stress fibers in a highly dynamic manner (Fig. 7 and not depicted). For an actin filament cross-linking protein, the fast association/dissociation rate of α-actinin was surprising but in agreement with the observed rapid association/dissociation rates of α-actinin to actin filaments in vitro (Goldmann and Isenberg, 1993).
Figure 7.
Dynamics of stress fiber components. Association/dissociation rates of GFP-actin were analyzed by FRAP from different types of contractile actin arrays. In addition, association/dissociation rates of MLC (GFP-MLC) and α-actinin (α-actinin–GFP) were analyzed from ventral stress fibers. (A) Time-lapse images before photobleaching (−20 s) and immediately after (+4 s) are shown together with selected time-lapse frames that demonstrate the rate of fluorescence recovery. The fourth frame (surrounded by wider white lines) represents the time point when approximately half of the fluorescence was recovered. (B) The rates of fluorescence recovery of individual filament bundles were analyzed by Leica software, half-recovery time was quantified from 8–9 recovery curves for each category, and kobs values were calculated. SEMs are indicated in the graph. d.s.f., dorsal stress fibers; v.s.f., ventral stress fibers Bars, 5 μm.
Discussion
We examined the assembly mechanisms of the three types of stress fibers in cultured U2OS cells by live cell microscopy methods and determined the dynamics of the most central stress fiber components by FRAP. Our analyses revealed the following: (1) Dorsal stress fibers are generated through formin (mDia1/DRF1)–driven actin polymerization at focal adhesions. (2) Myosin bundles can be subsequently incorporated into the α-actinin cross-linked actin bundles of dorsal stress fibers, although this incorporation typically occurs only after dorsal stress fibers have associated with transverse arcs at their proximal ends. (3) In contrast to dorsal stress fibers, transverse arcs form by the end-to-end annealing of cortical Arp2/3-nucleated actin bundles and myosin bundles. Experiments using blebbistatin demonstrated a critical role of myosin II activity in the transverse arc assembly and maintenance. (4) Ventral stress fibers, which are associated with focal adhesions at both ends, can be generated from the preexisting transverse arc/dorsal stress fiber network in U2OS cells. The latter also provides an explanation for how contractile actin bundles that are attached to substratum at their two ends can be generated in cells.
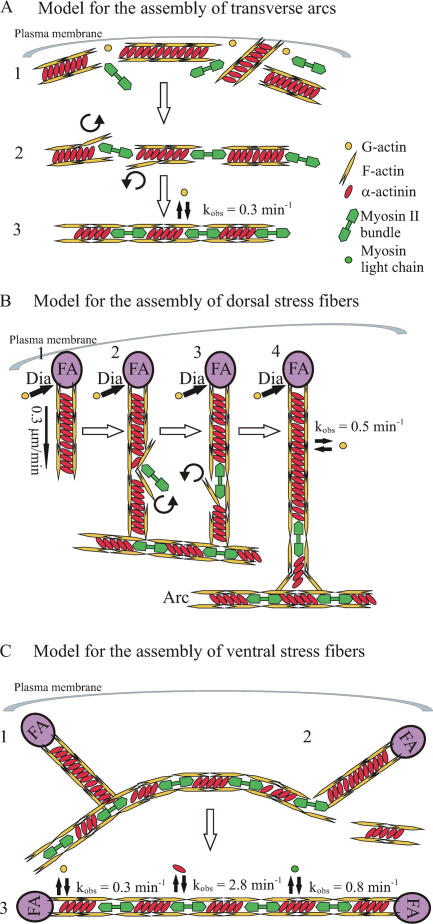
Models for dorsal stress fiber, transverse arc, and ventral stress fiber assembly based on the data obtained here are presented in Fig. 8. However, it is important to note that in addition to the model presented in Fig. 8 C, ventral stress fibers may also be generated by other mechanisms, such as the annealing of short focal adhesion–attached actin bundles (Zimerman et al., 2004). This is also supported by our observations showing that in Arp2/3 knockdown cells, two dorsal stress fibers growing from opposite sides of the lamella could fuse with each other to form a ventral stress fiber.
Figure 8.
