Abstract
Chromosomes move toward mitotic spindle poles by a Pacman-flux mechanism linked to microtubule depolymerization: chromosomes actively depolymerize attached microtubule plus ends (Pacman) while being reeled in to spindle poles by the continual poleward flow of tubulin subunits driven by minus-end depolymerization (flux). We report that Pacman-flux in Drosophila melanogaster incorporates the activities of three different microtubule severing enzymes, Spastin, Fidgetin, and Katanin. Spastin and Fidgetin are utilized to stimulate microtubule minus-end depolymerization and flux. Both proteins concentrate at centrosomes, where they catalyze the turnover of γ-tubulin, consistent with the hypothesis that they exert their influence by releasing stabilizing γ-tubulin ring complexes from minus ends. In contrast, Katanin appears to function primarily on anaphase chromosomes, where it stimulates microtubule plus-end depolymerization and Pacman-based chromatid motility. Collectively, these findings reveal novel and significant roles for microtubule severing within the spindle and broaden our understanding of the molecular machinery used to move chromosomes.
Introduction
The defining event of mitosis occurs during anaphase, when identical sister chromatids disjoin and separate toward opposite poles of the microtubule (MT)-based spindle. Anaphase chromatid-to-pole motion (anaphase A) is tightly linked to depolymerization of the opposite ends of chromosome-associated MTs. Chromosomes actively depolymerize MTs at their plus ends, thereby “chewing” their way poleward along MT tracks—a type of motility termed Pacman. At the same time, chromosome-associated MTs serve as traction fibers, which are drawn poleward via persistent depolymerization at their minus ends. This process, termed poleward flux because of the resulting poleward flow (flux) of MTs, reels in attached chromatids to spindle poles (for review see Mitchison and Salmon, 2001). Pacman and flux have been observed to occur simultaneously in diverse cell types and, in sum, account for the entire velocity of poleward chromosome motility (Mitchison and Salmon, 1992; Zhai et al., 1995; Maddox et al., 2002, 2003; Rogers et al., 2004; Civelekoglu-Scholey et al., 2006). Thus, we refer to the general translocation mechanism underlying anaphase A as Pacman-flux.
A large and growing set of proteins has been identified that bind to and modulate the functions of spindle MTs (Scholey et al., 2003) and thus could be incorporated into the Pacman-flux machinery that drives anaphase A. Among the most mysterious of the spindle binding proteins identified to date are MT severing enzymes (Vale, 1991; McNally and Vale, 1993). Proteins with the capacity to sever MTs have been found associated with spindles in a wide variety of systems, yet their specific mitotic functions remain largely unknown (McNally et al., 1996; Srayko et al., 2000; Errico et al., 2004; Svenson et al., 2005).
In particular, three conserved and closely related members of the AAA protein superfamily have been identified that may function by severing mitotic spindle MTs (Frickey and Lupas, 2004). The best characterized of these is Katanin, a heterodimer consisting of a 60-kD AAA catalytic subunit (p60) and an 80-kD targeting and regulatory subunit (p80; McNally and Vale, 1993; Hartman et al., 1998). Katanin has been found to target to centrosomes in diverse cell types (McNally et al., 1996; McNally and Thomas, 1998; Srayko et al., 2000), leading to the proposal that it contributes to MT minus-end depolymerization and flux (McNally and Vale, 1993; McNally et al., 1996; McNally and Thomas, 1998). However, Katanin's role in flux or chromosome motility has not been previously demonstrated. In addition, a Katanin homologue in Caenorhabditis elegans targets to meiotic chromosomes. Although this protein does not participate in mitosis, but instead is required for normal meiotic spindle assembly and dynamics (Srayko et al., 2000, 2006; McNally et al., 2006), it is conceivable that a similarly positioned MT severing protein in mitotic cells could contribute to Pacman-based chromosome motility.
A second AAA family member, Spastin, has also been found to sever MTs in cells and in vitro (Evans et al., 2005; Roll-Mecak and Vale, 2005). Spastin has been studied primarily for its role in neuronal development and function. Mutations in the Spastin gene are the major cause of hereditary spastic paraplegia, a disorder caused by the degeneration of subsets of neurons and hallmarked by the progressive weakening of lower extremities (Hazan et al., 1999). Loss-of-function mutations of the Spastin homologue in Drosophila melanogaster also cause behavioral abnormalities and perturb neuromuscular junctions and axonal MT arrays (Sherwood et al., 2004). Mitotic functions for the protein are unknown, but Spastin has been found to localize to centrosomes and spindle poles in vertebrate cells (Errico et al., 2004; Svenson et al., 2005), raising the possibility that it, too, could contribute to chromosome motility via the generation of poleward flux.
A third AAA protein family member, Fidgetin, groups closely with Spastin and Katanin by phylogenetic analysis (Frickey and Lupas, 2004) and thus may sever MTs as well, though this has not been demonstrated experimentally. Phenotypic analyses of Fidgetin mutant mice indicate important developmental functions for this protein. Mutants display a head-shaking or “fidget” phenotype stemming from defects in auditory development. They also develop small eyes, which are a manifestation of a cell cycle delay, suggesting a potential mitotic role for this protein (Cox et al., 2000). Fidgetin displays both cytoplasmic and nuclear localizations during interphase (Yang et al., 2005), but its mitotic localization and function have not been reported.
For this study, a series of live-cell analyses were performed to explore whether and how D. melanogaster orthologues of Katanin, Spastin, and Fidgetin contribute to mitotic spindle and chromosome dynamics. Our findings reveal that all three are incorporated into the anaphase Pacman-flux machinery used to separate chromosomes. Surprisingly, the functions of these proteins are segregated so that Spastin and Fidgetin stimulate MT minus-end depolymerization and flux, whereas Katanin stimulates plus-end depolymerization and Pacman-based anaphase A.
Results
The D. melanogaster genome encodes single orthologues of Spastin (Dm-Spastin; CG5977) and Fidgetin (Dm-Fidgetin; CG3326) and three potential Katanin p60s. The putative D. melanogaster Katanin p60 orthologues include the protein product of CG10229 (most similar to human Katanin p60 and referred to here as Dm-Kat60) and the more divergent protein products of CG1193 and CG10793 (Emes and Ponting, 2001; Kammermeier et al., 2003). This study reports on the mitotic functions of Dm-Spastin, Dm-Fidgetin, and Dm-Kat60 in D. melanogaster S2 cells (we found that the CG1193 protein had no impact on mitosis, and analyses of CG10793 were not performed). S2 cells were used for these studies because of their ready susceptibility to targeted protein knockdown by double-stranded (ds) RNAi and their amenability to live-cell visualization of mitotic spindle and chromosome dynamics (Goshima and Vale, 2003; Goshima et al., 2005; Maiato et al., 2005).
Only Dm-Spastin has been directly implicated as an MT severing enzyme in D. melanogaster cells. Overexpression of Dm-Spastin in S2 cells was found to cause a substantial loss of interphase MTs, which is typical of increased MT severing activity (McNally et al., 2000; Roll-Mecak and Vale, 2005). Using this same assay, we found that Dm-Fidgetin and Dm- Kat60 similarly disrupted MT arrays when overexpressed in S2 cells, consistent with the hypothesis that all three D. melanogaster proteins function as MT severing enzymes in cells (Fig. 1).
Figure 1.
Overexpression of Dm-Kat60, Dm-Spastin, or Dm-Fidgetin eliminates MTs in interphase S2 cells. (A–C) mRFP–α-tubulin expressing S2 cells were transiently transfected with full-length AAA-EGFP constructs. AAA refers specifically to Dm-Kat60, Dm-Spastin, and Dm-Fidgetin. Fluorescent MTs are pseudocolored green; expressed AAA fusion protein is pseudocolored red. Overexpression of any one of the AAA constructs visibly decreases the MT polymer mass. Bars, 5 μm. (D) Mean fluorescence intensities (+SD) of mRFP– α-tubulin for live whole cells after transient transfection with the indicated full-length AAA-EGFP construct. Red bars are tubulin fluorescence intensities of cells that visibly expressed fluorescent AAA protein. Gray bars are intensities of neighboring cells not visibly expressing fluorescent AAA protein. *, P < 0.001 versus controls. Numbers in bars are sample sizes. (E) Mean fluorescence intensities (+SD) of immunostained α-tubulin in wild-type S2 cells that had been transiently transfected with either EGFP or the full-length Fidgetin-EGFP construct. Gray and red bars are values of neighboring untransfected and transfected cells, respectively. The decrease of tubulin immunofluorescence of Fidgetin-EGFP expressing cells is significant when compared with either neighboring untransfected control cells or EGFP expressing control cells. Error bars indicate SD.
