Abstract
In late mitosis and early G1, replication origins are licensed for subsequent use by loading complexes of the minichromosome maintenance proteins 2–7 (Mcm2–7). The number of Mcm2–7 complexes loaded onto DNA greatly exceeds the number of replication origins used during S phase, but the function of the excess Mcm2–7 is unknown. Using Xenopus laevis egg extracts, we show that these excess Mcm2–7 complexes license additional dormant origins that do not fire during unperturbed S phases because of suppression by a caffeine-sensitive checkpoint pathway. Use of these additional origins can allow complete genome replication in the presence of replication inhibitors. These results suggest that metazoan replication origins are actually comprised of several candidate origins, most of which normally remain dormant unless cells experience replicative stress. Consistent with this model, using Caenorhabditis elegans, we show that partial RNAi-based knockdown of MCMs that has no observable effect under normal conditions causes lethality upon treatment with low, otherwise nontoxic, levels of the replication inhibitor hydroxyurea.
Introduction
In eukaryotes, precise duplication of the genome during S phase is achieved through the initiation of replication at numerous origins distributed throughout the DNA. In late mitosis and early G1, origins are licensed for replication by loading complexes of the minichromosome maintenance proteins 2–7 (Mcm2–7), thus, forming a “prereplicative complex” (Diffley, 2004; Blow and Dutta, 2005). Licensing involves the coordinated action of the origin recognition complex (ORC) Cdc6 and Cdt1 proteins (Gillespie et al., 2001), which probably act to clamp Mcm2–7 around the DNA. Mcm2–7 are displaced from replication origins as they initiate, and they have been suggested to provide a helicase activity to unwind DNA ahead of the replication fork (Labib and Diffley, 2001). To prevent replication origins from firing more than once in a single S phase, the ability to license new origins is prevented from late G1 through to midmitosis (Diffley, 2004; Blow and Dutta, 2005). This means that whatever the problems encountered by replication forks during S phase, only previously licensed sites can be used. If two converging replication forks stall irreversibly during S phase, the DNA between them will probably remain unreplicated, potentially leading to cell death or chromosome rearrangement. Therefore, it is crucial for cells to organize their replication machinery so as to minimize the risk of this happening.
Previous work in a range of eukaryotes, including Saccharomyces cerevisiae, humans, and Xenopus laevis, has demonstrated that Mcm2–7 complexes are loaded onto DNA in an ∼20-fold excess over the number of DNA-bound ORC molecules and over the number of replication origins (Burkhart et al., 1995; Lei et al., 1996; Rowles et al., 1996; Donovan et al., 1997; Mahbubani et al., 1997; Edwards et al., 2002). However, normal replication rates are maintained when the number of Mcm2–7 molecules is reduced to 1–2 per origin (Mahbubani et al., 1997; Edwards et al., 2002; Cortez et al., 2004; Oehlmann et al., 2004; Tsao et al., 2004). This observation, termed the “MCM paradox” (Hyrien et al., 2003), has led to speculation on the possible function of these extra complexes, particularly as MCM proteins do not exclusively colocalize with sites of DNA synthesis in S phase (Madine et al., 1995; Krude et al., 1996; Dimitrova et al., 1999).
There is evidence that Mcm2–7 complexes are found on DNA at significant distances from where ORC is bound (Ritzi et al., 1998; Edwards et al., 2002; Harvey and Newport, 2003; Danis et al., 2004). It has been suggested that these distant multiple MCM complexes could cooperatively pump double-stranded DNA and, thus, unwind it (Laskey and Madine, 2003). More recently, a role for excess MCM proteins in checkpoint activation has been proposed, based on the observation that cells partially depleted of Mcm7 display a reduction in replication checkpoint signaling (Cortez et al., 2004; Tsao et al., 2004). A third possibility that has been suggested is that excess Mcm2–7 can provide replication origins for use under certain contingencies, such as incomplete DNA replication (Lucas et al., 2000; Edwards et al., 2002; Hyrien et al., 2003; Blow and Dutta, 2005). These different proposals are not mutually exclusive.
In this study, we demonstrate a role for excess Mcm2–7 complexes in licensing “dormant” replication origins. These dormant origins are effectively suppressed during unperturbed DNA replication, but can support initiation when replication forks are stalled in response to a range of replicative stresses, including replication inhibition by aphidicolin, mitomycin C, etoposide, or actinomycin D. In support of this model, we demonstrate that in Caenorhabditis elegans, partial knockdown of MCMs induces hypersensitivity to otherwise nontoxic levels of hydroxyurea (HU).
Results
Minimally licensed chromatin replicates poorly in the presence of aphidicolin and caffeine
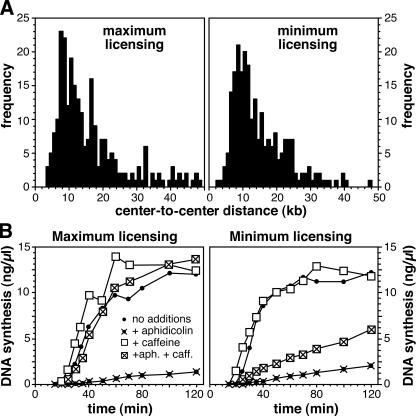
When demembranated sperm nuclei are incubated in X. laevis egg extracts, an average of 10–20 Mcm2–7 complexes are loaded onto each replication origin before entry into S phase (Mahbubani et al., 1997; Edwards et al., 2002; Oehlmann et al., 2004); we call this DNA “maximally licensed.” If the Cdt1 inhibitor geminin is added to the extract shortly after the sperm DNA, the number of DNA-bound MCM complexes can be limited to the minimum required to support approximately normal replication kinetics (Oehlmann et al., 2004); we call this DNA “minimally licensed.” To examine the role of excess Mcm2–7 complexes, we first investigated whether minimum licensing causes an alteration in the spacing between adjacent replication origins. Nascent DNA was labeled with biotin-16-dUTP during early S phase, after which the DNA was isolated, spread on microscope slides, and analyzed by fluorescence microscopy. Short fluorescent tracks were seen, which were caused by bidirectional replication from origins at the center of each track, and the spacing between replication origins was determined by measuring the distance between the centers of adjacent tracks (Herrick et al., 2000; Blow et al., 2001). Fig. 1 A shows that there was no significant difference in the average interorigin distance between the two samples (15.8 kb for maximum licensing; 17.1 kb for minimum licensing). Under normal circumstances, clusters of 3–7 adjacent origins initiate at similar times, with different clusters being activated at different stages of S phase (Blow et al., 2001). We observed no major difference between minimally and maximally licensed DNA in the clustering of replication origins, with an average of 6.1 origins per cluster for maximally licensed DNA and 4.8 for minimally licensed DNA. Therefore, we conclude that minimum licensing does not significantly change replication origin usage under normal conditions.
Figure 1.
Replication characteristics of minimally licensed DNA. (A) Sperm nuclei were incubated in X. laevis egg extract supplemented with biotin-16-dUTP. To produce minimally licensed DNA, extract was supplemented with geminin shortly after sperm addition. After 30 min, DNA was isolated and spread on glass slides, and the biotin-labeled tracks were detected with fluorescent antibodies. The distance between the center points of adjacent tracks of labeled DNA was measured. (B) Sperm nuclei were incubated in X. laevis egg extract containing α-[32P]dATP, plus or minus aphidicolin and/or caffeine. To produce minimally licensed DNA, extract was supplemented with geminin shortly after sperm addition. At the indicated times, the total amount of DNA synthesized was measured.
