Abstract
Proper sister chromatid cohesion is critical for maintaining genetic stability. San is a putative acetyltransferase that is important for sister chromatid cohesion in Drosophila melanogaster, but not in budding yeast. We showed that San is critical for sister chromatid cohesion in HeLa cells, suggesting that this mechanism may be conserved in metazoans. Furthermore, although a small fraction of San interacts with the NatA complex, San appears to mediate cohesion independently. San exhibits acetyltransferase activity in vitro, and its activity is required for sister chromatid cohesion in vivo. In the absence of San, Sgo1 localizes correctly throughout the cell cycle. However, cohesin is no longer detected at the mitotic centromeres. Furthermore, San localizes to the cytoplasm in interphase cells; thus, it may not gain access to chromosomes until mitosis. Moreover, in San-depleted cells, further depletion of Plk1 rescues the cohesion along the chromosome arms, but not at the centromeres. Collectively, San may be specifically required for the maintenance of the centromeric cohesion in mitosis.
Introduction
In eukaryotes, sister chromatid cohesion depends on the ringlike cohesin complex, consisting of four subunits (SMC1, SMC3, SCC3, and SCC1/MCD1/RAD21 or the α-kleisin). Cohesins are recruited to chromosomes before DNA replication, a process that requires the SCC2–SCC4 complex and the assembly of preRC (Ciosk et al., 2000; Toyoda et al., 2002; Gillespie and Hirano, 2004; Rollins et al., 2004; Takahashi et al., 2004; Watrin et al., 2006). However, the recruitment alone is not sufficient for sister chromatid cohesion because yeast mutants lacking the acetyltransferase Eco1/Ctf7 (or Eso1 in fission yeast) exhibit defective cohesion despite cohesins continuing to localize to the chromosomes (Skibbens et al., 1999; Toth et al., 1999; Tanaka et al., 2000).
The mechanism involving Eco1/Ctf7 seems to be conserved, as its orthologues have been identified in Drosophila melanogaster (Williams et al., 2003) and humans (Bellows et al., 2003; Hou and Zou, 2005). Eco1/Ctf7 family proteins exhibit acetyltransferase activity and modify several cohesion proteins in vitro (Ivanov et al., 2002; Bellows et al., 2003; Hou and Zou, 2005). However, the acetyltransferase activity may not be required for sister chromatid cohesion (Ivanov et al., 2002; Brands and Skibbens, 2005). It has been suggested that the interactions with other proteins, rather than the acetyltransferase activity, is important for sister chromatid cohesion. In budding yeast, Eco1/Ctf7 interacts genetically and physically with many proteins involved in DNA replication (Skibbens et al., 1999; Madril et al., 2001; Edwards et al., 2003; Kenna and Skibbens, 2003; Skibbens, 2004), and its physical interaction with PCNA is required for sister chromatid cohesion (Moldovan et al., 2006).
After it is established, sister chromatid cohesion is maintained until anaphase. In yeast, cohesins locate along the entire chromosome in S, G2, and M phase and hold sister chromatids together along their entire length. However, in higher eukaryotes, most of the cohesins are removed from the chromosome arms in prophase by the so-called “prophase pathway” (Waizenegger et al., 2000). This step of cohesin removal depends on Wapl (Gandhi et al., 2006; Kueng et al., 2006) and the phosphorylation of the SA1/2 subunit of the cohesin complex (Hauf et al., 2005), which is catalyzed by the pololike kinase 1 (Plk1) and aurora B kinase. The cohesins at the kinetochores and at some heterochromatin regions are protected from this prophase pathway. Proteins such as Sgo1, PP2A, and Bub1 have been implicated in this protection (Salic et al., 2004; Tang et al., 2004; Kitajima et al., 2005, 2006; Riedel et al., 2006; Tang et al., 2006). The second step of cohesin removal is catalyzed by separase, which cleaves the α-kleisin and triggers the final separation of sister chromatids in anaphase.
Interestingly, a genetic screen in D. melanogaster revealed a putative acetyltransferase called San (Williams et al., 2003). San is essential for life and the mutant exhibits apparent sister chromatid cohesion defects. In D. melanogaster, San associates with Nat1 and Ard1, both of which are subunits of the N-terminal acetyltransferase A (NatA), which is conserved from yeast to human (Polevoda and Sherman, 2003). NatA has been extensively characterized in yeast, and no cohesion phenotype has been reported. Furthermore, the closest yeast homologue to San is Nat5. Although Nat5 is also found in the yeast NatA complex, its deletion causes no detectable phenotype (Gautschi et al., 2003). Therefore, the requirement of San-like protein for sister chromatid cohesion is not conserved in budding yeast.
In this study, we found that depletion of San in HeLa cells also causes precocious sister chromatid separation. The depletion causes cohesin to dissociate from the centromeres in mitosis without affecting the localization of Sgo1. Different from the findings in D. melanogaster, most San proteins do not associate with the NatA complex and, unlike San, the NatA complex is not required for sister chromatid cohesion. In addition, recombinant San exhibits acetyltransferase activity on its own and is capable of modifying several chromosome-associated proteins. Rescue experiments indicate that the enzymatic activity of San is required for sister chromatid cohesion. Cells depleted of San exhibit normal Sgo1 localization, but no detectable cohesin complexes at the mitotic centromeres. Finally, depletion of Plk1 rescued sister chromatid cohesion in the San-depleted cells along the chromosome arms, but not at or near the centromeres. This result indicates that San is not required for the establishment of the cohesion along the chromosome arms. It is, however, necessary for centromeric cohesion in human cells.