Dm-Kat60, Dm-Spastin, and Dm-Fidgetin display distinct targeting within spindles
Because the localization of MT severing enzymes within D. melanogaster spindles had not been examined before this study, we raised antibodies against unique N-terminal domains of Dm-Kat60, Dm-Spastin, and Dm-Fidgetin (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200612011/DC1) to detect these proteins in S2 cells. Antibodies against each were found to react with a single band near the predicted molecular mass of the intended target on Western blots of S2 cell lysates (Fig. S1 B). The intensity of each band diminished substantially when the target protein was knocked down by 5–7 d of RNAi treatment, indicating the specificity of these antibodies (Fig. S2 A).
Immunofluorescence analyses revealed that all three severing proteins target to both centrosomes and chromosomes in D. melanogaster spindles (Fig. 2 A). In each case, RNAi specifically reduced or abolished the intensity of labeling of the intended target protein, supporting the specificity of this localization (Fig. S2 B). (Depletion of the targeted proteins by RNAi had no apparent affect on the expression or localization of any off-target protein tested [Fig. S2, C and D].) The centrosomal localization of these proteins was qualitatively similar and was not dependent on MTs (Fig. 2 A and Fig. S3).
Figure 2.
Dm-Kat60, Dm-Spastin, and Dm-Fidgetin display distinct targeting within spindles. (A) Immunolocalization of endogenous Dm-Kat60, Dm-Spastin, and Dm-Fidgetin during metaphase and anaphase. Cells were triple stained to show AAA proteins (green), MTs (red), and Cid (blue). AAA refers specifically to Dm-Kat60, Dm-Spastin, and Dm-Fidgetin. All three AAA proteins localize to the centrosomes (arrowheads). In addition, all three localize to chromosomes (arrows): Dm-Kat60 is dispersed widely on chromosomes, whereas Dm-Spastin and Dm-Fidgetin are typically located in fewer puncta near kinetochores. The chromosomal immunostaining of Dm-Kat60 remained constant from metaphase to anaphase, whereas that of Dm-Spastin and Dm-Fidgetin decreased. Similar staining patterns were observed in hundreds of cells from multiple experiments. Bar, 5 μm. (B) Immunolocalization of Dm-Kat60 in late anaphase spindles. In panel 1, MTs are blue, Dm-Kat60 is red, and Cid is green. Panels 2 and 3 are higher magnifications of the region boxed in 1. Dm-Kat60 remains dispersed over the anaphase chromosomes and is also found at kinetochores (arrows).
In contrast, their association with chromosomes was more complex. Dm-Kat60 was present on chromosomes throughout mitosis, where it appeared as dispersed bright puncta covering the chromosome mass (Fig. 2 A). During anaphase, some of these Dm-Kat60 puncta clearly targeted to kinetochores, as indicated by colocalization with the kinetochore marker, Cid (Fig. 2 B). In contrast, the chromosomal targeting of Dm-Spastin and Fidgetin was prominent only during prometaphase/metaphase and decreased substantially at the onset of anaphase. Before anaphase, both Dm-Spastin and Dm-Fidgetin appeared on chromosomes as distinct puncta near, but usually not overlapping with, kinetochores. Interestingly, of the three, only Dm-Kat60 associates with chromosomes in the absence of MTs, indicating a fundamental difference in the mechanisms targeting these proteins to chromosomes (Fig. S3).
Dm-Spastin and Dm-Fidgetin modify the interaction between centrosomes and spindle MTs
That Dm-Kat60, Dm-Spastin, and Dm-Fidgetin all target to mitotic centrosomes implies that this organelle is an important site for severing of spindle MTs. Indeed, MTs have been observed to detach from centrosomes in interphase and mitotic cells (Keating et al., 1997; Rusan and Wadsworth, 2005), and similar events have been inferred in mitotic spindles based on the displacement between minus ends and centrosomes (Mastronarde et al., 1993). Therefore, we initially assessed the roles of these MT severing enzymes in regulating interactions between centrosomes and spindle MTs. The depletion of Dm-Kat60, Dm-Spastin, or Dm-Fidgetin by RNAi produced no increase of gross defects in spindle structure (Fig. S4, B–D, available at http://www.jcb.org/cgi/content/full/jcb.200612011/DC1). However, the frequency of mitotic cells within RNAi-treated cultures did increase significantly, suggesting subtle perturbations of mitotic progression (Fig. S4 A).
To examine this issue in more detail, we developed an assay exploiting the mitotic phenotype resulting from the inhibition of the abnormal spindle protein (Asp; Saunders et al., 1997). Asp is thought to form an insoluble matrix that anchors possibly severed MT minus ends to centrosomes (Wakefield et al., 2001), and its depletion by RNAi results in an easily identifiable and highly penetrant phenotype in which centrosomes completely dissociate from the central spindle (do Carmo Avides and Glover, 1999; Morales-Mulia and Scholey, 2005). We reasoned that a decrease in MT severing activity at centrosomes would ameliorate the Asp RNAi phenotype: increasing the density of unsevered MTs at centrosomes would abrogate the need for the anchoring activity of Asp (Fig. 3 A). Indeed, codepletion of either Dm-Spastin or Dm-Fidgetin with Asp restored the ability of centrosomes to associate with spindles, consistent with the notion that these proteins actively sever and release MTs from centrosomes (Fig. 3, B–D). Analysis of Asp immunofluorescence in co-RNAi–treated cells was used to verify Asp depletion (Fig. S2 E). Thus, this rescue phenotype is not due to a decreased efficiency of Asp knockdown resulting from double-target RNAi treatments. In contrast, Dm-Kat60 RNAi had no noticeable effect on the Asp RNAi phenotype and thus likely influences mitosis by a mechanism that is different from the other severing proteins (Fig. 3, B–D).
Figure 3.
Dm-Spastin and Dm-Fidgetin regulate the interaction between centrosomes and spindle MTs. (A) Working model of Asp RNAi phenotype rescue by AAA co-RNAi. After nucleation from γ-TuRC (a) and severing by an AAA protein (b), the newly generated minus end of an MT (c) can be anchored in the pole matrix. Asp RNAi disrupts the pole matrix, allowing centrosomes to displace from minus ends; co-RNAi of AAA to inhibit severing might rescue this phenotype, preventing the release of MTs from γ-TuRC. (B) Major phenotypes of spindles after the indicated RNAi treatments. Asp RNAi causes centrosomes to become significantly displaced from spindle poles, producing a noticeable gap between centrosomes and the unfocused poles. Asp/Dm-Kat60 co-RNAi spindles still display the predominant Asp RNAi phenotype, but co-RNAi of Asp and either Dm-Spastin or Dm-Fidgetin results in spindles with centrosomes always fully attached (i.e., without visible separation) or closely positioned to the poles. (C) Co-RNAi of Asp with Dm-Spastin or Dm-Fidgetin significantly increased the percentage of metaphase spindles with attached centrosomes (i.e., lacking a visible separation between poles and centrosomes). Asterisks mark treatments with significantly different means than control. Numbers within bars are sample sizes, and error bars are SD. (D) The centrosome-to-pole distances of all metaphase spindles with unfocused poles, regardless of the state of centrosome attachment, were measured. Co-RNAi of Asp with Dm-Spastin or Dm-Fidgetin significantly decreased this distance. Error bars indicate SD.
Dm-Spastin and Dm-Fidgetin, but not Dm-Kat60, stimulate poleward flux in metaphase spindles
Severing of MT minus ends at centrosomes and/or spindle poles has been proposed to be a necessary prerequisite to flux-related minus-end depolymerization (McNally and Vale, 1993). Given the ability of Dm-Spastin and Dm-Fidgetin to disassociate MTs from centrosomes, we were particularly interested in assessing the impacts of these severing proteins on flux. As described above, flux results from the persistent depolymerization of MT minus ends, which is balanced during metaphase by polymerization at plus ends, allowing the spindle to maintain a constant length and chromosomes to remain at the spindle equator. During anaphase, the cessation of plus-end polymerization leaves the minus-end depolymerization unbalanced, so a pulling force is generated on chromosomes. To avoid ambiguity, we refer only the poleward movement of tubulin subunits (which occurs during both metaphase and anaphase) as flux.