Next, we examined whether excess Mcm2–7 complexes become important if replication forks are put under stress by supplementing extracts with low concentrations of the DNA polymerase inhibitor aphidicolin. Fig. 1 B (crosses and closed circles) shows that the replication of minimally and maximally licensed DNA was inhibited to similar extents by 10 μM aphidicolin. We have previously shown that, at this concentration, aphidicolin slows replication forks by approximately threefold, but that the major effect on replication is to induce an ATR-dependent checkpoint that suppresses further initiation events (Luciani et al., 2004). Therefore, we also supplemented extracts with caffeine, which is an ATR kinase inhibitor (Fig. 1 B, crossed boxes). Consistent with a previous study (Luciani et al., 2004), the addition of caffeine completely rescued the aphidicolin-induced inhibition of replication with maximally licensed DNA. In contrast, caffeine only partially rescued replication when minimally licensed DNA was replicated in the presence of aphidicolin. This suggests that excess Mcm2–7 complexes are required in some way to allow rescue of DNA replication after the inhibition of replication fork progression.
Maximally licensed chromatin can use extra origins of replication
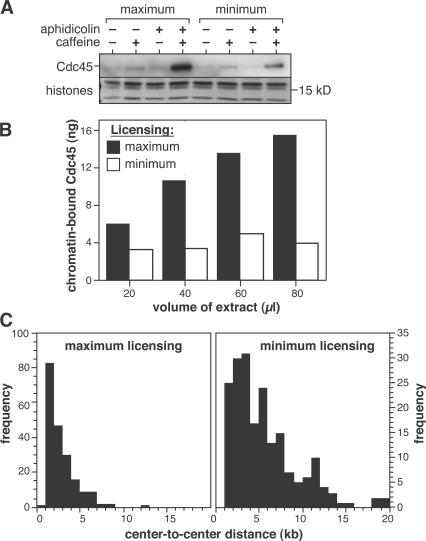
Replication forks in X. laevis normally progress at ∼10 bp/s (Callan, 1972; Mahbubani et al., 1992; Strausfeld et al., 1994; Walter and Newport, 1997), but are slowed approximately threefold by 10 μM aphidicolin (Luciani et al., 2004). Fork rate is not significantly affected by the presence of caffeine (Luciani et al., 2004). To replicate at normal rates in the presence of aphidicolin and caffeine, maximally licensed DNA must use more replication forks than normal. Minimally licensed DNA, in contrast, does not appear capable of using more replication forks. Therefore, we assessed the number of active forks by measuring the quantity of Cdc45 loaded onto chromatin. Cdc45 loads onto replication forks (probably binding Mcm2–7) just before initiation and moves with the forks as they elongate (Hopwood and Dalton, 1996; Aparicio et al., 1997, 1999; Zou and Stillman, 1998; Mimura and Takisawa, 1998). Fig. 2 A shows that both maximally and minimally licensed chromatin contained similar quantities of Cdc45 during an undisturbed S phase, as expected from their similar replication rates. Consistent with a previous study (Edwards et al., 2002), the amount of Cdc45 on maximally licensed chromatin increased ∼20-fold when extract was treated with a combination of aphidicolin and caffeine. In contrast, minimally licensed chromatin showed only a two- to threefold increase under similar conditions. A slight increase of chromatin-bound Cdc45 was also seen when both maximally and minimally licensed chromatin were treated with caffeine, which is consistent with the acceleration of the origin firing that caffeine causes (Marheineke and Hyrien, 2004; Shechter et al., 2004).
Figure 2.
Caffeine allows the initiation of extra forks on maximally licensed DNA. (A) Sperm nuclei were incubated in X. laevis egg extract, plus or minus aphidicolin and/or caffeine. To produce minimally licensed DNA, extract was supplemented with geminin shortly after sperm addition. At 40 min, chromatin was isolated and immunoblotted for Cdc45. Coomassie-stained histones are shown as a loading control. (B) Sperm nuclei (400 ng DNA) were incubated in different volumes of egg extract supplemented with 40 μM aphidicolin plus caffeine. To produce minimally licensed DNA, extract was supplemented with geminin shortly after sperm addition. At 40 min, chromatin was isolated and immunoblotted for Cdc45. (C) Sperm nuclei were incubated in egg extract supplemented with biotin-16-dUTP and caffeine. To produce minimally licensed DNA, extract was supplemented with geminin shortly after sperm addition. At 30 min, DNA was isolated and spread on glass slides, and the biotin-labeled tracks were detected with fluorescent antibodies. The distance between the center points of adjacent tracks of labeled DNA was measured.
Next, we performed quantitative immunoblotting to estimate the amount of Cdc45 loaded onto chromatin under these conditions (Fig. 2 B). A fixed quantity of X. laevis sperm nuclei was incubated in increasing quantities of egg extract supplemented with aphidicolin and caffeine. Extracts were optionally supplemented with geminin shortly after sperm addition to create minimally licensed chromatin. In the absence of added geminin (maximum licensing), the quantity of chromatin-bound Cdc45 increased with increasing extract volume (Fig. 2 B, shaded bars), caused, in part, by increased quantities of Mcm2–7 being loaded onto the DNA (Mahbubani et al., 1997). On the minimally licensed chromatin, however, Cdc45 remained at a fairly constant level of 3–5 ng (Fig. 2 B, open bars). This level of Cdc45 on minimally licensed chromatin corresponds to 1–2 molecules of Cdc45 per 10-kb DNA, or 1–2 molecules of Cdc45 per molecule of ORC (Rowles et al., 1996). If there is a single molecule of Cdc45 at each replication fork, these results suggest that under normal conditions a single molecule of ORC allows, on average, a single pair of replication forks to initiate; the number of forks can be increased under certain circumstances, but only if there are excess Mcm2–7 molecules present on the DNA.
These results suggest that maximally licensed DNA should contain an increased density of nascent strands under conditions where there are an increased number of forks. Because recent studies have shown that caffeine can accelerate origin firing (Marheineke and Hyrien, 2004; Shechter et al., 2004), we repeated the fluorescent fiber-labeling experiments using extracts supplemented with caffeine. Fig. 2 C (left) shows that when maximally licensed chromatin was replicated in extract containing caffeine the average origin spacing declined dramatically (compare with Fig. 1 A), with the observed spacing between nascent strands falling to near the resolution limit of this technique (1–2 kb). When the fiber-labeling experiments were repeated with minimally licensed chromatin, the reduction in spacing was much less pronounced than with maximally licensed DNA (Fig. 2 C, right). These data suggest that the extra Mcm2–7 molecules on maximally licensed DNA license dormant replication origins that can be used in the presence of caffeine.