Results
Depletion of San causes premature sister chromatid separation in HeLa cells
A single human cDNA (gi 13376735) was identified by BLAST search using the protein sequence of D. melanogaster San. The full-length cDNA, encoding a protein of 169 aa, was cloned from a human fetal thymus cDNA library (CLONTECH Laboratories, Inc.) by PCR and the sequence was confirmed by analyzing at least three different clones (unpublished data). This sequence is identical to the recently described human homologue of San (Arnesen et al., 2006). Full-length recombinant San was produced in Escherichia coli, polyclonal antibody was generated using the recombinant San as the antigen, and the antiserum was affinity purified before use. As shown in Fig. S1 (available at http://www.jcb.org/cgi/content/full/jcb.200701043/DC1), the antibody detected a single protein band of ∼20 kD in HTC116, 293T, and HeLa cell lysates, which comigrated with the in vitro–synthesized protein in wheat germ extract.
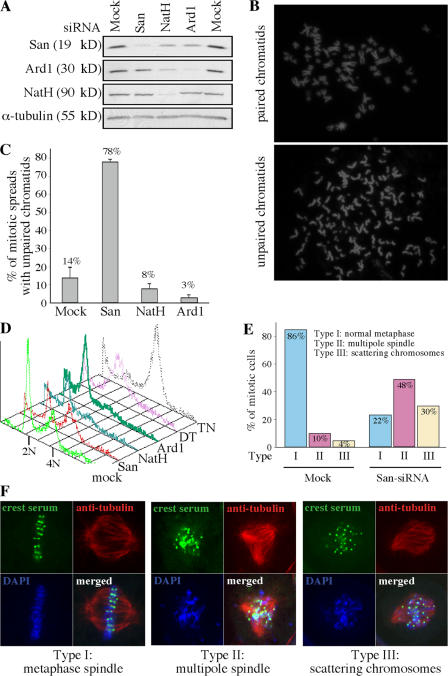
To investigate whether the loss of San causes any sister chromatid cohesion defect, we depleted endogenous San from HeLa cells by siRNA (Fig. 1 A). Mitotic chromosome spreads were prepared and analyzed. Remarkably, ∼78% of the spreads prepared from the San-depleted cells exhibited precocious chromatid separation, whereas only ∼14% of the mock-treated controls showed a similar phenotype (Fig. 1, B and C). In addition to the dramatic increase in the unpaired chromatids, San-siRNA cells also exhibited a noticeable delay in cell division. FACS analysis showed that San-siRNA cells of 4N DNA content accumulated from 15 to 60% (Fig. 1 D). Indirect immunofluorescent microscopy revealed that the incidences of multipole spindles (type II) and scattering chromosomes (type III) increased from 10 to 48% and from 4 to 30% of the mitotic cells, respectively (Fig. 1, E and F). The enrichment of 4N cells is likely caused by metaphase arrest because the level of cyclin B in these cells remained high, whereas the level of cyclin A was low (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200701043/DC1). All these phenotypes indicate that depletion of San causes premature sister chromatid separation in HeLa cells.
Figure 1.
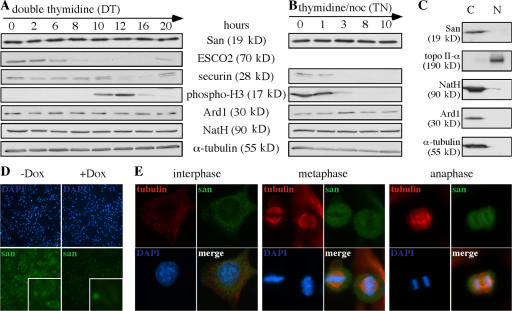
San is required for sister chromatid cohesion in HeLa cells. (A) San and the subunits of the NatA complex were effectively depleted by siRNA in HeLa cells. The levels of San, Ard1, and NatH were determined by immunoblot assay in siRNA- and mock-depleted cells. The level of α-tubulin was also blotted as a loading standard. (B) Chromosome spreads of paired (top) and unpaired chromatids (bottom) prepared from the aforementioned siRNA cells. (C) Quantification of the mitotic spreads with unpaired sister chromatids. The means of at least three independent trials are indicated above the bars. The error bars represent the SD. (D) FACS analysis of the HeLa cells depleted from San, NatH, and Ard1. As controls, cells arrested at the G1/S boundary by double-thymidine protocol (DT) and at c-metaphase by thymidine–nocodazole protocol (TN) were also analyzed. (E) Distribution of different types of mitotic cells observed in the mock- and San-siRNA cells. (F) Confocal images of the mitotic cells with multipole spindle and scattering chromosomes observed in San-siRNA cells. The kinetochores are stained with CREST serum (green). The microtubules are stained with a monoclonal anti–α-tubulin (red). DNA is stained with DAPI (blue).
Depletion of the NatA complex in HeLa cells causes lethality without detectable cohesion defects
In D. melanogaster, San was found exclusively in the NatA complex, implying a role of the N-terminal acetyltransferase activity in sister chromatid cohesion (Williams et al., 2003). To investigate any role of the human NatA complex in sister chromatid cohesion, we performed loss-of-function studies of the two subunits of the complex, NatH and Ard1 (Fluge et al., 2002; Arnesen et al., 2005). Depletion of Ard1 and NatH by siRNA caused a rapid loss of viability in 3 and 5 d, respectively (unpublished data). This was supported by the FACS analysis, which revealed many cells with DNA content smaller than 2N (Fig. 1 D). The lethality suggested that the depletion was effective. For the purpose of direct comparison with the result obtained in San-siRNA cells, we performed siRNA-depletion for both Ard1 and NatH and prepared chromosome spreads 5 d after the first transfection of the siRNA oligonucleotides. The depletion effect was further confirmed by Western blot (Fig. 1 A). In all cases, we did not observe any cohesion defects (Fig. 1 C). In fact, there were less unpaired sister chromatids than the mock-transfected controls. Therefore, it seems that the NatA complex is not involved in sister chromatid cohesion. However, we could not exclude the possibility that the essential function of the NatA complex, which causes lethality in its absence, overshadows its role in sister chromatid cohesion in these analyses.