Model for the assembly of transverse arcs, dorsal stress fibers, and ventral stress fibers. (A) Transverse arc. (1) Short actin bundles cross-linked by α-actinin are generated at the plasma membrane through nucleation by the Arp2/3 complex. (2) Actin bundles associate endwise with myosin II bundles close to the plasma membrane. (3) During centripetal flow, the α-actinin and myosin II bands are equalized in width, the entire bundle is straightened, and the width of bands is decreased, apparently as the result of a contraction of the transverse arc. (B) Dorsal stress fiber. (1) After formation of a focal adhesion (FA), short unipolar actin filaments are polymerized by mDia1/DRF1-dependent mechanism (Dia) from the focal adhesion at the rate of 0.3 μm/min. Polymerized actin filaments are simultaneously cross-linked by α-actinin. (2–4) The proximal end of a dorsal stress fiber is connected to the side of a transverse arc. When dorsal stress fiber has reached the length of several micrometers, myosin II can be occasionally incorporated into α-actinin cross-linked bundle, leading to a simultaneous displacement of α-actinin. (C) Ventral stress fiber. (1) Preassembled dorsal stress fibers and arcs interact with each other. (2) The transverse arc region that is not located between the two dorsal stress fibers is disconnected from the structure. (3) The transverse arc aligns between the two dorsal stress fibers, contracts, and subsequently forms a ventral stress fiber that is anchored to focal adhesions at both ends. The dynamics of actin, α-actinin, and MLC association to stress fibers are indicated in the figure. Note that α-actinin associates with stress fibers in a highly dynamic manner. This dynamic filament cross-linking may be essential for myosin incorporation into dorsal stress fibers as well as for contractility of mature stress fibers.
Together, our data provide a plausible explanation for the previous inconsistencies in the mechanism of stress fiber assembly. Earlier live cell microscopy analyses provided evidence that the initial site of stress fiber assembly involves discrete spots near the cell edge, followed by a unidirectional growth/stretching of actin bundles from this site (Wang, 1984). In contrast, other studies suggested that stress fibers are generated from preexisting short actin bundles, which assemble together to generate longer, contractile structures (Machesky and Hall, 1997; Zimerman et al., 2004). Our model proposing two distinct assembly mechanisms of stress fibers, formin-driven assembly at focal adhesions during dorsal stress fiber generation and endwise annealing of short myosin bundles and Arp2/3-nucleated actin bundles during transverse arc formation is in good agreement with these earlier, seemingly contradictory observations. Furthermore, our FRAP analysis provided direct evidence that dorsal stress fibers elongate through actin polymerization at focal adhesions and not through insertion of short actin filaments at the proximal ends of these structures as previously suggested (Heath and Holifield, 1993; Small and Resch, 2005).
Analysis of stress fiber component dynamics by FRAP demonstrated that actin filaments are relatively stable in all three types of actin bundles. However, our previous analyses demonstrated that stress fibers became more prominent and their turnover rates decreased when central regulators of actin filament depolymerization, actin depolymerizing factor/cofilin or cyclase-associated protein, were depleted from cells (Bertling et al., 2004; Hotulainen et al., 2005). This observation suggested that, despite slow turnover rates, the association and dissociation of actin monomers to and from stress fibers is promoted by actin binding proteins. Interestingly, the association of α-actinin with stress fibers is highly dynamic. We propose that a dynamic cross-linking of actin filaments during the formation of stress fibers and in mature stress fibers may be essential for the following reasons: First, the dynamic association of α-actinin may be necessary for the incorporation of myosin into dorsal stress fibers. Rapid exchange of this actin cross-linker would allow myosin motor domains to interact with actin filaments because myosin and α-actinin binding to actin filaments are mutually exclusive. Second, dynamic cross-linking could allow rotation of actin filaments to convert a unipolar noncontractile structure to a bipolar actin structure, which, together with a myosin bundle, is capable of contracting. Consistent with this hypothesis, it was recently shown that expression of a mutant α-actinin with defects in PI(4,5)P2 binding and slower association/dissociation rate to actin filaments in vivo results in the formation of abnormal stress fibers (Fraley et al., 2005). Finally, it is proposed that during stress fiber contraction, α-actinin must be progressively displaced to allow the movement of myosin toward the barbed ends on α-actinin cross-linked filaments (Peterson et al., 2004). Therefore, we suggest that dynamic cross-linking of actin filaments is also important for the contractility of stress fibers.