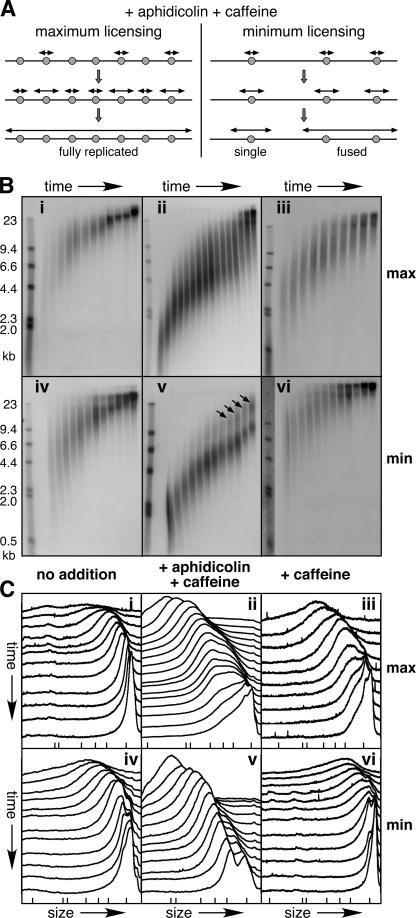
If this is correct, then nascent strands from these additional origins will very rapidly fuse into high molecular weight products that are observable on alkaline agarose gels (Fig. 3 A, left). On minimally licensed DNA, interorigin distances are larger, and nascent strand fusion should occur later (Fig. 3 A, right). To test this, maximally and minimally licensed DNA samples were pulsed with α-[32P]dATP at the beginning of S phase, and then chased with unlabeled dATP for different periods. Fig. 3 B (i and iv) shows that maximally and minimally licensed DNA gave very similar profiles in the absence of any drug treatment. Although the long smear of nascent strands makes an accurate rate determination impossible, the modal size of nascent strands increased at a rate of ∼25 nt/s. Within expected error, this is consistent with each strand being elongated by two forks, each moving at 10 nt/s, as previously reported (Callan, 1972; Mahbubani et al., 1992; Strausfeld et al., 1994; Walter and Newport, 1997; Luciani et al., 2004).
Figure 3.
Maximally licensed DNA can undergo premature nascent strand fusion in the presence of caffeine. (A) Model for the use of additional dormant origins in extract treated with aphidicolin and caffeine. A small segment (∼30 kb) of chromosomal DNA is shown, which normally supports initiation from three origins. Candidate replication origins with bound Mcm2–7 are shown as gray circles. Nascent strands are shown as double-headed arrows. On maximally licensed DNA (left), nascent strands initiate from dormant origins and rapidly fuse. On minimally licensed chromatin (right), there are no dormant origins so the first strand fusion events occur at the most closely spaced origins. (B and C) Sperm nuclei were incubated in X. laevis egg extract, plus or minus aphidicolin and/or caffeine. To produce minimally licensed DNA, extract was supplemented with geminin shortly after sperm addition. i–iii, maximally licensed DNA; iv–vi, minimally licensed DNA. i and iv, no additions; ii and v, plus caffeine and aphidicolin; iii and vi, plus caffeine. At the start of S phase, the extract was supplemented with α-[32P]dATP, which was chased with unlabeled dATP after either 2 min (i, iii, iv, and vi) or 5 min (ii and v). At different times thereafter DNA was isolated, separated by alkaline agarose gel electrophoresis, and autoradiographed. The times for i were every minute from 27–36 min. The times for ii were 31, 33, every minute from 35–45, 48, 51, and 54 min. The times for iii were every minute from 23–32 min. The times for iv were every minute from 23–30, 32, 34, 36, and 40 min. The times for v were every 2 min from 32–50, 55, and 60 min. The times for vi were every min from 23–30, 32, 34, 36, and 40 min. Molecular weight markers (λ-HindIII) are shown to the left (sizes in kilobases). The autoradiographs are shown in B. The x-ray film was then scanned and the density of label in each lane quantified. The scans for each lane are shown in C (earliest times at the top, slowest migration to the right). The migration of λ-HindIII is shown by ticks at the bottom.
In contrast, the behavior of maximally and minimally licensed DNA in the presence of aphidicolin and caffeine (Fig. 3 B, ii and v) showed a dramatic difference that was consistent with the model shown in Fig. 3 A. This is highlighted in Fig. 3 C, where the relative abundance of nascent strands in each sample lane of the corresponding alkaline gel has been plotted, with the earliest sample lane plotted nearest the top. Both maximally and minimally licensed nascent strands started off with a more uniform size than in the untreated sample because aphidicolin had limited the rate of elongation. During the first few minutes, both maximally and minimally licensed nascent strands increased in size at ∼6 nt/s (∼3 nt/s per fork). For the minimally licensed sample (Fig. 3, B [v] and C [v]), most strands maintained this rate over the time course. In later samples, however, some strands doubled in size, which is consistent with fusion between adjacent strands (Fig. 3 B, v, arrows). The larger strands first became visible at ∼10 kb, which is consistent with them being fusions between the smallest replicons of ∼5 kb seen in Fig. 1 A. A very different pattern was seen with maximally licensed DNA (Fig. 3, B [ii] and C [ii]). A smear of higher molecular weight strands built up much more rapidly than it did for the minimally licensed sample, so that by the middle of the time series the majority of the strands had shifted to the higher molecular weight form. By the end of the time series, the majority of the strands were above the exclusion limit of the gel. Unlike the case for the minimally licensed DNA, there was no obvious lower size limit at which the strand fusion events occurred. These results suggest that in the presence of aphidicolin and caffeine, maximally licensed DNA displays a dramatic reduction in replicon size, an effect that is not seen with the minimally licensed DNA. Note that, consistent with a previous work (Luciani et al., 2004), there was no sign of caffeine reducing fork stability, as we did not observe buildup of low molecular weight products.
When extracts were supplemented with caffeine alone, there was no obvious difference between maximally and minimally licensed DNA, with both samples yielding fully replicated product (Fig. 3 B, iii and vi). Our fiber-labeling experiments with caffeine (Fig. 2 C) predict that caffeine alone can increase the number of active forks when the DNA is maximally licensed. However, without the fork-slowing effect of aphidicolin, there is rapid fusion of the nascent strands arising from additional origins that would be dormant in the absence of caffeine. Once these initial fusion events have taken place in the maximally licensed sample, the maximally and minimally licensed samples behave similarly. This effect also explains why we do not see very high levels of Cdc45 loading in maximally licensed DNA treated with caffeine alone (Fig. 2 A).