Interestingly, siRNA-depletion of NatH reproducibly reduced the expression levels of both San and Ard1 (Fig. 1 A). It seems that the optimal protein levels of San and Ard1 depend on NatH. In addition, in approximately half of the depletion experiments, a reduction of San expression was also observed in the cells depleted of Ard1, such as the one shown in Fig. 1 A. This was likely caused by the observation that cells depleted of Ard1 were dying rapidly and San might be degraded in some experiments, depending on when the samples were harvested. In all cases, depletion of San did not affect the expression of Ard1 or NatH.
Over 80% of endogenous San is not associated with the NatA complex
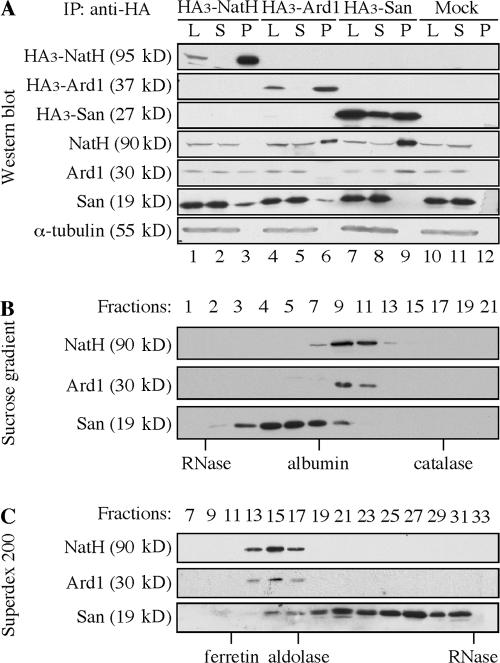
Because depletions of San and Ard1/NatH caused different phenotypes, we asked whether San associates with the NatA complex in HeLa cells. To this end, we fused HA3-tag to the N terminus of San and overexpressed it in 293T cells. The interactions between HA3–San and endogenous NatH and Ard1 were analyzed by coimmunoprecipitation with anti-HA beads. Both NatH and Ard1 precipitated with HA3-San and the amounts pulled down were >10% of their respective inputs (Fig. 2 A, lane 9). Similarly, both San and Ard1 coimmunoprecipitated with overexpressed HA3-NatH (Fig. 2 A, lane 3); and both San and NatH coimmunoprecipitated with overexpressed HA3-Ard1 (Fig. 2 A, lane 6). The interaction seemed to be stable, as it survived stringent washing conditions with up to 500 mM NaCl. A similar observation has recently been reported using 293 cells (Arnesen et al., 2006). However, the amounts of endogenous San pulled down by HA3-NatH and HA3-Ard1 were noticeably <10% of their respective inputs, despite the fact that the overexpressed HA3-NatH and HA3-Ard1 were virtually depleted by HA-beads (Fig. 2 A, lanes 2 and 5, respectively). This suggested that a significant pool of San might not associate with the NatA complex.
Figure 2.
The majority of San does not cofractionate with the NatA complex. (A) San interacts with the NatA complex in a coimmunoprecipitation assay. HA3-tagged NatH (lanes 1–3), Ard1 (lanes 4–6), and San (lanes 7–9) were individually expressed transiently in 293T cells. The tagged proteins were pulled down with anti-HA beads and the beads were washed in buffers containing 20 mM Tris, pH 8.0, 100–500 mM NaCl, 0.1% NP-40, and 10% glycerol. The proteins on anti-HA beads (P) and 10% of the lysates before (L) and after (S) immunoprecipitation were analyzed by immunoblot assay using antibodies to NatH, Ard1, and San. As a negative control, empty vector was also transfected and analyzed similarly (Mock). α-Tubulin was blotted as the loading control. 293T lysate (S100) was analyzed on a 0–10% sucrose gradient (B), and on a Superdex 200 gel filtration column (C). The standards are indicated below the lanes.
To further demonstrate whether San also exists outside of the NatA complex, we performed sucrose gradient analysis (Fig. 2 B) and size exclusion chromatography (Fig. 2 C) to examine whether endogenous San cofractionates with the NatA complex. In both analyses, NatH and Ard1 were detected in the same fractions. On the other hand, over 80% of San was found in the fractions excluding the NatA complex. This indicates that the majority of San either is not in the NatA complex or only weakly associates with the complex. Together with the loss-of-function studies, it appears that San mediates sister chromatid cohesion independent of the NatA complex.
San alone exhibits acetyltransferase activity in vitro
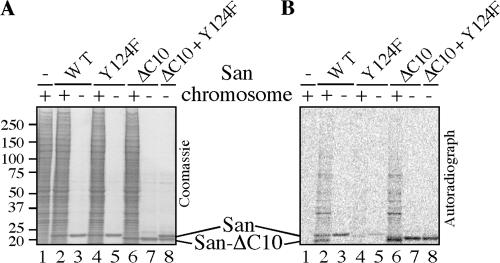
To determine whether San is an acetyltransferase independent of the NatA complex, we incubated purified chromosome pellet with recombinant San in the acetyltransferase activity assay. Immunoblot analysis failed to detect NatH and Ard1 in these chromosome pellets, which was expected because both proteins localize primarily in the cytoplasm. As expected, many proteins were acetylated in a San-dependent manner (Fig. 3, A and B, lane 2), although their identities have not yet been determined. Furthermore, San is also autoacetylated (Fig. 3, A and B, lane 3), which is a common feature of many acetyltransferases. Collectively, these results indicate that San, by itself, is an acetyltransferase.
Figure 3.