To test the importance of MT severing enzymes in flux, we monitored flux on metaphase spindles from control and RNAi-treated S2 cells using fluorescence speckle microscopy (Waterman-Storer et al., 1998) and photobleaching techniques (Fig. 4 A). Virtually identical results were obtained using either technique, so the data were pooled. In controls, flux was found to occur at a mean velocity of 0.81 ± 0.31 μm/min (Fig. 4 B), in good agreement with previous findings (Maiato et al., 2005).
Figure 4.
Dm-Spastin and Dm-Fidgetin, but not Dm-Kat60, stimulate poleward flux in metaphase spindles. (A) To visualize poleward flux, bars were photobleached across metaphase spindles of EGFP–α-tubulin expressing S2 cells, after RNAi treatment. Images are frames taken from time-lapse videos; the elapsed times (s) after photobleaching are indicated to the left or within the panels. Orange arrowheads mark the initial position of the photobleached bars, and blue arrowheads indicate their current positions. Kymographs were generated from the time-lapse recordings of these spindles to further illustrate the flux rates: a large angle between the tracks of bleached bar and pole indicates a relatively high flux rate (control and Dm-Kat60 RNAi), whereas more parallel tracks indicate decreased flux (Dm-Spastin and Dm-Fidgetin RNAi). t and d indicate the time and distance axes, respectively. (B) Mean flux rates (+SD) for metaphase cells after RNAi treatment. The flux rate after Dm-Kat60 RNAi is equivalent to control, whereas flux is significantly decreased after Dm-Spastin or Dm-Fidgetin RNAi. *, P < 0.05 versus controls. Numbers within bars are sample sizes.
RNAi of Dm-Spastin or Dm-Fidgetin significantly reduced the velocity of flux to 0.49 ± 0.29 and 0.48 ± 0.31 μm/min, respectively (Fig. 4 B). In the most severe cases, spindles lacking either of these proteins displayed a nearly complete cessation of flux. Unfortunately, simultaneous knock down of Dm- Spastin and Dm-Fidgetin resulted in a high degree of cell death, making it impossible to directly assess their combined impacts on flux or to determine whether they perform partially redundant functions. In contrast, RNAi of Dm-Kat60 had no discernible effect on metaphase flux, which continued at a mean velocity of 0.85 ± 0.33 μm/min (Fig. 4 B). Thus, Dm-Spastin and Dm-Fidgetin are flux regulators, whereas Dm-Kat60 is not.
Previously, we identified an MT-destabilizing Kinesin-13, KLP10A, as an important flux effector in D. melanogaster embryos (Rogers et al., 2004). This protein concentrates on centrosomes and spindle poles and likely catalyzes flux by directly stimulating the depolymerization of MT minus ends focused at poles. It is therefore conceivable that the depletion of Dm-Spastin or Dm-Fidgetin perturbs flux indirectly via the mislocalization of KLP10A. However, immunofluorescence performed on cells lacking Dm-Spastin or Dm-Fidgetin revealed no obvious alteration in KLP10A's localization (Fig. S2 D).
Dm-Spastin and Dm-Fidgetin stimulate the turnover of α-tubulin at the ends of spindle MTs
To further assess the impact of Dm-Kat60, Dm-Spastin, and Dm-Fidgetin on spindle MT dynamics, FRAP was used to measure and compare the turnover of fluorescent tubulin subunits at both MT ends in control RNAi cells. Regions at the spindle pole (MT minus ends) and the midzone (MT plus ends) of EGFP–α-tubulin containing metaphase spindles were photobleached, and recovery half-times (t 1/2) were calculated. Dm-Kat60 RNAi had no impact on tubulin turnover at either MT end (Fig. 5 A and Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200612011/DC1), consistent with its lack of impact on flux. In contrast, RNAi of either Dm-Spastin or Dm-Fidgetin significantly attenuated tubulin turnover at the spindle poles (Fig. 5 A, top and middle; and Fig. S5), consistent with the hypothesis that these proteins destabilize MT minus ends to promote flux. Somewhat surprisingly, depletion of these proteins also attenuated turnover at MT plus ends (Fig. 5 A, top and bottom; and Fig. S5). This finding is consistent with their association with preanaphase chromosomes (Fig. 2 A) and indicates that they play a more general role in regulating spindle MT dynamics, at least during metaphase. FRAP at MT ends, particularly plus ends, is likely driven by two nonexclusive mechanisms, flux and dynamic instability (Mitchison and Kirschner, 1984). Unfortunately, our assay was not sensitive enough to distinguish the relative contributions of each of these mechanisms to fluorescence recovery (although we believe that both contribute to EGFP–α-tubulin FRAP at plus ends).
Figure 5.
Dm-Spastin and Dm-Fidgetin stimulate the turnover of α-tubulin at MT ends and γ-tubulin at centrosomes. (A, top) FRAP of metaphase spindles of EGFP–α-tubulin expressing S2 cells after RNAi. One spindle pole region (MT minus ends) and one spindle equator region (MT plus ends) were photobleached, and their fluorescence recoveries were recorded. Images are frames from time-lapse recordings; the elapsed times after photobleaching are shown to the left. Fluorescent α-tubulin recovery after Dm-Kat60 RNAi is similar to control, whereas the recoveries after Dm-Spastin and Dm-Fidgetin RNAi are visibly slower. (middle) Mean fluorescence recovery half-times (t 1/2) for MT minus ends (left). Asterisks indicate treatments with statistically slower α-tubulin turnover rates compared with control. Percentage of fluorescence recoveries are shown (right). Numbers within bars are sample sizes, and error bars are SD. (bottom) Mean t 1/2 (+SD) of MT plus ends. (B, top) The two intensely fluorescent centrosomes of γ-tubulin–EGFP expressing S2 cells were photobleached, and their fluorescent recoveries were recorded. Selected time-point images and their postbleach elapsed times are shown. (middle) Mean t 1/2 (+SD) of γ-tubulin turnover at centrosomes. (bottom) Percentage of fluorescence recoveries is shown.
Dm-Spastin and Dm-Fidgetin catalyze the turnover of γ-tubulin at centrosomes
Severing of MTs near their minus ends has been proposed to stimulate flux by releasing minus ends from stabilizing caps formed by γ-tubulin ring complexes (γ-TuRCs; McNally and Thomas, 1998; Buster et al., 2002). If so, depletion of flux-stimulating Dm-Spastin or Dm-Fidgetin could also slow the turnover of γ-tubulin at centrosomes or elsewhere within the spindle (e.g., depolymerization of MT stubs that remain attached to γ-TuRCs after severing could disassemble the complex, causing γ-tubulin to be released from centrosomes or spindles). To test this hypothesis, FRAP analyses were performed in control and RNAi-treated S2 cells stably expressing γ-tubulin–EGFP. In these cells, γ-tubulin–EGFP fluorescence clearly concentrated at mitotic centrosomes but was difficult to visualize and measure with precision elsewhere within the spindle. Thus, we focused our analyses specifically on the turnover of γ-tubulin–EGFP associated with mitotic centrosomes.
After photobleaching in control cells, mitotic centrosome-associated γ-tubulin–EGFP fluorescence rapidly recovered (t 1/2 = 34 s) to a plateau of ∼40% of prebleach levels (Fig. 5 B and Fig. S5). This recovery showed a partial dependency on MTs: overnight treatment with colchicine (which depolymerizes MTs and arrests cells in mitosis) significantly reduced the FRAP recovery rate (t 1/2 = 60 s) in those cells that ultimately recovered to control levels. The recovery of γ-tubulin–GFP fluorescence, which, although attenuated, persists in the absence of MTs in some colchicine-treated cells, is consistent with the direct interchange between cytosolic and pericentriolar matrix-associated pools of the protein ( Khodjakov and Rieder, 1999).