The extra origins do not normally fire
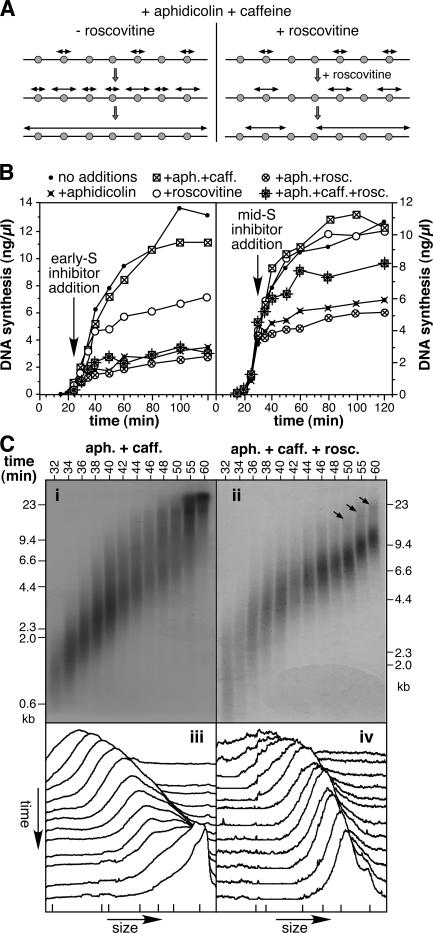
Our results so far suggest that excess Mcm2–7 complexes license dormant origins, which are not used under normal conditions, but which can fire to rescue DNA synthesis in response to replication fork inhibition. It is known that Cdks are required for the initiation of replication forks in the X. laevis system, but not for fork elongation (Strausfeld et al., 1994; Luciani et al., 2004). It is also known that Cdks act on individual origins very shortly before the origin fires (Strausfeld et al., 1994; Luciani et al., 2004). Therefore, we predicted that if Cdks were inhibited at the start of S phase, when only the very first origins had fired, dormant origin firing would be prevented and no rescue of DNA replication would take place. To inhibit Cdks, we used roscovitine, which is a purine analogue that inhibits initiation in the X. laevis system (Meijer et al., 1997; Luciani et al., 2004). A model of the predicted behavior is shown in Fig. 4 A. The model on the left shows initiation occurring at dormant origins when replication occurs in the presence of aphidicolin and caffeine. The model on the right shows that if roscovitine is added shortly after the first origins have fired, initiation cannot occur at the dormant origins, so the replication profile resembles that of minimally licensed DNA treated with aphidicolin and caffeine.
Figure 4.
Roscovitine blocks premature nascent strand fusion. (A) Model for the effect of roscovitine on dormant origin firing. The model is the same as the maximally licensed DNA shown in Fig. 3 A, except that roscovitine is added shortly after the first origins fire (right), thereby preventing the dormant origins from firing. (B) Sperm nuclei were incubated in X. laevis egg extract supplemented with α-[32P]dATP. At 25 min (left), or 30 min (right), extract was optionally supplemented with aphidicolin, caffeine, and/or roscovitine. At the indicated times, the total DNA synthesis was measured. (C) Sperm nuclei were incubated in X. laevis egg extract supplemented with aphidicolin and caffeine. At 25 min, the extract was supplemented with α-[32P]dATP minus (i and iii) or plus (ii and iv) roscovitine. At 30 min, extract was supplemented with unlabeled dATP. At the indicated times, DNA was isolated, separated by alkaline agarose gel electrophoresis, and autoradiographed. End-labeled λ-HindIII was run as molecular weight standards. The autoradiographs are shown in i and ii. The x-ray film was then scanned and the density of the label in each lane was quantified. The scans for each lane are shown in iii and iv (earliest times at the top; slowest migration to the right). The migration of λ-HindIII is shown by ticks at the bottom.
Fig. 4 (B and C) show experimental results that support this model. Fig. 4 B (left) shows the effect on DNA replication when different combinations of caffeine, aphidicolin, and roscovitine were added to X. laevis egg extract at the start of S phase. Consistent with previous work (Luciani et al., 2004), the addition of roscovitine in early S phase allowed existing forks to elongate, but the replication rate subsequently trailed off as these clustered nascent strands fused with one another. As expected for maximally licensed DNA, aphidicolin inhibited replication and this inhibition could be completely rescued by the addition of caffeine. However, when roscovitine was added together with aphidicolin and caffeine, there was no rescue of replication. A related experiment is shown in Fig. 4 B (right), the difference being that the inhibitors were added later in S phase. At this time, addition of roscovitine alone had no significant effect on replication kinetics, suggesting that virtually all the normal replication origins had already fired by the time the inhibitors were added. As before, inhibition of replication by aphidicolin could be rescued by coaddition of caffeine, and this rescue was substantially dependent on further Cdk activity. Some roscovitine-resistant rescue by caffeine was observed, possibly reflecting additional recovery pathways, such as the restart of previously stalled forks. The significance of this version of the experiment is that a requirement for Cdks is seen at a late stage in S phase when most origins have already initiated, suggesting that complete replication is dependent on the use of origins that would never normally have fired (dormant origins).
To confirm our interpretation of these results, we performed alkaline gel analysis of the nascent strands. Nascent DNA was labeled with α-[32P]dATP at the beginning of S phase, in the presence of aphidicolin and caffeine. The 32P was then chased with unlabeled dATP, while, at the same time, extract was supplemented minus (Fig. 4 C, i and iii) or plus roscovitine (Fig. 4 C, ii and iv). At different times thereafter, labeled DNA was analyzed on alkaline gels. The sample treated with aphidicolin and caffeine (Fig. 4 C, i) closely resembled the corresponding sample shown in Fig. 3 B (ii), with nascent DNA increasing rapidly in size so that most migrated at the exclusion limit of the gel at the end of the time series. Addition of roscovitine (Fig. 4 C, ii and iv) almost completely inhibited the increase in modal strand length, so that most strands increased in size at a constant rate of ∼6 nt/s (∼3 nt/s per fork). At later time points, a small number of strands migrated in a higher molecular weight smear at approximately twice the size of the majority, which is consistent with the fusion of replicons 5–10 kb in size (Fig. 4 C, ii, arrows). This profile resembles that of minimally licensed DNA treated with aphidicolin plus caffeine (Fig. 3, B [v] and C [v]). As predicted, by inhibiting Cdk-dependent initiation of dormant origins in maximally licensed DNA, the pattern of nascent strand labeling is made to resemble that of minimally licensed DNA, in which dormant origins do not exist.
Dormant origins are activated in response to a range of replication stresses
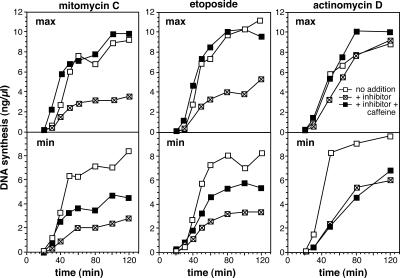
We next investigated whether the activation of dormant origins in maximally licensed DNA is useful in recovering from any replication-induced stresses other than aphidicolin (Fig. 5). Both mitomycin C (a cross-linking agent) and etoposide (a topoisomerase II inhibitor) induced a block to replication that could only be efficiently rescued by caffeine when excess Mcm2–7 were present on the DNA. This is similar to the results with aphidicolin (Fig. 1 A) and suggests a general requirement for dormant origins in rescue of DNA synthesis after replication block. Actinomycin D, which acts both as a DNA intercalator and an inhibitor of the primase component of DNA polymerase α showed an even more dramatic effect. Replication was only slightly inhibited when maximally licensed DNA was treated with 4 ng/μl actinomycin D, but the inhibition was much more severe on minimally licensed DNA. Although the small amount of inhibition of maximally licensed DNA was fully reversed by caffeine, virtually no rescue with caffeine was observed with minimally licensed DNA. This result is explained by the inability of actinomycin D to activate a strong ATR-dependent checkpoint response, despite strong inhibition of replication (Hekmat-Nejad et al., 2000). Dormant origins in such cases can fire and promote significant recovery from replication inhibition without the need for checkpoint alleviation by caffeine. Consistent with this interpretation, treatment of X. laevis egg extracts with actinomycin D greatly enhances Cdc45 loading (Edwards et al., 2002).
Figure 5.