Recombinant San possesses acetyltransferase activity. In a 10-μl assay, ∼1 μg of recombinant San (wild type or mutant, as indicated above the lanes) was used in the presence of ∼1 mM 14C-acetyl-CoA. (A) After a 60-min incubation at 30°C, SDS-PAGE was performed and the gel was dried after Coomassie blue staining. (B) The same gel was also exposed to a phosphorimaging screen to detect the protein bands labeled by 14C. The molecular weight standards (in kilodaltons) are marked on the left of the Coomassie blue staining images. Chromosome pellet (4 μl) was used as the substrate in lanes 1, 2, 4, and 6.
To generate a mutant San defective in acetyltransferase activity, we substituted tyrosine-124 with phenylalanine because this tyrosine is conserved among several well-characterized acetyltransferases. Based on the atomic structure studies, mutational analyses, and/or enzymatic kinetics studies of serotonin N-acetyltransferase (SNAT), aminoglycoside 6′-N-acetyltransferase (ACC6), and HPA1/2 (Angus-Hill et al., 1999; Hickman et al., 1999; Wybenga-Groot et al., 1999), this tyrosine is required for catalysis. Furthermore, SNAT is predicted to be the closest structural homologue of San by 3D-PSSM (Kelley et al., 2000). As expected, this mutation reduced the level of autoacetylation by approximately threefold, as determined by quantifying the 14C signals of lanes 3 and 5 in Fig. 3 B. Similarly, the Y124F mutant only slightly increased the level of labeling above the background (Fig. 3 B, compare lane 4 with 1), and the 14C signals of lane 4 in the area above San were reduced by approximately ninefold. The difference in reduction between the autoacetylation and the acetylation of the chromosome substrates can be explained if the autoacetylation occurs via an intramolecular mechanism. Unlike an intermolecular mechanism, the reaction rate of an intramolecular mechanism will not be affected by the diminishing concentration of the substrate. Therefore, during the 1-h incubation time, the autoacetylation could be faster and more complete than the acetylation of the chromosome substrates. To determine whether autoacetylation occurs via an intramolecular mechanism, we constructed a mutant San lacking the C-terminal 10 aa (ΔC10). This mutant migrates faster than the full-length San on SDS-PAGE and remains active in vitro (Fig. 3 B, lanes 6 and 7). The mutant also rescued sister chromatid cohesion when expressed in the San-depleted cells (unpublished data). When incubated with ΔC10, the catalytic-defective Y124F mutant remained unlabeled (Fig. 3 B, lane 8), indicating that the autoacetylation, indeed, occurs via an intramolecular mechanism. Therefore, we have constructed a catalytic-defective San mutant by a single substitution at the conserved tyrosine-124.
The Y124F mutant does not rescue cohesion when expressed near the physiological levels
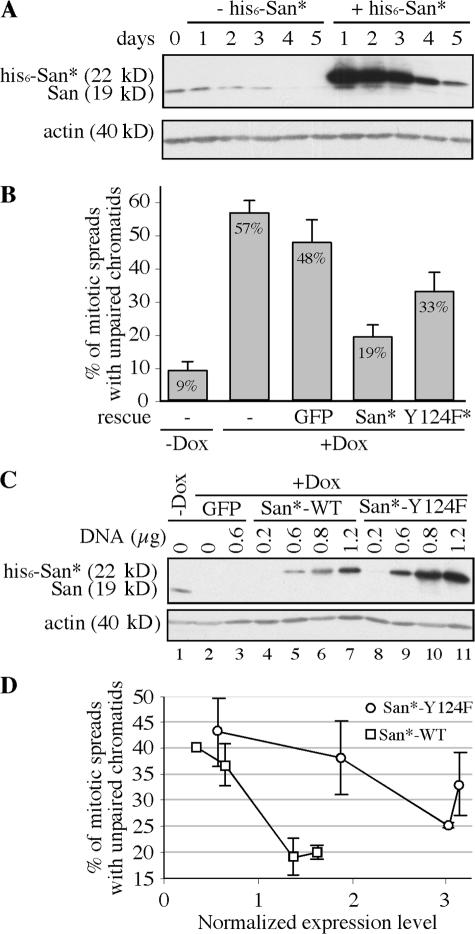
To determine whether the acetyltransferase activity of San is required for sister chromatid cohesion, we performed a rescue experiment. To this end, we first constructed a stable cell line where the expression of a shRNA targeting San was induced by doxycycline. The design of the shRNA is identical to the siRNA oligonucleotides that successfully deplete San in HeLa cells (Fig. 1 A). After screening ∼200 stable lines, one San-shRNA cell line that was integrated with three copies of shRNA units was isolated. After induction for ∼4–5 d, the level of endogenous San was reduced ∼90% (Fig. 4 A). The depletion increased the percentage of the mitotic spreads with unpaired sister chromatids from 9 to 57% (Fig. 4 B; P = 3 × 10−5; n = 5 in a paired t test). This, again, confirmed that San is required for sister chromatid cohesion.
Figure 4.
The Y124F mutant partially rescues the sister chromatid cohesion defect. (A) The inducible San-shRNA cell line and the scrambled rescue construct. The depletion effect of San-shRNA was analyzed by immunoblot assay over 5 d after doxycycline induction. Endogenous San was depleted in ∼4–5 d. The kinetics of depletion was not affected by the expression of the scrambled rescue construct (his6-San*). (B) Quantification of the rescue effect by San* and Y124F*. The means and the SDs (represented by error bars) of the percentage of the mitotic spreads with unpaired chromosomes were calculated using the data from five independent trials. (C) Immunoblot showing the levels of endogenous San and the rescue constructs at the time the cells were harvested. Actin was blotted as the loading controls. The amounts of transfected rescue constructs were indicated. (D) Rescue of sister chromatid cohesion by the wild-type and Y124F mutant San at different expression levels. The normalized expression level is determined by quantification of the Western blot shown in C and represents the fold of difference comparing to the expression level of the endogenous San. The error bars indicate the SD, which was calculated basing on three to five independent trials.