In addition to stress fibers, multicellular organisms display other types of contractile actin filament bundles, such as myofibrils in muscle cells. The assembly of myofibrils is generated through intermediates, namely, premyofibrils and nascent myofibrils. Premyofibrils consist of actin, α-actinin, and nonmuscle myosin II and, interestingly, the assembly of premyofibrils visualized with α-actinin–GFP closely resembled the assembly of transverse arcs described in this study. Similar to transverse arcs, formation of premyofibrils begins at the spreading edges of the myocytes (Dabiri et al., 1997). We speculate that a molecular assembly mechanism similar to the transverse arc assembly pathway revealed here may also contribute to the formation of the periodic contractile actin structures of muscle cells.
Together, these studies form a framework explaining how contractile actin stress fibers are assembled in cultured U2OS cells. In the future, it will be important to examine whether stress fibers in other cell types are generated through similar or different mechanisms. Recent studies have also shown that mechanical stretching induces changes in the morphology and composition of stress fibers (Kaunas et al., 2005; Yoshigi et al., 2005). Therefore, how actin polymerization in focal adhesions and myosin incorporation into actin filament bundles are regulated during maturation and stretching of a ventral stress fiber provides important challenges for future research.
Materials and methods
Cell culture and immunofluorescence microscopy
Human osteosarcoma (U2OS) cells (a gift from T. Mäkelä and T. Vallenius, University of Helsinki, Helsinki, Finland) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone), 2 mM l-glutamine, penicillin, and streptomycin (Sigma-Aldrich). For immunofluorescence, the U2OS cells were plated 3 h before fixation on coverslips precoated with 10 μg/ml fibronectin. Immunofluorescence was performed as described previously (Vartiainen et al., 2000). As an exception, for the mDia1/DRF1 stainings, cells were permeabilized with 0.05% Saponin. Vinculin was visualized with a monoclonal anti-vinculin antibody (dilution 1:140; Sigma-Aldrich); mDia1/DRF1 with monoclonal anti-mDia1 antibody (dilution 1:100; BD Biosciences); p34 with rabbit polyclonal anti-p34 antibody (dilution 1:50; Upstate Biotechnology); α-actinin with monoclonal α-actinin antibody (dilution 1:50; Sigma-Aldrich); myosin II with rabbit anti–nonmuscle myosin antibodies (dilution 1:60; Biomedical Technologies); and secondary antibodies conjugated to FITC, rhodamine, or Cy5 (Invitrogen). F-actin was visualized with Alexa 488 or 568 phalloidin (dilution 1:100; Invitrogen). Images were acquired through a charge-coupled device camera (DP70; Olympus) on a microscope (AX70 Provis; Olympus). For the image acquirement, the AnalySIS software (Olympus) and PlanApo 60×/1.40 (oil) or UPlanApo 100×/1.35 (oil) objectives (Olympus) were used. (−)-Blebbistatin was obtained from Sigma-Aldrich.
Plasmid construction and transient transfections
Human GFP–β-actin plasmid (Choidas et al., 1998) was a gift from M. Bähler (Westfalian Wilhelms-University, Münster, Germany). YFP–β-actin was purchased from CLONTECH Laboratories, Inc. Human nonmuscle α-actinin-1 cDNA cloned to the 5′-end of the humanized S65T version of GFP (Edlund et al., 2001) was a gift from C. Otey (University of North Carolina School of Medicine, Chapel Hill, NC). The cDNA encoding α-actinin was amplified by PCR (primers 5′-GCCGCTCGAGATGGACCATTATGATTCTCAGCA-3′ and 5′-GCCGGAATTCCGAGGTCACTCTCGCCGTACAG-3′), and the PCR fragment was subcloned into the XhoI–EcoRI sites of the pECFP-N1 and pEYFP-N1 vectors (CLONTECH Laboratories, Inc.) to generate plasmids expressing α-actinin–CFP and –YFP fusion proteins. PCR fragment was verified by sequencing. GFP-MLC (Peterson et al., 2004) was a gift from L. Peterson (University of North Carolina School of Medicine, Chapel Hill, NC). To create YFP-MLC, MLC cDNA was excised with XhoI and BamHI from GFP-MLC plasmid and ligated into the corresponding sites of the pEYFP-C1 vector (CLONTECH Laboratories, Inc.). Zyxin-GFP plasmid (Rottner et al., 2001) was a gift from K. Rottner (German Research Center for Biotechnology, Braunschweig, Germany). The zyxin cDNA was transferred from zyxin-GFP into pECFP-N1 vector (CLONTECH Laboratories, Inc.) using the EcoRI and BamHI restriction sites to generate zyxin-CFP plasmid. To generate the GFP-mDia1 Ile810Ala mutation construct, the GFP-mDia1 construct (a gift from N. Watanabe, Kyoto University, Kyoto, Japan; Higashida et al., 2004) was mutated by inverse PCR by using primers 5′-CCTTTTTGGGTTCATTCCGCATGCCCTA-3′ and 5′-CTGAGAGATTCTGCGCTGTCTTTGAATC-3′. This generated two nucleotide changes to the mDia1 target sequence (TCAATCTTT sequence was mutated to TCAGCCTTT), which altered isoleucine 810 residue to alanine. mDia1 Ile810 is comparable to Bni1p Ile1341 (Xu et al., 2004). Myc-tagged Scar1-W and -WA fragments were gifts from L. Machesky (University of Birmingham, Birmingham, UK; Machesky and Insall, 1998). Transient transfection of U2OS cells was performed with FuGENE6 (Roche) using 25% of the recommended DNA and FuGENE amounts according to the manufacturer's instructions.