Similar to colchicine treatment, depletion of either Dm-Spastin or Dm-Fidgetin significantly decreased the turnover of γ-tubulin at centrosomes, whereas Dm-Kat60 did not (Fig. 5 B and Fig. S5). Specifically, the FRAP t 1/2 in Dm-Spastin or Dm-Fidgetin RNAi-treated cells was more than double that of control cells (34 s for controls, 75 s for Dm-Spastin RNAi, and 78 s for Dm-Fidgetin RNAi). The overall similarity of these results supports the notion that Dm-Spastin and Dm-Fidgetin influence the turnover of centrosomal γ-tubulin by a mechanism involving MTs.
Dm-Spastin and Dm-Fidgetin regulate the number of plus ends in preanaphase spindles
If Dm-Spastin and Dm-Fidgetin catalyze the release of MT minus ends from their nucleating γ-TuRCs and/or centrosomes, then depletion of these proteins could also decrease the nucleation of new MTs and thus reduce the number of polymerizing plus ends within the spindle. To test this hypothesis, live analyses were performed on control and RNAi-treated cells expressing the plus-end tracking protein EGFP-EB1, which appears as comets on polymerizing plus ends (Rogers et al., 2002). As expected, the depletion of Dm-Spastin and Dm-Fidgetin, but not Dm-Kat60, significantly reduced the number of EGFP-EB1 comets near spindle poles and also within the interior of preanaphase spindles (Fig. 6 A). In a corroborating line of study, we found that the depletion of Dm-Spastin and Dm-Fidgetin slightly reduced the MT polymer mass within spindles relative to cytoplasmic tubulin; Dm-Kat60 depletion had no measurable effect (Fig. 6 B). This decrease alone is not likely the cause of the observed reductions in flux velocity, as flux continued at a statistically normal rate when MT polymer mass was reduced to a similar extent by treatment with low levels of colchicine (flux rate of untreated cells, 0.83 ± 0.22 μm/min; rate of colchicine-treated cells, 0.93 ± 0.18 μm/min; treatment was 50 μM colchicine for ∼30 min).
Figure 6.
Dm-Spastin or Dm-Fidgetin RNAi decreases the number of polymerizing plus ends and the fraction of total tubulin incorporated into the MT array of spindles. (A, left) Single confocal section of an EB1-EGFP expressing S2 cell before and after processing to enhance contrast of EB1-EGFP comets. The pole and equator regions used for EB1-EGFP comet counting are outlined with dashed lines. (right) Densities of EB1-EGFP at spindle poles (black bars) and equators (gray bars) after RNAi treatment. *, P < 0.05 versus controls. Numbers within parentheses are sample sizes, and error bars are SD. (B, left) A z-series projection of an EGFP–α-tubulin expressing cell. Total fluorescence is measured within region 1 (enclosing the metaphase spindle) and region 2 (the remainder of the cell) to calculate the spindle polymer/cytoplasmic fluorescence ratio. (The brightness of this image was increased to improve its visibility; for fluorescence measurements, saturated images were avoided.) (right) Ratios of tubulin polymer/monomer fluorescence in metaphase S2 cells after RNAi. Numbers within bars are sample sizes.
Dm-Kat60, Dm-Spastin, and Dm-Fidgetin are all required for chromosome segregation during anaphase A
We have shown that Dm-Spastin and Dm-Fidgetin are regulators of flux, at least during metaphase. If they persist in this function through anaphase, then depletion of these proteins should also cause a commensurate decrease in anaphase A velocity by diminishing the flux component of the Pacman-flux mechanism. Additionally, although Dm-Kat60 does not appear to influence flux, its association with anaphase chromosomes/kinetochores raises the possibility that it, too, contributes to anaphase A, but by a different mechanism (e.g., the stimulation of Pacman-associated MT plus-end depolymerization). To determine whether these proteins are generally involved in anaphase A, we used 4D spinning-disk confocal microscopy to visualize anaphase A chromosome movements in live RNAi-treated S2 cells stably expressing EGFP–α-tubulin. S2 cell spindles contain prominent bundles of kinetochore MTs, and chromosomes appear at the ends of these as EGFP-free dots. The motions of the negatively stained chromosomes can be tracked during anaphase (Fig. 7 A and Videos 1–4, available at http://www.jcb.org/cgi/content/full/jcb.200612011/DC1). In controls, chromosomes moved poleward at a mean velocity of 1.03 ± 0.28 μm/min (Fig. 7 B), consistent with earlier findings (Goshima and Vale, 2005; Maiato et al., 2005).
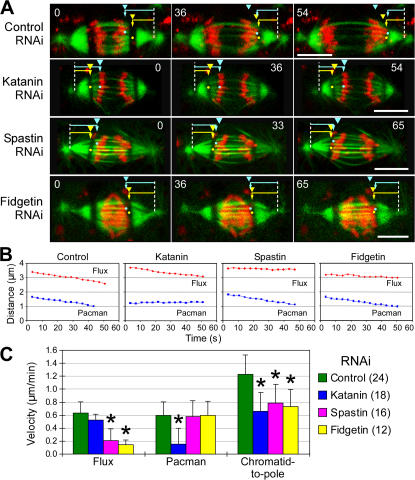
Figure 7.
Dm-Kat60, Dm-Spastin, and Dm-Fidgetin are all required for proper chromosome segregation during anaphase A. (A) After RNAi, EGFP–α-tubulin expressing S2 cells were recorded from metaphase until late anaphase as single confocal sections. Chromosomes are visible as black dots at the ends of the prominent kinetochore fibers. In the last frames, yellow dots mark the positions of the chromosomes, and blue dots the centrosome position, in a half spindle. Numbers are elapsed time after the first frame. (B) Anaphase chromatid-to-pole velocities (+SD) after RNAi. *, P < 0.05 versus controls. (C) Representative plots of chromatid-to-pole distance versus time taken from individual chromosomes in A.
Surprisingly, the depletion of any of these proteins caused a significant attenuation of anaphase A rate. These effects were remarkably similar regardless of the RNAi target, resulting in ∼50% reduction in the poleward velocity of chromatids: this motility was slowed to 0.47 ± 0.25 μm/min by Dm-Kat60 RNAi, to 0.48 ± 0.19 μm/min by Dm-Spastin RNAi, and to 0.43 ± 0.12 μm/min by Dm-Fidgetin RNAi (Fig. 7, B and C). A common manifestation of slowed anaphase A was the decondensation of chromosomes while still some distance from the poles, indicating that chromosome motion was reduced to such an extent that decondensation initiated before segregation was completed.
Dm-Kat60 contributes to anaphase A by stimulating MT plus-end depolymerization and Pacman
Finally, to specify the contributions of Dm-Spastin, Dm-Fidgetin, and Dm-Kat60 to anaphase A, we photobleached lines across spindles of live S2 cells containing both EGFP–α-tubulin and Hoechst-labeled chromosomes. The rate at which chromosomes moved toward the bleach mark corresponds to Pacman, whereas the poleward translocation of the bleach mark corresponds to flux (Fig. 8 A and Videos 5–7, available at http://www.jcb.org/cgi/content/full/jcb.200612011/DC1). In controls, Pacman and flux each accounted for ∼50% of the velocity of poleward chromatid motility (Fig. 8 C). After the depletion of Dm-Kat60, flux rate was normal but very little Pacman was observed (Fig. 8, B and C). Thus, Dm-Kat60 is involved in the stimulation MT plus-end depolymerization. (Dm-Kat60 RNAi did not influence the localization of several kinetochore/centromere-associated regulators of MT dynamics [Fig. S2 D]; thus, the inhibition of Pacman does not result indirectly from gross structural perturbations of kinetochore or the loss of other Pacman effectors from kinetochores.)
Figure 8.
Dm-Kat60 stimulates Pacman, whereas Dm-Spastin and Dm-Fidgetin drive poleward flux. (A) Frames of time-lapse recordings of anaphase EGFP–α-tubulin expressing S2 cells in which chromosomes were Hoechst stained just before visualization, and bars were photobleached across each half spindle. Yellow arrowheads mark the initial position of bleach marks; yellow arrows and dots mark their current positions. Blue arrowheads mark the initial positions of the leading edge of a selected chromosome; blue arrows and dots mark the current positions of the chromosome. The control spindle displays both poleward flux and Pacman motility (manifested as chromosomes overtaking bleach marks), but the Dm-Kat60 RNAi spindle has visibly diminished Pacman motility, and the Dm-Spastin and Dm-Fidgetin spindles have diminished flux. (B) Representative plots of distance versus time of individual chromosomes measured from the RNAi-treated cells in A. Red lines represent flux, and blue lines represent Pacman motility (measured as the approach of chromosomes to bleach marks). (C) Mean flux, Pacman, and chromatid-to-pole velocities (+SD) for anaphase spindles. Statistical differences from the control are marked with asterisks. Numbers in parentheses are sample sizes.