Using this conditional shRNA cell line, we tested whether sister chromatid cohesion could be rescued by the wild-type or Y124F mutant San. The Y124F mutant was chosen because it is based on an established substitution that has been shown to specifically affect catalysis in several acetyltransferases. The cells were transfected with the scrambled rescue constructs (his6-San* and his6-San-Y124F*) at the time of induction. After two rounds of transfections with 0.8 μg of the rescue construct, cells were harvested on day five. As shown in Fig. 4 B, transfecting GFP did not rescue the cohesion defect (48 vs. 57%; P = 0.09; n = 5). On the other hand, expression of the wild-type San significantly reduced the cohesion defect from 48 to 19% (P = 0.002; n = 5). The <100% transfection efficiency was likely responsible for not reducing the defects to the background level of 9% detected in the uninduced cells. On the other hand, the Y124F mutant only partially reduced the defects to 33%, which is not significantly different from 48% obtained with GFP (P = 0.06; n = 5), but significantly greater than 19% achieved by the wild type (P = 0.004; n = 5).
The partial rescue is most likely caused by the residual activity of the Y124F mutant described in Fig. 3, A and B. The effect of this residual activity might be further augmented by the high expression level of this mutant (Fig. 4 C, compare lane 6 with 10). We titrated the amount of transfected DNA from 0.2–1.2 μg and measured the rescue effects (Fig. 4 D). Within this range of DNA, we did not detect any variations in transfection efficiency by transfecting GFP under the same vector (unpublished data). With the same amount of DNA, the expression level of the Y124F mutant was reproducibly higher than that of the wild-type San, perhaps caused by a difference in mRNA or protein stability. To compare the effects of the rescue constructs expressing at the same level, we measured the signals of San and actin with a densitometry reader, calculated the ratio of the signals of San to actin, normalized the ratios derived from the rescue constructs to the ratio of endogenous San, and plotted the rescue effects of the wild-type and mutant San against their normalized expression levels (Fig. 4 D). When the wild-type San was expressed at a slightly higher rate than the endogenous San, sister chromatid cohesion was rescued with only ∼20% the spreads exhibiting unpaired chromatids. At the same expression level, the Y124F mutant had little, if any, effect. When the expression of Y124F increased to approximately threefold of the endogenous San, we observed a rescue effect similar to that achieved by the wild-type San expressing near the endogenous level. Therefore, the Y124F mutant is approximately threefold less effective at rescuing sister chromatid cohesion in San-siRNA cells. Remarkably, the Y124F mutant is also threefold less active than the wild-type San, as determined by autoacetylation (Fig. 3 B). Therefore, we conclude that the acetyltransferase activity of San is required for its function in mediating stable sister chromatid cohesion.
San localizes to the cytoplasm and its expression levels are unchanged during the cell cycle
Although the results from the aforementioned experiments indicate that San and its acetyltransferase activity are required for stable sister chromatid cohesion in HeLa cells, the mechanism remains elusive. Delineating the regulation of San may shed some light on this. To this end, we analyzed the expression levels of San in different phases of the cell cycle. HeLa cells were synchronized at the G1/S transition and c-metaphase by double-thymidine and thymidine–nocodazole treatment, respectively. Next, the cultures were synchronously released into the cell cycle. Cells were withdrawn from the cultures at various time points, and the levels of San and an array of cell cycle markers were determined by immunoblot assay. As shown in Fig. 5 (A and B), the expression levels of San are constant throughout the cell cycle.
Figure 5.
Cell cycle regulation and cellular localization of San. (A and B) The protein level of San is unchanged throughout the cell cycle. HeLa cells were synchronized at the G1–S transition by the double thymidine arrest protocol (A) and at c-metaphase by the thymidine–nocodazole arrest protocol (B) and released into the cell cycle. ESCO2, phospho-histone H3, and securin were blotted as the markers for different stages of the cell cycle. Specifically, the degradation of ESCO2 (Hou and Zou, 2005) commences at late S phase (6 h after release from DT); histone H3 becomes phosphorylated in mitosis (10–12 h after release from DT and 0–3 h after release from TN); and securin is degraded in anaphase and early G1 (12–16 h after release from DT and 3–10 h after release from TN). (C) San is detected in the cytoplasmic fraction. HeLa cells were fractionated into the cytoplasm (C) and the nuclear (N) fractions. Topoisomerase II and α-tubulin are markers for the nuclear and cytoplasm fractions, respectively. As expected, NatA and Ard1 were also detected in the cytoplasm (Arnesen et al., 2005). (D) The antibody to San detected San and its cytoplasmic location (insets) in the uninduced (−Dox), but not in the induced (+Dox), San-shRNA cells. San-shRNA cells were incubated under induced and uninduced conditions for 5 d before being analyzed. (E) The localization of San, as determined by immunofluorescent microscopy. The localization of San in HeLa cells was analyzed in the cells at interphase, metaphase, and anaphase.
Next, we tested whether the cellular localization of San is regulated. We performed cellular fractionation and detect San only in the cytoplasmic fraction (Fig. 5 C). To confirm this localization, we also performed indirect immunofluorescence microscopy using our affinity-purified polyclonal antibody. As shown in Fig. 5 D, the antibody is specific to San because it detects strong signals in the uninduced San-shRNA cells (−Dox), but only weak signals upon induction (+Dox). Enlarged images are shown in the insets, and, consistent with the cellular fractionation, San signals were detected in the cytoplasm in the uninduced cells. On the contrary, the weak background signals distributed uniformly in the induced cells. Using this antibody, we examined the localization of San in various phases of the cell cycle. San localizes to the cytoplasm in interphase and is excluded from chromosomes in metaphase and anaphase (Fig. 5 E). Similar observations were confirmed with HA3-tagged San, which was transiently overexpressed in HeLa cells (unpublished data). Although we could not exclude the possibility that a small undetectable fraction of San is inside the nucleus, the cytoplasmic localization implies that San may facilitate sister chromatid cohesion either directly and only during mitosis after the breakdown of the nuclear envelope or indirectly in interphase by acetylating a cytoplasmic factor, which is shuttled into the nucleus.