siRNA treatment and Western blotting
For the siRNA experiments, 1,500 ng of preannealed 3′ Alexa Fluor 488–labeled mDia1/DRF1 siRNA (target sequence AAAGGCAGAGCCACACTTCCT [Unsworth et al., 2004]; QIAGEN) or p34 siRNA (target sequence AAGGAACTTCAGGCACATGGT) oligonucleotide duplexes were transfected into cells on 6-well plates by using GeneSilencer's siRNA-transfection reagent (Gene Therapy Systems) according to the manufacturer's recommendations. After 44 or 68 h, the cells were detached with trypsin-EDTA, diluted, and plated on precoated (10 μg/ml fibronectin) glass-bottomed dishes (MatTek) for the live cell imaging. Analysis of the cells was performed 46–52 h after transfection. The cells for Western blotting were washed three times with cold PBS, scraped, and lysed with PBS containing 1% Triton X-100 and 0.3 mM PMSF. Total protein concentrations were measured using Bradford reagent (Sigma-Aldrich). Anti-mDia1 antibody (BD Biosciences) was used at a dilution of 1:500, anti-p34 (Upstate Biotechnology) at a dilution of 1:500, and anti-actin AC-15 antibody (Sigma-Aldrich) at a dilution of 1:10,000.
Live cell microscopy
Transfected cells were replated on 10 μg/ml fibronectin–coated glass-bottomed dishes. Normal growth medium was used as imaging medium. The time-lapse images were acquired with an inverted microscope (IX-71; Olympus) equipped with a Polychrome IV monochromator (TILL Photonics) with the appropriate filters, heated sample environment (+37°C), and CO2 control. UApo 40×/1.35 (oil) objective (Olympus) with 1.6× magnification or PLAPON 60×O TIRFM 60×/1.45 (oil) objective (Olympus) were used. The software used for the image acquirement was TILL Vision 4 (TILL Photonics). Total internal reflectance fluorescence analysis was performed with the same microscope setup using a 488-nm laser line. The cameras used for the study were Imago QE (TILL Photonics) and Andor iXon (Andor). Time-lapse videos were deconvoluted by AutoQuant AutoDeblur 2D Blind Deconvolution (AutoQuant Imaging, Inc.).
FRAP
To analyze the association/dissociation rates of stress fiber components, U2OS cells expressing various GFP constructs were grown for 24 h on glass-bottomed dishes or 3–5 h on glass-bottomed dishes precoated with 10 μg/ml fibronectin. Normal growth medium was used as imaging medium. Confocal imaging was performed on a confocal microscope (TCS SP2 AOBS; Leica) equipped with Leica Confocal Software (Lite 2.61.1537), heating (+37°C), and CO2 control. For GFP imaging, a 488-nm line and a HCX PL APO 63×/1.4–0.6 (oil) objective were used. After three prebleach scans of an entire image, five bleaching scans (3.9 s each) with 100% intensity of 476 nm (15 mW), 488 nm (70 mW), and 496 nm (15 mW) laser lines over the region of interest (2.5 × 20 μm) were performed. After bleaching, the fluorescence recovery was monitored 10 times every 3.9 s and 15 times every 20 s. The recovery of the GFP intensity was measured by Leica Confocal Software. The intensity of the bleached area was normalized to a neighboring nonbleached area to diminish the error caused by normal photobleaching during the monitoring period. Bleached and control areas used for measurements were also outlined to contain only one stress fiber to diminish fast intensity recovery caused by the diffusion of soluble proteins. The values of intensity versus time (min) were charted in a scatter plot, the recovery half-time (t1/2) was measured from the plots, and the kobs values (first-order rate constant) were calculated by using the equation kobs = 0.693/t1/2.