In contrast, RNAi of Dm-Spastin or Dm-Fidgetin had no impact on Pacman motility but, as expected, dramatically reduced the flux velocity (Fig. 8, B and C), consistent with the hypothesis that their impact on anaphase A stems entirely from their role in the stimulation of MT minus-end depolymerization and resulting influence on flux. Thus, all three proteins are incorporated into the Pacman-flux machinery that drives anaphase A. Dm-Spastin and Dm-Fidgetin are required for MT minus-end depolymerization and poleward flux, whereas Dm-Katanin is required for MT plus-end depolymerization and Pacman-based motility.
Discussion
The results of this study show that three closely related MT severing enzymes, Dm-Kat60, Dm-Spastin, and Dm-Fidgetin, are important for mitosis in D. melanogaster S2 cells. Interestingly, the activity of these proteins is segregated both spatially and temporally, allowing them to perform complementary functions throughout the spindle. This is most apparent during anaphase A, when all three are integrated into the Pacman-flux machinery used to move chromosomes.
Dm-Spastin and Dm-Fidgetin, but not Dm-Katanin, influence centrosome–MT attachments and poleward flux during mitosis
Dm-Spastin and Dm-Fidgetin emerge from this study as new regulators of poleward MT flux. Specifically, inhibition of either protein results in a significant reduction in flux velocity. In addition, we have found that both proteins similarly promote the turnover of α-tubulin at spindle poles and γ-tubulin at centrosomes. In sum, these data are consistent with a general model for flux and chromosome motility, in which Dm-Spastin and Dm-Fidgetin function to release MT minus ends from their nucleating γ-TuRCs, which are believed to cap and stabilize MT ends (Wiese and Zheng, 2000). In turn, severing exposes minus ends to depolymerization by spindle pole–associated Kinesin-13 (KLP10A in D. melanogaster), which has been shown to also contribute to flux (Rogers et al., 2004; Fig. 9 A). During anaphase A, the MT minus-end depolymerization of flux “reels in” chromosomes to the poles.
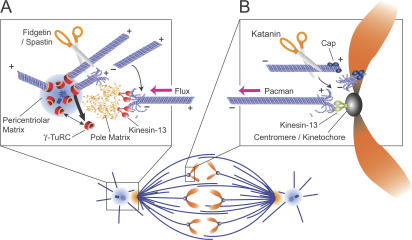
Figure 9.
Model: Katanin, Spastin, and Fidgetin function distinctly to drive Pacman-flux chromosome motility during anaphase. (A) At centrosomes, Spastin and Fidgetin sever MTs from the protective γ-TuRCs associated with centrosomes/poles. The newly generated MT ends can now disassemble; minus-end disassembly of the released MT is promoted by Kinesin-13 at the poles. Plus-end disassembly eliminates the short remaining MT and allows γ-TuRCs turnover at centrosomes. (B) At chromosomes, Katanin severs near the plus ends of spindle MTs, generating new free ends. This activity would remove any protective caps stabilizing plus ends. Kinesin-13 at centromeres/kinetochores then stimulates minus-end disassembly, enabling chromosomes to move poleward by Pacman.
Based on our proposal that Dm-Spastin and Dm-Fidgetin work in concert with KLP10A to promote flux, one would expect many similarities in the phenotypes resulting from the inhibition of these proteins. Indeed, as in Dm-Spastin or Dm-Fidgetin RNAi–treated cells, depletion of KLP10A also inhibits flux and slows anaphase A (unpublished data). A notable difference, however, is that KLP10A RNAi induces spindle elongation (Rogers et al., 2004; Goshima et al., 2005; Laycock et al., 2006), whereas Dm-Spastin or Dm-Fidgetin RNAi does not. One possible explanation for this apparent inconsistency stems from the fact that spindles probably elongate as a result of continued plus-end polymerization and MT sliding when minus-end depolymerization (i.e., flux) is decreased after KLP10A RNAi. Thus, the puzzling absence of spindle elongation after Dm-Spastin or Dm-Fidgetin RNAi might be explained by the observation that plus-end polymerization is also significantly decreased in these cells (Fig. 5 A, bottom).
Although our data demonstrate roles for Dm-Spastin and Dm-Fidgetin in regulating flux and MT–centrosome interaction (i.e., catalyzing the turnover of γ-tubulin and regulating Asp-mediated attachments with MT minus ends), it is currently unclear whether centrosomes are the sole or even primary site of action of these proteins in the spindle. Indeed, the presence of centrosomes is not required for spindle assembly, flux, and chromosome segregation in some D. melanogaster cell types (Bonaccorsi et al., 2000; Goshima and Vale, 2003; Mahoney et al., 2006) and other systems such as oocyte spindles (Sawin and Mitchison, 1994; Heald et al., 1997; Khodjakov et al., 2000). Additionally, even in centrosome-containing cells, the majority of MT minus ends are often positioned at a distance from centrosomes, and many spindle MTs are thought to arise from noncentrosomal sources (e.g., chromosomes/kinetochores; Khodjakov et al., 2000; Goshima and Vale, 2003; Maiato et al., 2004; Mahoney et al., 2006). These MTs may still be capped by cytoplasmic γ-TuRCs, and it is conceivable that severing within the spindle (i.e., away from centrosomes) is required for their normal dynamics and flux. Thus, although our model (Fig. 9 A) depicts Dm-Spastin and Dm-Fidgetin as functioning only at centrosomes (where these proteins concentrate), this may be an oversimplification.
Why both Dm-Spastin and Dm-Fidgetin would be used for the same task is unclear. At present, we have no clear evidence for a functional or physical interaction between these proteins. Each protein might sever a distinct subset of centrosomal MTs, but to our knowledge this would be unprecedented, and we consider it unlikely. Unfortunately, coinhibition of these proteins by RNAi causes a high degree of cell death, making it difficult to assess this possibility. Alternatively, a degree of functional redundancy may explain why a small portion of D. melanogaster carrying null mutations in the spastin gene survive to adulthood (Sherwood et al., 2004). Genetic analysis of the relationship between these proteins should be revealing and may help answer this question.
It is notable that Dm-Kat60 also localizes to centrosomes but performs no obvious function there, at least based on the assays used here. However, Dm-Kat60 RNAi does impact the mitotic index (Fig. S4 A), which is likely indicative of subtle preanaphase Dm-Kat60 activities, which are beyond the sensitivity of our visualization techniques.
MT severing by Dm-Katanin stimulates plus-end depolymerization and Pacman by anaphase chromosomes
Although Dm-Kat60 does not appear to function at centrosomes (Figs. 3 and 5), our data indicate that this protein plays an important role in moving anaphase chromosomes (Figs. 7 and 8). Dm-Kat60, which localizes to both chromosome arms and kinetochores, functions during anaphase to stimulate the depolymerization of MT plus ends, thereby moving chromosomes by a Pacman mechanism. We propose that Dm-Katanin functions in this regard by uncapping MT plus ends—much the same as Dm-Spastin and Dm-Fidgetin do at minus ends—and exposing them to depolymerization by centromere/kinetochore-associated Kinesin-13, which is also required for Pacman (Fig. 9 B). Although Pacman-inhibiting plus-end caps have not been identified, several MT-stabilizing microtubule-associated proteins (such as the plus-end tracking proteins CLASP, EB1, and CLIP-190) associate with kinetochore-associated MT plus ends (Dujardin et al., 1998; Rogers et al., 2002; Dzhindzhev et al., 2005). Whether the association of these proteins with plus ends inhibits depolymerization by Kinesin-13s is unknown.
Additionally, severing by Katanin could uncap plus ends associated with chromosome arms. A vertebrate kinesin, XKLP1, which targets to chromosome arms, has been shown to bind and stabilize MT plus ends and would probably resist Pacman motility (Bringmann et al., 2004). The D. melanogaster genome encodes several potential XKLP1 homologues, and it will be interesting to see whether Katanin has an antagonistic relationship with any of these.