Sgo1, but not cohesin, localizes to the centromeres in San-depleted HeLa cells
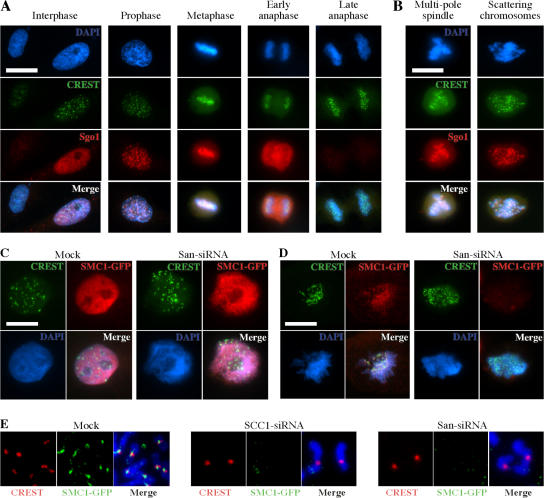
Studies in D. melanogaster suggested that San might be involved in the centromeric cohesion. To investigate whether the same is true in HeLa cells, we analyzed whether the localization of Sgo1, which is one of the factors required for cohesion at the centromeres, was affected by depletion of San. Using an antibody described previously (Tang et al., 2004), we examined the localization of Sgo1 in the San-shRNA cell line under both the induced and uninduced conditions. We first confirmed the previous finding that Sgo1 localizes to the centromeres. As shown in Fig. 6 A, the nuclear localization of shugoshin was detected in some, but not all, interphase cells. This is consistent with the fact that Sgo1 is a substrate of the anaphase-promoting complex (APC/C) and is degraded only in early G1 cells (Salic et al., 2004). In prophase cells, punctuated staining of Sgo1 was detected, which roughly colocalized with the kinetochores illuminated by crest serum. Centromeric Sgo1 was also detected in metaphase cells. However, in early anaphase, although the overall levels of Sgo1 remained high, the signals on chromosomes had greatly diminished. By late anaphase or telophase, Sgo1 was barely detectable. Similar analysis revealed the same dynamics of Sgo1 in cells depleted of San. As shown in Fig. 6 B, we detected strong centromeric Sgo1 signals in the cells with either multipolar spindles or scattering chromosomes (Fig. 6 B), indicating that, as in D. melanogaster, Sgo1 localizes to the centromeres in a San- independent manner.
Figure 6.
Depletion of San causes centromeric cohesion defects. (A) Shugoshin localizes to the centromeres in HeLa cells. Shugoshin localization at various stages of the cell cycle was analyzed by immunofluorescent microscopy. The cells used in this experiment were the San-shRNA cells without induction. (B) Shugoshin localizes to the centromeres in San-shRNA cells under induction. Images shown are San-depleted cells with multipole spindle and scattering chromosomes. (C) Immunofluorescent staining of the interphase SMC1-GFP HeLa cells. The cells were extracted before fixation. (D) Immunofluorescent staining of the mitotic SMC1-GFP cells. The cells were extracted before fixation. (E) Immunofluorescent staining of the mitotic spreads prepared from the SMC1-GFP cells. SCC1 was also depleted by siRNA to serve as a control. The signal of SMC1-GFP appears sandwiched between the signals of CREST. These signals were undetectable in both SCC1- and San-siRNA cells. Bars, 5 μm.
Next, we determined whether the centromeric localization of the cohesin complexes was affected by depletion of San. To this end, we used a stable cell line expressing a C-terminal GFP-tagged SMC1. The tagged SMC1 is a good reporter for the cohesin dynamics for the following reasons. First, the expression of SMC1-GFP is well below the endogenous level of SMC1 (Fig. S3 A, lane 1, available at http://www.jcb.org/cgi/content/full/jcb.200701043/DC1). Second, SMC1-GFP interacts with SCC1 in a coimmunoprecipitation assay (Fig. S3 A, lane 2) and cosediments with the cohesin complex on a sucrose gradient (Fig. S3 B). Third, SMC1-GFP localizes to chromosomes in interphase, and most of them disassociate from chromosomes in mitosis (Fig. 6 C and Fig. S3 C). Fourth, the chromosome localization of SMC-GFP depends on the presence of SCC1 (Fig. 6 E). Finally, SMC1-GFP can be detected at or near the centromeres in mitosis (Fig. 6, D and E). All of these observations are expected for the cohesin complex. As shown in Fig. S3 E, we were able to effectively deplete San in this cell line without affecting the protein levels of SCC1 and SMC1-GFP. The depletion increased the percentage of the mitotic spreads with unpaired chromatids from 2 to 58%. Next, the depleted cells were extracted with detergent and fixed to analyze the localization of SMC1-GFP. As shown in Fig. 6 C, SMC1-GFP localized to chromosomes during interphase in both mock and San siRNA-depleted cells. Therefore, San is not required for the overall association of the cohesin complex with chromosomes. On the other hand, SMC1-GFP was no longer detected on the mitotic chromosomes in the absence of San (Fig. 6 D). By analyzing the mitotic chromosome spreads (Fig. 6 E), cohesin was no longer detected at the centromeres in San-siRNA cells. Therefore, it seems that San is required for the mitotic localization of cohesin at the centromeres.