Elongation rate measurements
Stress fiber elongation and arc flow rates were measured from GFP-actin videos acquired using TILL Photonics imaging system. The length of dorsal stress fibers (μm) was measured from every 10th time-lapse frame (5 min) by Bitplane Imaris suite software (Bitplane AG). The length versus time was plotted in a chart, and k (length change in μm/min) was calculated from the linear trend line (Excel; Microsoft). Polymerization rates from focal adhesions were measured by bleaching the region of interest as described in the previous section, and the elongation of bright stress fiber region from the focal adhesion was measured similarly to the elongation of an entire stress fiber.
Online supplemental material
Fig. S1 shows that the overexpression of a loss-of-function mDia1/DRF1 mutant induces abnormal dorsal stress fiber morphology and accumulation of α-actinin in these structures. Fig. S2 shows that expression of Scar-WA in U2OS cells leads to a loss of lamellipodia and transverse arcs. Video 1 displays actin dynamics in U2OS cells visualized by GFP-actin. Video 2 shows actin dynamics in U2OS cells visualized with GFP-actin. Video 3 shows ventral stress fiber assembly visualized with YFP-actin and zyxin-CFP. Video 4 shows dorsal stress fiber elongation visualized by zyxin-CFP and α-actinin–YFP in U2OS cells. Video 5 displays dorsal stress fibers and arcs visualized by α-actinin–YFP in U2OS cells. Video 6 shows dorsal stress fibers and arcs visualized by zyxin-CFP and α-actinin–YFP in mDia1/DRF1 siRNA–transfected U2OS cells. Video 7 shows dorsal stress fibers and arcs visualized by zyxin-CFP and α-actinin–YFP in mDia1/DRF1 siRNA–transfected U2OS cells. Video 8 demonstrates arc assembly from short α-actinin cross-linked actin filament bundles generated at the leading lamellipodium. Video 9 shows dorsal stress fibers visualized by GFP-actin in a p34 siRNA–transfected U2OS cells. Video 10 shows the effect of blebbistatin treatment in U2OS cells expressing GFP-actin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200511093/DC1.
Supplementary Material
Acknowledgments
The authors are grateful to Tomi Mäkelä and Tea Vallenius for providing U2OS cells and Art Alberts, Martin Bähler, Laura Machesky, Gerard Marriott, Carol Otey, Lynda Peterson, Klemens Rottner, and Naoki Watanabe for constructs and antibodies. Petri Auvinen, Mikko Frilander, Bruce Goode, Keith Kozminski, Oscar Puig, and J. Victor Small are acknowledged for discussions and critical reading of the manuscript. Risto Hotulainen is acknowledged for the statistical support and Johan Peränen for the help with movie conversions. We thank Kimmo Tanhuanpää for the excellent maintenance of the microscopy unit and Miia Koskinen for outstanding technical assistance.
This study was supported by grants from the Academy of Finland (SA 212657 and SA 203655) and the Emil Aaltonen Foundation.
Abbreviation used in this paper: MLC, myosin light chain.