Minimally licensed DNA is sensitive to a range of replication inhibitors. Sperm nuclei were incubated in X. laevis egg extract supplemented with α-[32P]dATP plus or minus caffeine. To produce minimally licensed DNA, extract was supplemented with geminin shortly after sperm addition. At 5 min, extract was optionally supplemented with 500 μM mitomycin C, 200 μM etoposide, or 4 ng/μl actinomycin D. At the indicated times, the total amount of DNA synthesized was measured.
Excess Mcm2–7 are required for C. elegans to survive in HU
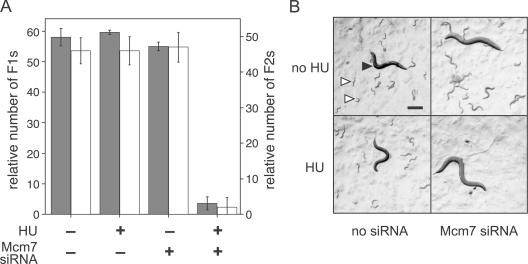
The large amounts of replicative stress that we need to use in the X. laevis cell-free system activate global checkpoint pathways that prevent replication from completing. To test the physiological significance of our findings, we turned to an in vivo system, where the effect of much lower levels of replicative stress can be reliably assessed. We took advantage of the ability to lower specific gene expression in C. elegans by providing siRNA-expressing bacteria as a food source. We first mixed different ratios of bacteria expressing MCM7 siRNA and bacteria expressing empty vector to determine the maximum quantity of MCM7 siRNA that had no observable effect on the C. elegans life cycle (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200602108/DC1). This creates a state that is functionally equivalent to the minimally licensed state in X. laevis. We also determined a low concentration of HU (9.5 mM), which had no observable effect on the C. elegans life cycle (Table S1). HU lowers dNTP pools by inhibiting ribonucleotide reductase and thereby inhibits DNA synthesis. Fig. 6 A shows that treatment with a combination of the MCM7 siRNA and 9.5 mM of HU completely abrogated C. elegans proliferation. There was a dramatic reduction in the number of adult worms in the first generation that had fed on the siRNA. The few adult worms in this generation appeared to be sterile (not depicted), and produced virtually no eggs or larvae (Fig. 6 B). Similar results were obtained using knockdown of MCM5 and MCM6 (Tables S2 and S3). These experiments show that a reduction of Mcm2–7 proteins causes a dramatic hypersensitivity to HU, while showing no obvious defect in the absence of replicative stress. Although there are many possible explanations for what is happening in these C. elegans experiments, the results are fully consistent with the results obtained in X. laevis and the idea that excess Mcm2–7 license dormant origins that are not used during undisturbed S phases.
Figure 6.
Knockdown of Mcm7 in C. elegans causes hypersensitivity to HU. C. elegans were grown on plates plus or minus bacteria expressing anti-Mcm7 siRNA (2%) and ±9.5 mM HU. After 7 d, plates were examined for worm growth. (A) The number of F1 (first) and F2 (second) generation worms in standardized areas was counted, ±SD. (B) Representative picture of a plate, in each case showing one adult F1 worm. Closed arrowhead, F1 adult; open arrowheads, F2 larvae. Note the absence of F2 worms in the sample containing both HU and anti-Mcm7 siRNA. Bar, 0.2 mm.
Discussion
The results presented in this work provide a solution to the MCM paradox by showing that excess Mcm2–7 license many dormant replication origins that are not normally used because of suppression by a caffeine-sensitive pathway. In X. laevis, there are ∼10 dormant origins for each active origin, and when dormant origin activation is permitted by treatment with caffeine, the average origin spacing drops 5- to 10-fold. The caffeine sensitivity suggests the involvement of an ATR-dependent checkpoint in suppressing the activation of dormant origins. This conclusion is consistent with results in mammalian cells, which show that suppression of ATM–ATR or Chk1 checkpoint pathways leads to a dramatic reduction in the average spacing between adjacent replication origins (unpublished data). We also show that these dormant origins can be used to allow complete replication under conditions of replicative stress, and we propose that this is at least part of the reason for their existence.
Detailed analysis of this effect has been performed in X. laevis egg extracts, but we have also tested a key consequence of this model by lowering MCM levels in C. elegans to operationally mimic the minimally licensed state. Reduction of MCM levels causes a dramatic hypersensitivity to HU, while showing no obvious defect in the absence of replicative stress. There is also published evidence that this effect may operate in mammals. DNA fiber autoradiography studies have shown an increase in the density of replication origins after replication has been inhibited by FdUrd, which is a thymidylate synthase inhibitor (Ockey and Saffhill, 1976; Taylor, 1977), or by treatment with ultraviolet light (Griffiths and Ling, 1985, 1987, 1989; Painter, 1985). Similarly, reduced nucleotide availability has been shown to increase the frequency of initiation at origins that are not normally used (Anglana et al., 2003). Therefore, we believe that dormant origins are present in many metazoans, where they can potentially provide a role in responding to replicative stresses.
A model for the behavior of dormant origins
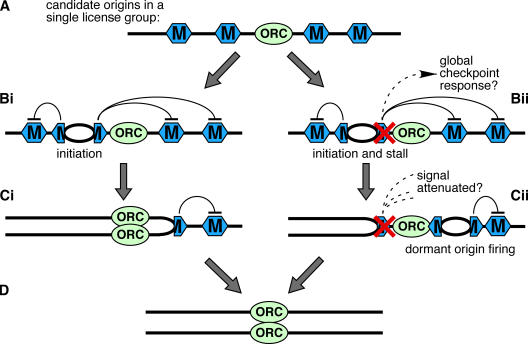
Our results suggest that classically defined replication origins are actually comprised of several candidate origins, most of which are not normally used, as outlined in Fig. 7. It is known from work in the X. laevis system that replication forks initiate from sites where Mcm2–7, and not ORC, are bound (Hua and Newport, 1998; Rowles et al., 1999; Edwards et al., 2002; Harvey and Newport, 2003; Danis et al., 2004). Based on previous studies, Mcm2–7 may be spread several kilobases around the ORC that loaded them, to form what we term a “license group” (Ritzi et al., 1998; Edwards et al., 2002; Harvey and Newport, 2003; Danis et al., 2004). Each site containing bound Mcm2–7 is a candidate replication origin that can potentially initiate replication (Fig. 7 A). The wide dispersal of Mcm2–7 increases the usefulness of these candidate origins in dealing with replicative stress.
Figure 7.
Model for the use of dormant origins on chromosomal DNA. (A) A small segment of chromosomal DNA is shown with a single molecule of ORC bound to it. Distributed around the ORC are several Mcm2–7 double hexamers (hexagons), which each represent a candidate origin. The collection of Mcm2–7 around the ORC are designated a single license group. (Bi) Once initiation occurs in a given license group it generates a weak caffeine-sensitive checkpoint signal that inhibits initiation from any other Mcm2–7 within that license group. (Ci) As the fork replicates the DNA, uninitiated Mcm2–7 at dormant origins are displaced from the DNA. (Bii) The rightward fork stalls, potentially generating a strong global checkpoint signal. (Cii) One of the dormant origins within the license group escapes inhibition and initiates, thus, allowing DNA to the right of the stalled fork to be replicated. The escape from inhibition may be stochastic, or may occur as a result of attenuation of the checkpoint signal after DNA repair. (D) Completion of DNA replication.