Depletion of Plk1 in the San-depleted cells rescues sister chromatid cohesion at the chromosome arms, but not centromeres
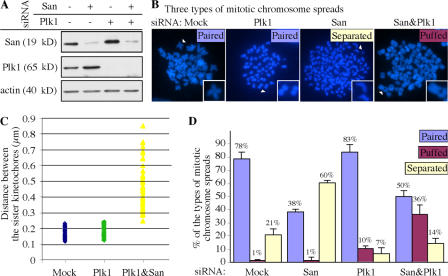
To determine whether San is specifically involved in centromeric cohesion or is also required for cohesion at chromosome arms, we inactivated the prophase pathway so that cohesion at chromosome arms could be examined in mitosis. To this end, both Plk1 and San were depleted separately and simultaneously (Fig. 7 A), and the resulting chromosome spreads were analyzed. We observed three major types of mitotic chromosome spreads. The “paired” type contained mostly the paired chromatids. This type could be further classified into two subtypes. One consisted of the butterfly-shaped chromosomes observed mostly in the mock-treated samples (Fig. 7 B, Mock). The other subtype had tightly paired chromosomes with cohesion along their entire length. This subtype is observed in the Plk1-depleted samples (Fig. 7 B, Plk1). The “separated” type contained mostly completely separated chromatids observed in the San-depleted samples (Fig. 7 B, San). In the double-depleted samples, many of the spreads contained chromatids that were separated only in the middle of the chromosomes (Fig. 7 B, San&Plk1). This type of spread was called the “puffed” type. Remarkably, the separated region always contained the centromeres, as revealed by staining the spreads with the crest serum and Sgo1, which illuminate the kinetochores (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200701043/DC1). The distances between the kinetochores of these puffed chromatids in the San&Plk1-depleted samples were significantly extended compared with the paired chromatids in the mock and Plk1-depleted samples (Fig. 7 C). Therefore, the “puffed” type represents the chromosomes with defective cohesion specifically at their centromeres.
Figure 7.
Depletion of Plk1 rescued the cohesion at chromosome arms, but not at centromeres, in San-depleted cells. (A) San and Plk1 were effectively depleted by siRNA. The immunoblot shows the level of San and Plk1 after RNAi depletion. The level of actin serves as the loading control. (B) Examples of three types of the mitotic chromosome spreads observed in the depletion experiments described in A. The chromosomes marked with the white arrowheads are enlarged by twofold and displayed in the insets. (C) Comparison of the distance between the sister kinetochores in various siRNA-depletion cells. The distance was measured between the crest signals between two paired or puffed chromatids (Fig. S4). Completely separated chromatids were not analyzed. The medians of the distances in mock-, Plk1-, and Plk1&San-depleted cells are 0.18, 0.20, and 0.43 μm, respectively. For each depletion, 60 chromosomes were measured. (D) Distribution of the types of mitotic spreads observed. The means of at least four independent trials are indicated above the bars. The error bars represent the SD. Fig. S4 is available at available at http://www.jcb.org/cgi/content/full/jcb.200701043/DC1.
As expected, Plk1 depletion resulted in 83% of the mitotic spreads displaying mostly tightly paired chromosomes (Fig. 7 D). This depletion also reproducibly caused 10% of the “puffed” type, which may reflect a background level of defective cohesion at the centromeres in HeLa cells. In the absence of cohesion at the arms, this background defect would have contributed to the “separated” type observed in 21% of the mock-treated cells. In addition, we also detected ∼7% of the “separated” type in the Plk1 depletion samples. These cells grossly failed to establish or maintain sister chromatid cohesion. Remarkably, double depletion of San and Plk1 significantly reduced the “separated” type from 60 to 14%in the San-depleted samples (P = 0.001; n = 3). At the same time, the “puffed” type significantly increased from 10% (the background level in the Plk1-depleted cells) to 36% (P = 0.03; n = 3) and the “paired” type slightly increased from 38% (mostly the butterfly-shaped chromosomes) to 50% (mostly the tightly paired chromosomes; P = 0.08; n = 3). The rescue of cohesion at the chromosome arms indicates that San is not required for establishment or maintenance of cohesion at these regions. On the other hand, the failure to rescue the centromeric cohesion suggests that San is necessary for the establishment and/or maintenance of the cohesion at the centromeres.
Discussion
San is required for sister chromatid cohesion in metazoans
Depletion of San causes sister chromatids to separate prematurely. This result can be interpreted in two ways. The simplest explanation is that San is required for sister chromatid cohesion. Alternatively, depletion of San may cause prolonged mitotic arrest, which allows enough time for the prophase pathway to remove even centromeric cohesion. We consider the second explanation unlikely for the following two reasons: first, the actual length of mitotic arrest was <12 h in our experiments because we removed most of the loosely attached mitotic cells by a shake-off during a medium change 12 h before harvesting the cells; second, the prophase pathway mainly depends on Plk1 to remove cohesins from chromosomes. However, in the double-depletion experiment, cohesion between the chromosome arms is rescued, whereas the centromeres remain separated (Fig. 7 D). This directly argues against any significant role of the prophase pathway in mediating the premature chromatid separation in cells depleted of San. Collectively, these data strongly support that San is required for stable sister chromatid cohesion. Because the homologue of San is required for cohesion in D. melanogaster, but not in budding yeast, this function of San may be conserved among only metazoans.