References
- Bear, J.E., T.M. Svitkina, M. Krause, D.A. Schafer, J.J. Loureiro, G.A. Strasser, I.V. Maly, O.Y. Chaga, J.A. Cooper, G.G. Borisy, and F.B. Gertler. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521. [DOI] [PubMed] [Google Scholar]
- Bertling, E., P. Hotulainen, P.K. Mattila, T. Matilainen, M. Salminen, and P. Lappalainen. 2004. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell. 15:2324–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choidas, A., A. Jungbluth, A. Sechi, J. Murphy, A. Ullrich, and G. Marriott. 1998. The suitability and application of a GFP-actin fusion protein for long-term imaging of the organization and dynamics of the cytoskeleton in mammalian cells. Eur. J. Cell Biol. 77:81–90. [DOI] [PubMed] [Google Scholar]
- Cramer, L.P., M. Siebert, and T.J. Mitchison. 1997. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 136:1287–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri, G.A., K.K. Turnacioglu, J.M. Sanger, and J.W. Sanger. 1997. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc. Natl. Acad. Sci. USA. 94:9493–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund, M., M.A. Lotano, and C.A. Otey. 2001. Dynamics of alpha-actinin in focal adhesions and stress-fibers visualized with alpha-actinin-green fluorescent protein. Cell Motil. Cytoskeleton. 48:190–200. [DOI] [PubMed] [Google Scholar]
- Fraley, T.S., C.B. Pereira, T.C. Tran, C. Singleton, and J.A. Greenwood. 2005. Phosphoinositide binding regulates alpha-actinin dynamics: mechanism for modulating cytoskeletal remodeling. J. Biol. Chem. 280:15479–15482. [DOI] [PubMed] [Google Scholar]
- Goldmann, W.H., and G. Isenberg. 1993. Analysis of filamin and alpha-actinin binding to actin by the stopped flow method. FEBS Lett. 336:408–410. [DOI] [PubMed] [Google Scholar]
- Heath, J.P. 1983. Behaviour and structure of the leading lamella in moving fibroblasts. I. Occurrence and centripetal movement of arc-shaped microfilament bundles beneath the dorsal cell surface. J. Cell Sci. 60:331–354. [DOI] [PubMed] [Google Scholar]
- Heath, J.P., and B.F. Holifield. 1993. On the mechanisms of cortical actin flow and its role in cytoskeletal organisation of fibroblasts. Symp. Soc. Exp. Biol. 47:35–56. [PubMed] [Google Scholar]
- Higashida, C., T. Miyoshi, A. Fujita, F. Oceguera-Yanez, J. Monypenny, Y. Andou, S. Narumiya, and N. Watanabe. 2004. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 303:2007–2010. [DOI] [PubMed] [Google Scholar]
- Hotulainen, P., E. Paunola, M.K. Vartiainen, and P. Lappalainen. 2005. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 16:649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A.B., and A. Hall. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269. [DOI] [PubMed] [Google Scholar]
- Kaunas, R., P. Nguyen, S. Usami, and S. Chien. 2005. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc. Natl. Acad. Sci. USA. 102:15895–15900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff, E., J.C. Simpson, C.B. Leberfinger, I.M. Otto, T. Doerks, P. Bork, U.R. Rapp, T. Raabe, and R. Pepperkok. 2001. The Spir actin organizers are involved in vesicle transport processes. Curr. Biol. 11:1963–1968. [DOI] [PubMed] [Google Scholar]
- Kovar, D.R., E.S. Harris, R. Mahaffy, H.N. Higgs, and T.D. Pollard. 2006. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 124:423–435. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., and A. Hall. 1997. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J. Cell Biol. 138:913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M., and R.H. Insall. 1998. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8:1347–1356. [DOI] [PubMed] [Google Scholar]
- Mullins, R.D., J.A. Heuser, and T.D. Pollard. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 95:6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson-Dykstra, S., H.N. Higgs, and E.S. Harris. 2005. Actin dynamics: growth from dendritic branches. Curr. Biol. 15:R346–R357. [DOI] [PubMed] [Google Scholar]
- Pantaloni, D., C. Le Clainche, and M.F. Carlier. 2001. Mechanism of actin-based motility. Science. 292:1502–1506. [DOI] [PubMed] [Google Scholar]
- Pellegrin, S., and H. Mellor. 2005. The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol. 15:129–133. [DOI] [PubMed] [Google Scholar]