Fig. 7 (Bi–Ci) shows events occurring in the absence of replicative stress. Initiation occurs at one of the candidate origins (the primary origin). Consistent with previous studies (Edwards et al., 2002; Marheineke and Hyrien, 2004; Shechter et al., 2004), our results with caffeine suggest a role for ATM–ATR in preventing more than one origin in a license group from initiating replication. We speculate that once initiation has taken place at the primary origin, ATM–ATR becomes activated, perhaps locally, to inhibit initiation at the remaining candidate origins in the same license group (Fig. 7 Bi). To prevent initiation at these dormant origins, this inhibition need not be absolute or prolonged, as the forks rapidly replicate the surrounding DNA, thereby displacing other bound Mcm2–7 molecules and eliminating the dormant origins (Fig. 7, Ci and D). Abolishing this inhibitory signal with caffeine in the absence of replication stress causes a decrease in origin spacing, but does not significantly increase overall replication rates. In contrast to Shechter et al. (2004), we find that if we restrict our analysis to extracts that replicate DNA efficiently (as should be the case if extracts recapitulate the behavior of intact eggs), inhibition of ATM and ATR kinases does not greatly accelerate the replication-timing program (Luciani et al., 2004; this study). Dormant origins are therefore not just late-firing origins, as confirmed by our experiments with roscovitine (Fig. 4), and the replication-timing program appears to operate independently of checkpoint signals (unpublished data). Our results do not rule out a role for Mcm2–7 in checkpoint activation; indeed, the recently reported phosphorylation of Mcm2–7 proteins by ATR (Cortez et al., 2004; Tsao et al., 2004) may contribute to the inhibition of dormant origins that we propose.
Fig. 7 (Bii–Cii) shows events occurring at an origin where one of the forks experiences replicative stress and is stalled or slowed. In Fig. 7 Bii, the rightward fork stalls so that the dormant origins in the license group are not eliminated. If enough forks stall (as occurred in our experiments using replication inhibitors in X. laevis), a global checkpoint signal is activated that represses initiation at replication origins throughout the genome. However, the use of dormant origins may be more physiologically important at levels of fork stalling that do not strongly activate this global checkpoint response. Fig. 7 Cii shows that after a fork has stalled, one of the nearby dormant origins could undergo initiation, ultimately allowing all of the DNA in the region to be replicated (Fig. 7 D). There are several potential explanations for how the dormant origin might escape the inhibitory signal emitted from the stalled fork. One possibility, mentioned in the previous paragraph, is that the inhibitory signal may only be weak (e.g., if only a few forks have stalled and there is no global suppression of initiation), so that in a stochastic fashion the extended lifespan of the dormant origin provides a sufficient length of time for it to undergo initiation. Another possibility is that DNA repair processes operating at the stalled fork attenuate the checkpoint signal. A third possibility is that certain types of replication inhibition (such as that induced by actinomycin D) may not robustly activate the checkpoint system, thereby leaving dormant origins free to fire. These controls would allow dormant origins to be used only when nearby forks encounter problems, thus, lowering the probability that two converging forks will stall irreversibly and leave the DNA between them unable to replicate.
Our model emphasizes the counterintuitive nature of the intra–S phase checkpoint that globally blocks further origin firing in response to replication stress because under these conditions it might seem better for cells to respond by using more origins, rather than fewer. One possible explanation is that the global intra–S phase checkpoint provides a strategy for responding to severe replicative stresses that affect many replication forks but are only transient.
What is a replication origin?
Our model suggests that under normal conditions the firing of a primary origin represses other origins within a license group. Thus, maximally and minimally licensed DNA have the same overall replication kinetics and the same average origin spacing. Origin spacing is therefore largely independent of the number of Mcm2–7 complexes bound to DNA and is determined by the location of ORC binding. In X. laevis, regular ORC spacing (Blow et al., 2001; Oehlmann et al., 2004), coupled with the repression of excess Mcm2–7 within license groups, can reliably support complete genome replication, with dormant origins used as a backup only if problems are encountered by forks originating from primary origins. This is reminiscent of the “Jesuit model” for selection of replication initiation sites, which suggests that DNA contains many potential initiation sites, but some sites are repressed, while others are activated (DePamphilis, 1999).
Replication origins in metazoans can take two different forms (DePamphilis, 1999; Gilbert, 2001; Machida et al., 2005). At some replication origins, such as the origin near the lamin B2 gene, replication initiates reproducibly at a tightly defined locus; at other origins, such as the origin near the dihydrofolate reductase gene, replication can initiate at many potential sites distributed throughout an initiation zone. In the case of origins with large initiation zones, such as the dihydrofolate reductase origin, each zone may consist of multiple sites containing bound Mcm2–7 (Alexandrow et al., 2002), with only one of these candidate origins initiating during S phase. It is possible that the tightly localized origins also have multiple Mcm2–7 complexes associated with them, but that some mechanism reproducibly selects one of them to initiate and form the primary origin.
It remains unclear how certain candidate origins might be selected for preferential initiation, though some difference in the underlying chromatin would be an attractive possibility (Machida et al., 2005). In the case of X. laevis, initiation can occur at some variable distance away from where ORC is bound (Harvey and Newport, 2003) and can be directed toward specific DNA sequences by DNA-bound transcription factors (Danis et al., 2004). Because this occurs without changing the localization of ORC, it presumably happens by increasing the initiation probability of certain candidate origins. Interestingly, transcription factor binding induced histone acetylation at the favored origins (Danis et al., 2004), supporting the case for chromatin modification being involved in candidate origin selection. Somatic cells undergo a transition called the origin decision point (ODP) in the middle of G1, where initiation is directed to occur at specific preferred sequences rather than at more randomly distributed sites (Wu and Gilbert, 1996). Because the ODP occurs after Mcm2–7 have bound to DNA and the origins have been functionally licensed (Dimitrova et al., 2002), it is possible that the ODP represents the stage when certain candidate origins are selected to preferentially support initiation.
Materials and methods
Preparation and use of X. laevis egg extracts
X. laevis egg extracts were prepared as previously described (Chong et al., 1997) and supplemented with 250 μg/ml cycloheximide, 25 mM phosphocreatine, 15 μg/ml creatine phosphokinase, and 300 μM CaCl2 before use. Demembranated X. laevis sperm nuclei (Chong et al., 1997) were added to extract to a final concentration of 10–15 ng DNA/μl and incubated at 23°C. For replication assays, extracts were supplemented with α-[32P]dATP, and DNA synthesis was measured by trichloroacetic acid precipitation, as previously described (Chong et al., 1997). To obtain minimally licensed DNA samples, recombinant geminin lacking the destruction box (McGarry and Kirschner, 1998; Tada et al., 2001) was added to a final concentration of 10 ng/μl. The appropriate geminin addition time was determined for each extract to produce minimal licensing, as previously described (Oehlmann et al., 2004). Chromatin was isolated and subjected to Western blot analysis, as previously described (Oehlmann et al., 2004).