We cannot rule out the possibility that D. melanogaster Katanin directly stimulates the depolymerization of kinetochore-associated MT plus ends. Indeed, it could conceivably supplant chromosome-associated Kinesin-13s in some systems, potentially explaining why the Kinesin-13 KLP59C does not appear to play a direct role in chromosome motility in S2 cells (Goshima and Vale, 2005) even though it drives Pacman in D. melanogaster embryos (Rogers et al., 2004). However, we have recently identified another Kinesin-13 that is needed for Pacman in S2 cells (unpublished data), making it unlikely that Dm-Katanin directly depolymerizes plus ends.
It is notable that Dm-Spastin and Dm-Fidgetin also target to chromosomes before anaphase, where they may function similarly to Dm-Katanin. FRAP analysis indicates that both proteins normally enhance the turnover of chromosome-associated plus ends on preanaphase spindles (Fig. 5 A, bottom). Why the chromosome activity of these proteins is down-regulated at the onset of anaphase while Katanin, which associates with chromosome throughout mitosis, is up-regulated is unclear. The loss of Spastin and Fidgetin from chromosomes may result from the underlying dependence of this targeting on MTs. Both are released from chromosomes in the presence of colchicine, and alterations in MT dynamics that accompany the onset of anaphase may have the same effect. Alternatively, Katanin's activity may be up- or down-regulated by phosphorylation. Indeed, the primary sequence of Dm-Kat60 contains several putative CDK1 phosphorylation motifs (unpublished data). Finally, Katanin's severing activity may be negatively regulated by MT-coating microtubule-associated proteins, as demonstrated in other systems (Vale, 1991; McNally et al., 2002; Qiang et al., 2006).
Conservation of mechanism
Interestingly, Katanin does not appear to target to chromosomes or kinetochores in many cell types. In fact, the only system besides D. melanogaster in which a Katanin homologue has been reported to associate with chromosomes is C. elegans, which does not use Katanin for mitosis (Srayko et al., 2000). This raises the question of whether the mitotic functions of MT severing proteins, particularly Katanin, are conserved throughout phylogeny. In this regard, we note that several additional Katanin p60 homologues whose functions have not yet been analyzed have been identified within vertebrate and invertebrate genomes (Emes and Ponting, 2001; Frickey and Lupas, 2004; see Results). Any of these could target to chromosomes and stimulate Pacman-based anaphase A. Moreover, a recent yeast two-hybrid study has shown that Fidgetin associates with the protein kinase A anchoring protein, AKAP95, which targets to chromosomes throughout mitosis (Yang et al., 2006). D. melanogaster contain no obvious AKAP95 homologue, perhaps explaining why Fidgetin does not impact Pacman in this system. Future studies to examine the possible mitotic functions of vertebrate Fidgetin and Katanin homologues would address this question.
In closing, we suggest a general mechanism in which appropriately positioned and tightly regulated MT severing proteins provide a means to rapidly create free MT ends, which are then exposed to the actions of other regulatory proteins. During anaphase, such an activity works in close coordination with Kinesin-13s, stimulating poleward chromatid motility by a combined Pacman-flux mechanism (Kwon et al., 2004). In other instances, the creation of free ends could have a very different impact on MT behaviors. Future analyses examining the interactions between severing proteins and Kinesin-13s, as well as other regulators of MT dynamics, will help test this proposal.
Materials and methods
S2 cell culture
S2 cells stably transfected with an EGFP–α-tubulin construct (under control of the copper-inducible pMT promoter or the constitutive pAc5.1 promoter [Invitrogen]) or with an EB1–EGFP construct (controlled by the pMT promoter) were a gift from R. Vale (University of California, San Francisco, San Francisco, CA). Cells were cultured according to published methods (Rogers et al., 2002).
dsRNAi
Sequences to be used for RNAi were selected by alignment of mRNAs to identify 500–600-bp regions for each protein that displayed minimal homology with other proteins in the FlyBase database. Selected sequences are as follows: Dm-Kat60 (CG10229), nucleotides 1885–2448 (in 3′ UTR of NM_080258); Dm-Spastin (CG5977), 2831–3389 (in 3′ UTR of NM_170115); Dm-Fidgetin (CG3326), 150–671 (in CDS of NM_134919); Asp (CG6875), 4207–4872 (in CDS of NM_079764). DNA templates for RNA synthesis were obtained by PCR of D. melanogaster ESTs (Drosophila Genomics Research Center) or S2 cell cDNA using the primers listed in the following paragraph. dsRNA was generated using commercial transcription kits (Megascript T7 [Ambion] or Ribomax T7 [Promega]) according to the manufacturers' instructions. For RNAi, S2 cells were treated on day 0, 2, and 4 by incubating for 1 h in 1 ml serum-free Schneider cell medium (Invitrogen) with 20 μg dsRNA, followed by addition of 1 ml Schneider medium containing 20% heat-inactivated FBS. Cells were replated and analyzed on day 5.
Each primer for generating dsRNA was preceded with T7 sequence (taatacgactcactataggg). The gene-specific sequences used for primers are as follows (listed as 5′ to 3′ for both forward/reverse primers): control, atggataagttgtcgatcg/accaggttcacatgcttgcg (template = pBluescript SK+; Stratagene); Asp, ctacatctgcgcgaggttacc/agcccttcgcttcatctcg; Dm-Fidgetin, tgctgcgctcaaggatcac/ttcgagctcacagttcgcttg; Dm-Kat60, gaatggctagcgattgtagg/atctctgcctgcactaaactatg; Dm-Spastin, cgttgtttaac caccatgtcc/acaccagatccatacgcacc.
Antibody production
GST and maltose binding protein fusions of the N-terminal regions of each protein displayed in Fig. S1 were bacterially expressed and purified using glutathione–Sepharose or amylose resins. Polyclonal antibodies were generated against the GST fusions (ProteinTech). Antibodies were affinity purified from sera using their respective maltose binding protein fusion proteins coupled to Affigel resin (Bio-Rad Laboratories). In addition, affinity-purified antibodies were preabsorbed with resin-bound fusions of N-terminal regions of the other AAA proteins to eliminate cross-reactivity.
Immunofluorescence microscopy
After RNAi, S2 cells were plated on concanavalin A–coated coverslips to stimulate cell spreading for microscopy (Rogers et al., 2002). Cells were fixed in −20°C methanol for 20 min and blocked with 5% normal goat serum in PBS containing 0.1% Triton X-100. Primary antibodies (against the Dm-AAA proteins or α-tubulin [DM1a; Sigma-Aldrich], γ-tubulin [GTU-88; Sigma-Aldrich], phospho-histone H3 [Upstate Biotechnology], or centromere marker Cid [a gift from G. Karpen, University of California, Berkeley, Berkeley, CA]) were applied at 1–20 μg ml−1 final concentrations in blocking buffer. Fluorescent secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 7.5 μg ml−1. DNA was stained with 1 μg ml−1 propidium iodide, 5 μM Draq5, or 0.3 μg ml−1 Hoechst 33258. Specimens were imaged using an Ultraview spinning-disk confocal system (PerkinElmer) mounted on an inverted microscope (Eclipse TE 300; Nikon) with a 100×, 1.4 NA objective and captured with a digital camera (Orca ER; Hamamatsu). Most images are displayed as maximum intensity projections of the captured z stacks. For experiments requiring MT disruption, cells were treated with 30 μM colchicine for 16 h just before fixation.
In vivo severing assay
Two similar approaches were used to assay severing activity. For Fig. 1 D, S2 cells (constitutively expressing mRFP–α-tubulin) were transiently transfected with a copper-inducible gene encoding full-length Dm-AAA protein fused to EGFP. Transfected cells were induced with 500 μM CuSO4 for 8–12 h and then imaged with the system described. Images of live cells were captured digitally with identical system settings (exposure time, gain, etc.). Using ImageJ (NIH), fluorescence intensities for both EGFP and mRFP were measured from entire cells expressing Dm-AAA-EGFP and neighboring control cells not visibly expressing EGFP.