- Peterson, L.J., Z. Rajfur, A.S. Maddox, C.D. Freel, Y. Chen, M. Edlund, C. Otey, and K. Burridge. 2004. Simultaneous stretching and contraction of stress fibers in vivo. Mol. Biol. Cell. 15:3497–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, M.E., J.E. Heuser, E. Kerkhoff, and R.D. Mullins. 2005. Drosophila Spire is an actin nucleation factor. Nature. 433:382–388. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J., and A. Hall. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 70:389–399. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J., M.A. Schwartz, K. Burridge, R.A. Firtel, M.H. Ginsberg, G. Borisy, J.T. Parsons, and A.R. Horwitz. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. [DOI] [PubMed] [Google Scholar]
- Romero, S., C. Le Clainche, D. Didry, C. Egile, D. Pantaloni, and M.F. Carlier. 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 119:419–429. [DOI] [PubMed] [Google Scholar]
- Rottner, K., M. Krause, M. Gimona, J.V. Small, and J. Wehland. 2001. Zyxin is not colocalized with vasodilator-stimulated phosphoprotein (VASP) at lamellipodial tips and exhibits different dynamics to vinculin, paxillin, and VASP in focal adhesions. Mol. Biol. Cell. 12:3103–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., J. Limouze, C.A. Combs, A.F. Straight, and J.R. Sellers. 2005. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry. 44:584–588. [DOI] [PubMed] [Google Scholar]
- Schirenbeck, A., T. Bretschneider, R. Arasada, M. Schleicher, and J. Faix. 2005. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 7:619–625. [DOI] [PubMed] [Google Scholar]
- Schumacher, N., J.M. Borawski, C.B. Leberfinger, M. Gessler, and E. Kerkhoff. 2004. Overlapping expression pattern of the actin organizers Spir-1 and formin-2 in the developing mouse nervous system and the adult brain. Gene Expr. Patterns. 4:249–255. [DOI] [PubMed] [Google Scholar]
- Small, J.V., and G.P. Resch. 2005. The comings and goings of actin: coupling protrusion and retraction in cell motility. Curr. Opin. Cell Biol. 17:517–523. [DOI] [PubMed] [Google Scholar]
- Small, J.V., K. Rottner, I. Kaverina, and K.I. Anderson. 1998. Assembling an actin cytoskeleton for cell attachment and movement. Biochim. Biophys. Acta. 1404:271–281. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., A. Cheung, J. Limouze, I. Chen, N.J. Westwood, J.R. Sellers, and T.J. Mitchison. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 299:1743–1747. [DOI] [PubMed] [Google Scholar]
- Svitkina, T.M., and G.G. Borisy. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145:1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T.M., E.A. Bulanova, O.Y. Chaga, D.M. Vignjevic, S. Kojima, J.M. Vasiliev, and G.G. Borisy. 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga, T., E. Sahai, P. Chardin, F. McCormick, S.A. Courtneidge, and A.S. Alberts. 2000. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell. 5:13–25. [DOI] [PubMed] [Google Scholar]
- Unsworth, K.E., M. Way, M. McNiven, L. Machesky, and D.W. Holden. 2004. Analysis of the mechanisms of Salmonella-induced actin assembly during invasion of host cells and intracellular replication. Cell. Microbiol. 6:1041–1055. [DOI] [PubMed] [Google Scholar]
- Vartiainen, M., P.J. Ojala, P. Auvinen, J. Peränen, and P. Lappalainen. 2000. Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol. Cell. Biol. 20:1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.L. 1984. Reorganization of actin filament bundles in living fibroblasts. J. Cell Biol. 99:1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, N., T. Kato, A. Fujita, T. Ishizaki, and S. Narumiya. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136–143. [DOI] [PubMed] [Google Scholar]
- Welch, M.D., and R.D. Mullins. 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18:247–288. [DOI] [PubMed] [Google Scholar]
- Xu, Y., J.B. Moseley, I. Sagot, F. Poy, D. Pellman, B.L. Goode, and M.J. Eck. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723. [DOI] [PubMed] [Google Scholar]
- Yasuda, S., F. Oceguera-Yanez, T. Kato, M. Okamoto, S. Yonemura, Y. Terada, T. Ishizaki, and S. Narumiya. 2004. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 428:767–771. [DOI] [PubMed] [Google Scholar]
- Yoshigi, M., L.M. Hoffman, C.C. Jensen, H.J. Yost, and M.C. Beckerle. 2005. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 171:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, S.H. 2004. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 16:99–105. [DOI] [PubMed] [Google Scholar]
- Zimerman, B., T. Volberg, and B. Geiger. 2004. Early molecular events in the assembly of the focal adhesion-stress fiber complex during fibroblast spreading. Cell Motil. Cytoskeleton. 58:143–159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.