Reagents and antibodies
Aphidicolin (Sigma-Aldrich) was dissolved in DMSO at 18 mM and used at a final concentration of 10 μM in extract. Caffeine (Sigma-Aldrich) was dissolved in H20 at 100 mM and was used at a final concentration of 5 mM. Roscovitine (Calbiochem) was dissolved in DMSO at 400 mM and used at a final concentration of 500 μM. Mitomycin C was dissolved in H20 at 5 mM and used at a final concentration of 500 μM. Etoposide was dissolved in DMSO at 25 mM and used at a final concentration of 200 μM. Actinomycin D was dissolved in DMSO at 1 μg/μl and used at a final concentration of 4 ng/μl. Anti-Cdc45 antibody (Mimura and Takisawa, 1998) was a gift from H. Takisawa (Osaka University, Osaka, Japan). Geminin was a gift from A. Ferenbach (University of Dundee, Dundee, Scotland).
DNA fiber labeling
X. laevis sperm DNA was incubated in extract supplemented with 50 μM biotin-16-dUTP (Roche) under the desired conditions. Reactions (20 μl) were stopped by resuspension in 350 μl of ice-cold NIB (50 mM Hepes-KOH, pH 7.6, 50 mM KCl, 5 mM MgCl2, 2 mM DTT, 0.5 mM spermidine-3HCl, 0.15 mM spermine-4HCl, 100 μM PMSF, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin) containing 30 μM aphidicolin. The resuspended extract was underlayered with 100 μl NIB containing 30 μM aphidicolin and 10% (wt/vol) sucrose and spun at 2,000 g in a swinging bucket rotor for 5 min at 4°C. The supernatant was removed to leave only the sucrose cushion, and the top of the cushion was then washed twice with 100 μl NIB plus 30 μM aphidicolin. The top part of the cushion was carefully removed to leave 40 μl, which was resuspended in 400 μl ice-cold PBS plus 0.1% (vol/vol) Triton X-100. This was underlayered with PBS plus 10% sucrose and spun like the resuspended extract. The supernatant was removed and the top of the cushion was washed twice with 100 μl PBS. The cushion was carefully removed to leave the pelleted nuclei, which were gently resuspended in 50 μl PBS before freezing on dry ice.
DNA was spread on glass slides according to the previously described standard conditions (Blow et al., 2001). Slides were rehydrated with HPLC pure water and incubated for 2 h in blocking solution containing PBS, 1% (wt/vol) BSA, and 0.05% (vol/vol) TWEEN 20. Slides were incubated with anti-biotin (B7653; Sigma-Aldrich) at 20 μg/ml in this buffer for 2 h, washed extensively (10 changes of buffer over 2 h), and labeled with 1 μg/ml cy3-conjugated anti–mouse antibody (715-165-150; Jackson ImmunoResearch Laboratories) in the same buffer for 2 h. Samples were washed extensively (10 changes of buffer over 2 h), and then washed twice in PBS and DNA stained with YOYO (Y-3601 diluted at 1:10,000 from a 1-mM stock; Invitrogen) for 10 min. Samples were rinsed five times in PBS and mounted in Vectashield. Images were captured using a microscope (LSM510-META; Carl Zeiss MicroImaging, Inc.) and measurements were made using the associated LSM software. Random fields were selected using YOYO staining to ensure that only single DNA fibers, and not fiber bundles, were scored. Fields rich in unambiguous single fibers were recorded using a 100×, NA 1.4, lens for optimal resolution and sensitivity. For each sample a minimum of 200 measurements was made.
Pulse-chase experiments
Pulse-chase experiments were performed by incubating X. laevis sperm DNA in interphase egg extract, with inhibitors added as was appropriate. At the beginning of S phase (the exact timing being determined independently for each extract), the extract was supplemented with 0.2 mCi/ml α-[32P]dATP (from a stock of 10 mCi/ml and 3,000 Ci/mmol; GE Healthcare). After a 2-min pulse (5 min for samples containing aphidicolin), the radioactive nucleotide was chased by addition of 2.5 mM unlabeled Mg-dATP. Aliquots (10 μl) were subsequently removed from the sample at the desired time points, reactions were stopped, and the DNA was processed for electrophoresis on 0.7% agarose gels, as previously described (Luciani et al., 2004). HindIII-digested λ-phage DNA (New England Biolabs) was end-labeled with α-[32P]dATP and loaded for comparison (12.5 ng per lane). Alkaline agarose gels were exposed to autoradiography film (Kodak), which was scanned using a scanner (CanoScan 9900F; Canon) and Photoshop software (Adobe). Lane densities were analyzed using GelEval software (FrogDance Software).
C. elegans work
C. elegans RNAi feeding was performed as previously described, using MCM5 (R10E4.4), MCM6 (ZK632.1), and MCM7 (F32D1.10) from the genome-wide RNAi library (Fraser et al., 2000; Kamath et al., 2003). MCM RNAi-expressing bacteria were titrated with bacteria expressing RNAi against GFP. Three P0 worms were placed on RNAi plates containing 100 μg/ml ampicillin, 5 mM IPTG, and the indicated concentration of HU at the L4 larval stage, and the resulting adult worms were taken off those plates 40 h later. After 4 d, the number of adult F1 worms was scored, and sterile worms were determined by the absence of eggs in their body. The number and viability of F2 animals was scored 2–3 d later. At this stage, under wild-type conditions, plates were already starved because of excessive F2 larvae. Plates were viewed with a microscope (Stemi SV11; Carl Zeiss MicroImaging, Inc.) using a 20× objective. Representative images were captured using Openlab 3.1.2 software (Improvision) and exported as tiff files.
Online supplemental material
Tables S1–S3 show the number of C. elegans adult F1 worms observed at different HU concentrations and different amounts of siRNA directed against MCM7 (Table S1), MCM5 (Table S2), and MCM6 (Table S3). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200602108/DC1.
Supplementary Material
Acknowledgments
Thanks to Andrew Ferenbach for recombinant geminin, and Neil Perkins, Anatoliy Li, and Margret Michalski-Blow for comments on the manuscript.
This work was supported by Cancer Research UK (CR-UK) grants C303/A3135 and STU063/001 to J.J. Blow and CR-UK grant CDA A5991 to A. Gartner.
The authors have no financial conflicts of interest in this work.
M.G. Luciani's present address is Dept. of Medicine, Vienna General Hospital, 1090 Vienna, Austria.
M. Oehlmann's present address is Dept. of Biochemistry, National University of Ireland, Galway, Ireland.
Abbreviations used in this paper: HU, hydroxyurea; MCM, minichromosome maintenance; ODP, origin decision point; ORC, origin recognition complex.