For Fig. 1 E, wild-type S2 cells were transiently transfected with plasmids encoding EGFP or full-length Fidgetin-EGFP. After induction, cells were fixed (4% paraformaldehyde, 0.14% glutaraldehyde, 1 μM taxol, 0.1% Triton X-100, 1 mM MgCl2, 1 mM EGTA, and 80 mM Pipes, pH 6.8; 15 min, 24°C) and immunostained with DM1a to visualize MTs, and digital micrographs were captured as described. Polymer fluorescence intensity was calculated by subtracting the mean cytosolic fluorescence intensity of a cell (calculated from measurements made in several cytoplasmic regions devoid of MTs) from its total mean fluorescence intensity (measured from the entire cell).
Measurement of flux and anaphase chromatid-to-pole rates
Two techniques were used to measure poleward flux rates of preanaphase spindles: (1) fluorescence speckle microscopy and (2) the tracking of marks photobleached onto uniformly fluorescent spindle MTs. Because the flux rates produced by each technique were in good agreement for several experiments, the data were pooled.
To measure flux rates by fluorescence speckle microscopy, RNAi-treated cells expressing EGFP–α-tubulin (under control of the inducible pMT promoter) were plated on concanavalin A–coated microwell dishes as described. The leaky basal expression of the pMT promoter sometimes produces a low EGFP–α-tubulin titer necessary for speckling of spindle MTs. Speckled MTs were visualized with the Ultraview spinning-disk confocal system, and images were captured at 2- or 5-s intervals as single optical sections. Only cells with spindle orientations near perpendicular to the light path were analyzed. Using MetaMorph (Universal Imaging Corp.), images were processed with the high-sharpen and low-pass functions, and kymographs were generated from prominent speckles in each half-spindle. Each kymograph included the spindle pole, which served as a fiduciary point relative to which the rates of fluxing speckles were measured. Flux rates were calculated from the angles between the tracks of pole and speckles measured by MetaMorph.
Poleward MT flux rates were also measured on spindles of S2 cells constitutively expressing EGFP–α-tubulin (under control of the pAc5.1 promoter). Narrow rectangular regions were photobleached across the fluorescent bipolar spindles of RNAi-treated S2 cells using a confocal system (TCS SP2; Leica) on an inverted microscope (DMIRE2 [Leica]; Plan Apo 63× objective, 1.4 NA). Time-lapse videos of the photobleached spindles were captured with 3–6-s frame intervals. The movement of the bleach mark through the spindle was measured with the MetaMorph calipers tool, and flux rate was calculated from the change of distance between bleach mark and spindle pole as a function of time.
Chromatid-to-pole rates were measured in RNAi-treated anaphase S2 cells constitutively expressing EGFP–α-tubulin and vital stained with Hoechst 33258. Images of the Hoechst-stained chromosomes on fluorescently tagged spindle MTs were captured at 3–5-s intervals, and the chromatid anaphase rates were measured from the translocation distance through time. Alternatively, anaphase chromatids were tracked in EGFP– α-tubulin expressing cells by their negatively stained profiles at the ends of prominent kinetochore fibers.
FRAP
To measure α-tubulin turnover at MT ends, rectangular regions at the pole (MT minus ends) and spindle equator (MT plus ends) of RNAi-treated S2 cells stably expressing EGFP–α-tubulin were photobleached using the confocal system described in the previous section, and time-lapse videos of the bleached cells were immediately recorded. To measure γ-tubulin turnover at centrosomes, the two large fluorescent spots of S2 cells stably expressing γ-tubulin–EGFP were photobleached and their subsequent recoveries recorded. The half-time for fluorescence recovery (t 1/2) of each bleached region was measured from the plots of fluorescence recovery (corrected for postbleach fluorescence loss because of imaging). For those cases when the percentage of fluorescence recovery was <25%, the corresponding t 1/2 values were not included in calculating the mean t 1/2 for a treatment. Photobleaching was not observed to adversely affect cells; for example, photobleached cells were sometimes observed to proceed to anaphase.
Measurement of EB1-EGFP densities and tubulin polymer/monomer fluorescence ratios
Densities of comets of a fluorescently tagged +Tip protein, EB1-EGFP, were measured within metaphase spindles of RNAi-treated S2 cells by capturing each spindle as a z series of 1-μm optical sections with a spinning-disk confocal microscope (see Immunofluorescence microscopy), and images were processed by sequentially applying the “convolve” and “smooth” functions of ImageJ. Two spindle pole regions (each extending 1.25 μm from the tip of a pole toward the metaphase plate) and a single spindle equator region (extending 1.25 μm to each side of the metaphase plate) were delimited in each spindle. Fluorescent puncta of EB1-EGFP were counted within each z section of each region and then totaled. (Comet totals for the two pole regions were combined.) Densities were calculated by dividing EB1 comet totals by the region volumes (calculated by multiplying region areas [obtained from ImageJ] by the number of sections).
Total fluorescence of EGFP–α-tubulin within spindles and total fluorescence of EGFP–α-tubulin within the remainder of the cytoplasm were measured from projections of z series of confocal digital images of metaphase S2 cells using ImageJ. Ratios of these values were calculated and used to estimate proportions of tubulin distributed between polymer and soluble fractions. This estimation assumes that spindle fluorescence is primarily due to MT polymer and that fluorescence outside the spindle is primarily due to soluble tubulin.
Image processing and data analysis
Datasets were saved as stacks of TIFF files, and time-lapse series were saved as AVI videos. Datasets were processed and analyzed with MetaMorph or ImageJ as described. When fluorescence intensities were to be quantitated (Fig. 1), the digital images were recorded with identical settings of microscope and Ultraview software. The statistical differences between treatments were analyzed using either a one-way nonparametric analysis of variance (Kruskal-Wallis) for multiple group comparisons or a nonparametric t test (Mann-Whitney) for two group comparisons (SigmaStat, Systat Software, or GraphPad Prism; GraphPad Software). Measurement means were taken to be statistically different if P < 0.05.
Online supplemental material
Supplemental figures show production of mono-specific antibodies against the AAA proteins (Fig. S1), verification of target AAA protein knockdown after RNAi (Fig. S2), immunolocalization of AAA proteins after colchicine treatment (Fig. S3), analysis of spindle phenotypes after AAA RNAi (Fig. S4), and representative fluorescence recovery curves of photobleached α-tubulin or γ-tubulin of RNAi-treated spindles (Fig. S5). Videos 1–4 are recordings of live EGFP–α-tubulin expressing, anaphase S2 cells after RNAi with control, Dm-Katanin, Dm-Spastin, or Dm-Fidgetin dsRNA, respectively. Videos 5–7 are recordings of live EGFP–α-tubulin expressing, anaphase S2 cells after RNAi to knock down control, Dm-Katanin, or Dm-Spastin, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200612011/DC1.
Supplementary Material
Acknowledgments
The stable S2 cell lines expressing EGFP–α-tubulin or EB1-EGFP are much-appreciated gifts from the laboratory of Ron Vale. Thanks to the members of the Sharp laboratory, Frank McNally (University California, Davis), J.-Y. Huang (University of Newcastle), and J.R. McIntosh (University of Colorado) for useful discussion and insightful comments.
Research described in this article was supported by Philip Morris USA, Inc., Philip Morris International, and National Institutes of Health grant R01 GM65940 to D.J. Sharp. D.J. Sharp is a scholar of the Leukemia and Lymphoma Society.
Abbreviations used in this paper: ds, double-stranded; γ-TuRC, γ-tubulin ring complex; MT, microtubule.
References
- Bonaccorsi, S., M.G. Giansanti, and M. Gatti. 2000. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2:54–56. [DOI] [PubMed] [Google Scholar]
- Bringmann, H., G. Skiniotis, A. Spilker, S. Kandels-Lewis, I. Vernos, and T. Surrey. 2004. A kinesin-like motor inhibits microtubule dynamic instability. Science. 303:1519–1522. [DOI] [PubMed] [Google Scholar]
- Buster, D., K. McNally, and F.J. McNally. 2002. Katanin inhibition prevents the redistribution of gamma-tubulin at mitosis. J. Cell Sci. 115:1083–1092. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey, G., D.J. Sharp, A. Mogilner, and J.M. Scholey. 2006. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys. J. 90:3966–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, G.A., C.L. Mahaffey, A. Nystuen, V.A. Letts, and W.N. Frankel. 2000. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat. Genet. 26:198–202. [DOI] [PubMed] [Google Scholar]
- do Carmo Avides, M., and D.M. Glover. 1999. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science. 283:1733–1735. [DOI] [PubMed] [Google Scholar]
- Dujardin, D., U.I. Wacker, A. Moreau, T.A. Schroer, J.E. Rickard, and J.R. De Mey. 1998. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 141:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev, N.S., S.L. Rogers, R.D. Vale, and H. Ohkura. 2005. Distinct mechanisms govern the localisation of Drosophila CLIP-190 to unattached kinetochores and microtubule plus-ends. J. Cell Sci. 118:3781–3790. [DOI] [PubMed] [Google Scholar]
- Emes, R.D., and C.P. Ponting. 2001. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Genet. 10:2813–2820. [DOI] [PubMed] [Google Scholar]
- Errico, A., P. Claudiani, M. D'Addio, and E.I. Rugarli. 2004. Spastin interacts with the centrosomal protein NA14, and is enriched in the spindle pole, the midbody and the distal axon. Hum. Mol. Genet. 13:2121–2132. [DOI] [PubMed] [Google Scholar]
- Evans, K.J., E.R. Gomes, S.M. Reisenweber, G.G. Gundersen, and B.P. Lauring. 2005. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 168:599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey, T., and A.N. Lupas. 2004. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 146:2–10. [DOI] [PubMed] [Google Scholar]
- Goshima, G., and R.D. Vale. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and R.D. Vale. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 16:3896–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., R. Wollman, N. Stuurman, J.M. Scholey, and R.D. Vale. 2005. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988. [DOI] [PubMed] [Google Scholar]
- Hartman, J.J., J. Mahr, K. McNally, K. Okawa, A. Iwamatsu, S. Thomas, S. Cheesman, J. Heuser, R.D. Vale, and F.J. McNally. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93:277–287. [DOI] [PubMed] [Google Scholar]
- Hazan, J., N. Fonknechten, D. Mavel, C. Paternotte, D. Samson, F. Artiguenave, C.S. Davoine, C. Cruaud, A. Durr, P. Wincker, et al. 1999. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 23:296–303. [DOI] [PubMed] [Google Scholar]
- Heald, R., R. Tournebize, A. Habermann, E. Karsenti, and A. Hyman. 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier, L., J. Spring, M. Stierwald, J.M. Burgunder, and H. Reichert. 2003. Identification of the Drosophila melanogaster homolog of the human spastin gene. Dev. Genes Evol. 213:412–415. [DOI] [PubMed] [Google Scholar]
- Keating, T.J., J.G. Peloquin, V.I. Rodionov, D. Momcilovic, and G.G. Borisy. 1997. Microtubule release from the centrosome. Proc. Natl. Acad. Sci. USA. 94:5078–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and C.L. Rieder. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., R.W. Cole, B.R. Oakley, and C.L. Rieder. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67. [DOI] [PubMed] [Google Scholar]
- Kwon, M., S. Morales-Mulia, I. Brust-Mascher, G.C. Rogers, D.J. Sharp, and J.M. Scholey. 2004. The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell. 15:219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laycock, J.E., M.S. Savoian, and D.M. Glover. 2006. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J. Cell Sci. 119:2354–2361. [DOI] [PubMed] [Google Scholar]
- Maddox, P., A. Desai, K. Oegema, T.J. Mitchison, and E.D. Salmon. 2002. Poleward microtubule flux is a major component of spindle dynamics and anaphase A in mitotic Drosophila embryos. Curr. Biol. 12:1670–1674. [DOI] [PubMed] [Google Scholar]
- Maddox, P., A. Straight, P. Coughlin, T.J. Mitchison, and E.D. Salmon. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, N.M., G. Goshima, A.D. Douglass, and R.D. Vale. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569. [DOI] [PubMed] [Google Scholar]
- Maiato, H., C.L. Rieder, and A. Khodjakov. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., A. Khodjakov, and C.L. Rieder. 2005. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7:42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N., K.L. McDonald, R. Ding, and J.R. McIntosh. 1993. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, F.J., and R.D. Vale. 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 75:419–429. [DOI] [PubMed] [Google Scholar]
- McNally, F.J., and S. Thomas. 1998. Katanin is responsible for the M-phase microtubule-severing activity in Xenopus eggs. Mol. Biol. Cell. 9:1847–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, F.J., K. Okawa, A. Iwamatsu, and R.D. Vale. 1996. Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J. Cell Sci. 109:561–567. [DOI] [PubMed] [Google Scholar]
- McNally, K.P., O.A. Bazirgan, and F.J. McNally. 2000. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J. Cell Sci. 113:1623–1633. [DOI] [PubMed] [Google Scholar]
- McNally, K.P., D. Buster, and F.J. McNally. 2002. Katanin-mediated microtubule severing can be regulated by multiple mechanisms. Cell Motil. Cytoskeleton. 53:337–349. [DOI] [PubMed] [Google Scholar]
- McNally, K., A. Audhya, K. Oegema, and F.J. McNally. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T., and M. Kirschner. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J., and E.D. Salmon. 1992. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 119:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J., and E.D. Salmon. 2001. Mitosis: a history of division. Nat. Cell Biol. 3:E17–E21. [DOI] [PubMed] [Google Scholar]
- Morales-Mulia, S., and J.M. Scholey. 2005. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 16:3176–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang, L., W. Yu, A. Andreadis, M. Luo, and P.W. Baas. 2006. Tau protects microtubules in the axon from severing by katanin. J. Neurosci. 26:3120–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.C., S.L. Rogers, T.A. Schwimmer, S.C. Ems-McClung, C.E. Walczak, R.D. Vale, J.M. Scholey, and D.J. Sharp. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. [DOI] [PubMed] [Google Scholar]
- Rogers, S.L., G.C. Rogers, D.J. Sharp, and R.D. Vale. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak, A., and R.D. Vale. 2005. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 15:650–655. [DOI] [PubMed] [Google Scholar]
- Rusan, N.M., and P. Wadsworth. 2005. Centrosome fragments and microtubules are transported asymmetrically away from division plane in anaphase. J. Cell Biol. 168:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, W., V. Lengyel, and M.A. Hoyt. 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 8:1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K.E., and T.J. Mitchison. 1994. Microtubule flux in mitosis is independent of chromosomes, centrosomes, and antiparallel microtubules. Mol. Biol. Cell. 5:217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey, J.M., I. Brust-Mascher, and A. Mogilner. 2003. Cell division. Nature. 422:746–752. [DOI] [PubMed] [Google Scholar]
- Sherwood, N.T., Q. Sun, M. Xue, B. Zhang, and K. Zinn. 2004. Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2:e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko, M., D.W. Buster, O.A. Bazirgan, F.J. McNally, and P.E. Mains. 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Srayko, M., E.T. O'Toole, A.A. Hyman, and T. Muller-Reichert. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. [DOI] [PubMed] [Google Scholar]
- Svenson, I.K., M.T. Kloos, A. Jacon, C. Gallione, A.C. Horton, M.A. Pericak-Vance, M.D. Ehlers, and D.A. Marchuk. 2005. Subcellular localization of spastin: implications for the pathogenesis of hereditary spastic paraplegia. Neurogenetics. 6:135–141. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. 1991. Severing of stable microtubules by a mitotically activated protein in Xenopus egg extracts. Cell. 64:827–839. [DOI] [PubMed] [Google Scholar]
- Wakefield, J.G., S. Bonaccorsi, and M. Gatti. 2001. The Drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 153:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., A. Desai, J.C. Bulinski, and E.D. Salmon. 1998. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 8:1227–1230. [DOI] [PubMed] [Google Scholar]
- Wiese, C., and Y. Zheng. 2000. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2:358–364. [DOI] [PubMed] [Google Scholar]
- Yang, Y., C.L. Mahaffey, N. Berube, A. Nystuen, and W.N. Frankel. 2005. Functional characterization of fidgetin, an AAA-family protein mutated in fidget mice. Exp. Cell Res. 304:50–58. [DOI] [PubMed] [Google Scholar]
- Yang, Y., C.L. Mahaffey, N. Berube, and W.N. Frankel. 2006. Interaction between fidgetin and protein kinase A-anchoring protein AKAP95 is critical for palatogenesis in the mouse. J. Biol. Chem. 281:22352–22359. [DOI] [PubMed] [Google Scholar]
- Zhai, Y., P.J. Kronebusch, and G.G. Borisy. 1995. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 131:721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.