References
- Alexandrow, M.G., M. Ritzi, A. Pemov, and J.L. Hamlin. 2002. A potential role for mini-chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 277:2702–2708. [DOI] [PubMed] [Google Scholar]
- Anglana, M., F. Apiou, A. Bensimon, and M. Debatisse. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 114:385–394. [DOI] [PubMed] [Google Scholar]
- Aparicio, O.M., D.M. Weinstein, and S.P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 91:59–69. [DOI] [PubMed] [Google Scholar]
- Aparicio, O.M., A.M. Stout, and S.P. Bell. 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. USA. 96:9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow, J.J., and A. Dutta. 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6:476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow, J.J., P.J. Gillespie, D. Francis, and D.A. Jackson. 2001. Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J. Cell Biol. 152:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart, R., D. Schulte, D. Hu, C. Musahl, F. Gohring, and R. Knippers. 1995. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur. J. Biochem. 228:431–438. [PubMed] [Google Scholar]
- Callan, H.G. 1972. Replication of DNA in the chromosomes of eukaryotes. Proc. R. Soc. Lond. B. Biol. Sci. 181:19–41. [DOI] [PubMed] [Google Scholar]
- Chong, J.P., P. Thömmes, A. Rowles, H.M. Mahbubani, and J.J. Blow. 1997. Characterization of the Xenopus replication licensing system. Methods Enzymol. 283:549–564. [DOI] [PubMed] [Google Scholar]
- Cortez, D., G. Glick, and S.J. Elledge. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 101:10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis, E., K. Brodolin, S. Menut, D. Maiorano, C. Girard-Reydet, and M. Mechali. 2004. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 6:721–730. [DOI] [PubMed] [Google Scholar]
- DePamphilis, M.L. 1999. Replication origins in metazoan chromosomes: fact or fiction? Bioessays. 21:5–16. [DOI] [PubMed] [Google Scholar]
- Diffley, J.F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14:R778–R786. [DOI] [PubMed] [Google Scholar]
- Dimitrova, D.S., I.T. Todorov, T. Melendy, and D.M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova, D.S., T.A. Prokhorova, J.J. Blow, I.T. Todorov, and D.M. Gilbert. 2002. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 115:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan, S., J. Harwood, L.S. Drury, and J.F. Diffley. 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 94:5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M.C., A.V. Tutter, C. Cvetic, C.H. Gilbert, T.A. Prokhorova, and J.C. Walter. 2002. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277:33049–33057. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., R.S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408:325–330. [DOI] [PubMed] [Google Scholar]
- Gilbert, D.M. 2001. Making sense of eukaryotic DNA replication origins. Science. 294:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, P.J., A. Li, and J.J. Blow. 2001. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, T.D., and S.Y. Ling. 1985. Effect of ultraviolet light on thymidine incorporation, DNA chain elongation and replicon initiation in wild-type and excision-deficient Chinese hamster ovary cells. Biochim. Biophys. Acta. 826:121–128. [DOI] [PubMed] [Google Scholar]
- Griffiths, T.D., and S.Y. Ling. 1987. Activation of alternative sites of replicon initiation in Chinese hamster cells exposed to ultraviolet light. Mutat. Res. 184:39–46. [DOI] [PubMed] [Google Scholar]
- Griffiths, T.D., and S.Y. Ling. 1989. Effects of UV light on DNA chain growth and replicon initiation in human cells. Mutat. Res. 218:87–94. [DOI] [PubMed] [Google Scholar]
- Harvey, K.J., and J. Newport. 2003. CpG methylation of DNA restricts prereplication complex assembly in Xenopus egg extracts. Mol. Cell. Biol. 23:6769–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Nejad, M., Z. You, M.C. Yee, J.W. Newport, and K.A. Cimprich. 2000. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 10:1565–1573. [DOI] [PubMed] [Google Scholar]
- Herrick, J., P. Stanislawski, O. Hyrien, and A. Bensimon. 2000. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J. Mol. Biol. 300:1133–1142. [DOI] [PubMed] [Google Scholar]
- Hopwood, B., and S. Dalton. 1996. Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc. Natl. Acad. Sci. USA. 93:12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, X.H., and J. Newport. 1998. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol. 140:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien, O., K. Marheineke, and A. Goldar. 2003. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 25:116–125. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., A.G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. [DOI] [PubMed] [Google Scholar]
- Krude, T., C. Musahl, R.A. Laskey, and R. Knippers. 1996. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J. Cell Sci. 109:309–318. [DOI] [PubMed] [Google Scholar]
- Labib, K., and J.F. Diffley. 2001. Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev. 11:64–70. [DOI] [PubMed] [Google Scholar]
- Laskey, R.A., and M.A. Madine. 2003. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 4:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, M., Y. Kawasaki, and B.K. Tye. 1996. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5081–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, I., M. Chevrier-Miller, J.M. Sogo, and O. Hyrien. 2000. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 296:769–786. [DOI] [PubMed] [Google Scholar]
- Luciani, M.G., M. Oehlmann, and J.J. Blow. 2004. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 117:6019–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, Y.J., J.L. Hamlin, and A. Dutta. 2005. Right place, right time, and only once: replication initiation in metazoans. Cell. 123:13–24. [DOI] [PubMed] [Google Scholar]
- Madine, M.A., C.Y. Khoo, A.D. Mills, C. Musahl, and R.A. Laskey. 1995. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr. Biol. 5:1270–1279. [DOI] [PubMed] [Google Scholar]
- Mahbubani, H.M., T. Paull, J.K. Elder, and J.J. Blow. 1992. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 20:1457–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani, H.M., J.P. Chong, S. Chevalier, P. Thömmes, and J.J. Blow. 1997. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J. Cell Biol. 136:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marheineke, K., and O. Hyrien. 2004. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J. Biol. Chem. 279:28071–28081. [DOI] [PubMed] [Google Scholar]
- McGarry, T.J., and M.W. Kirschner. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 93:1043–1053. [DOI] [PubMed] [Google Scholar]
- Meijer, L., A. Borgne, O. Mulner, J.P.J. Chong, J.J. Blow, N. Inagaki, M. Inagaki, J.G. Delcros, and J.P. Moulinoux. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527–536. [DOI] [PubMed] [Google Scholar]
- Mimura, S., and H. Takisawa. 1998. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 17:5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockey, C.H., and R. Saffhill. 1976. The comparative effects of short-term DNA inhibition on replicon synthesis in mammalian cells. Exp. Cell Res. 103:361–373. [DOI] [PubMed] [Google Scholar]
- Oehlmann, M., A.J. Score, and J.J. Blow. 2004. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J. Cell Biol. 165:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter, R.B. 1985. Inhibition and recovery of DNA synthesis in human cells after exposure to ultraviolet light. Mutat. Res. 145:63–69. [DOI] [PubMed] [Google Scholar]
- Ritzi, M., M. Baack, C. Musahl, P. Romanowski, R.A. Laskey, and R. Knippers. 1998. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 273:24543–24549. [DOI] [PubMed] [Google Scholar]
- Rowles, A., J.P. Chong, L. Brown, M. Howell, G.I. Evan, and J.J. Blow. 1996. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 87:287–296. [DOI] [PubMed] [Google Scholar]
- Rowles, A., S. Tada, and J.J. Blow. 1999. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 112:2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6:648–655. [DOI] [PubMed] [Google Scholar]
- Strausfeld, U.P., M. Howell, R. Rempel, J.L. Maller, T. Hunt, and J.J. Blow. 1994. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr. Biol. 4:876–883. [DOI] [PubMed] [Google Scholar]
- Tada, S., A. Li, D. Maiorano, M. Mechali, and J.J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.H. 1977. Increase in DNA replication sites in cells held at the beginning of S phase. Chromosoma. 62:291–300. [DOI] [PubMed] [Google Scholar]
- Tsao, C.C., C. Geisen, and R.T. Abraham. 2004. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 23:4660–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, J., and J.W. Newport. 1997. Regulation of replicon size in Xenopus egg extracts. Science. 275:993–995. [DOI] [PubMed] [Google Scholar]
- Wu, J.R., and D.M. Gilbert. 1996. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science. 271:1270–1272. [DOI] [PubMed] [Google Scholar]
- Zou, L., and B. Stillman. 1998. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 280:593–596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.