The acetyltransferase activity of San is required for sister chromatid cohesion
The implication of San in sister chromatid cohesion again raises the question of whether the activity itself is required. To test this, we generated a catalytic-defective mutant San by a single mutation at the conserved tyrosine-124. Based on the extensive studies of several established acetyltransferases, this conserved tyrosine is involved in catalysis (Angus-Hill et al., 1999; Hickman et al., 1999; Wybenga-Groot et al., 1999). As expected, the Y124F substitution reduces the autoacetylation of San about threefold. Because the autoacetylation occurs via an intramolecular mechanism (Fig. 3), a reduction of autoacetylation indicates a reduction in the enzymatic activity rather than substrate interaction. Furthermore, although the residual activity is less desirable for the rescue experiment, it does indicate that the substitution does not grossly disrupt the conformation of San. Consistent with the notion that the acetyltransferase activity of San is required for sister chromatid cohesion, the Y124F mutant, even expressed at a higher level, failed to rescue the cohesion as efficiently as the wild-type San (Fig. 4). Collectively, our results strongly suggest that the acetyltransferase activity of San is required for its function in sister chromatid cohesion.
San is required for centromeric cohesion by stabilizing cohesin at the centromeres in a Sgo1-independent manner
In HeLa cells, the localization of Sgo1 appears unchanged after the depletion of San (Fig. 6 B). This is similar to what was reported in D. melanogaster (Williams et al., 2003). Nonetheless, the centromeric cohesion is compromised based on the “puffed” phenotype in cells depleted of both San and Plk1 and the lack of cohesin at the mitotic centromeres in cells depleted of San. How does San mediate the centromeric cohesion? One possibility is that San is required for the establishment of sister chromatid cohesion specifically at the centromeres. In the absence of San, the centromeric cohesion may never be properly established, thus cannot be rescued by inactivating the prophase pathway. This scenario is attractive because genetic studies suggest that acetyltransferase Eco1/Ctf7 may be involved in the establishment of sister chromatid cohesion during S phase. It is possible that in metazoans, the establishment of cohesion may use two different acetyltransferases. However, San seems to localize to the cytoplasm in interphase cells (Fig. 5). This localization is also conserved in D. melanogaster (Williams et al., 2003). In contrast to San, the members of the Eco1/Ctf7 family localize directly on interphase chromosomes in yeast and human cells (Toth et al., 1999; Tanaka et al., 2001; Hou and Zou, 2005). Therefore, it is unlikely that San plays a direct role in the establishment of sister chromatid cohesion in interphase. It may, however, modify a cohesion establishment factor in the cytoplasm, which is then shuttled into the nucleus.
Alternatively, San may be involved in the maintenance of the centromeric cohesion in mitosis. This is consistent with the apparently normal cohesin localization in interphase cells (Fig. 6 C). If this is the case, the epistasis of the San and Plk1 depletions suggests that San may function downstream of Plk1. However, because Plk1 seems to phosphorylate SA2 directly to remove cohesin from chromosomes (Hauf et al., 2005), a linear pathway, which places San between Plk1 and cohesin, becomes less plausible. On the other hand, San and Plk1 may work independently to stabilize cohesins on the mitotic centromeres, in which both the dephosphorylated status of SA2 and the acetylated-status of a San substrate are required for stable centromeric cohesion. This scenario will satisfy the epistasis described in Fig. 7. Interestingly, a recent study demonstrated that a phosphorylation of histone H3 may be required for sister chromatid cohesion in a Sgo1-independent manner (Dai et al., 2006). It is possible that San may also mediate this histone phosphorylation event. The identification of the “cohesion substrate” of San will ultimately shed light on the mechanism.
Materials and methods
Antibodies
The polyclonal rabbit antibody to human San was raised by Genemed Synthesis, Inc. using His-tagged recombinant San as the antigen. The resulting crude serum was affinity-purified before use in immunoblot and indirect immunofluorescent staining. Antibodies to NatH and Ard1 were provided by J.R. Lillehaug (University of Bergen, Bergen, Norway; Arnesen et al., 2005). Antibody to Sgo1 was a gift from H. Yu (University of Texas Southwestern Medical Center, Dallas, TX; Tang et al., 2004). Antibodies to securin (Zou et al., 1999) and ESCO2 (Hou and Zou, 2005) have been previously described. Antibodies to cyclin B1, cyclin A2, α-tubulin, phospho-H3, and topoisomerase II-α were purchased from Santa Cruz Biotechnology. The CREST serum was purchased from Immunovision. FITC, Cy3, and Alexa Fluor 488–labeled secondary antibodies were purchased from Invitrogen.
Cell culture, cell cycle synchronization, cellular fractionation
HeLa and 293T cells were grown in DME, whereas HCT116 cells were cultured in McCoy's 5A, both supplemented with 10% FBS. The GFP-SMC1–stable HeLa cell line was maintained in DME containing 0.5 mg/ml G418 (Sigma-Aldrich). HeLa cells were synchronized at G1/S or c-metaphase by double thymidine block or thymidine–nocodazole arrest, respectively (Fang et al., 1998). The cellular fractionation was performed as previously described (Mendez and Stillman, 2000).
Transient transfection of siRNA oligonucleotides
To knockdown San and NatH transiently in HeLa cells, two consecutive transfections were performed on days one and two by the calcium phosphate method, and cells were harvested on day five for analysis. To deplete Plk1, a previously described RNA duplex (Kraft et al., 2003) was introduced into HeLa cells on day four. The RNA oligonucleotides (sense/antisense) synthesized by Thermo Fisher Scientific to deplete NatH, Ard1, and San are ACCUUGGCUAUGAAAGGActt/AUCCUUUCAUAGCCAAGGUtt, UGGGAAGAUUGUGGGGUActt/AUACCCCACAAUCUUCCCAtt, and GACAAGUUCUACAAUUAGtt/AUCCUUGUAGAACUUGUCAtt, respectively. To deplete San from the SMC1-GFP cells, we used GGCUAGGAAUAGGAACUAAtt/UUAGUUCCUAUUCCUAGCCtt.
San shRNA-inducible stable cell line and the rescue experiment
The DNA oligonucleotides GATCCCGTGACAAGTTCTACAATTAGTTCAAGAGAATCCTTGTAGAACTTGTCATTTTTA and AGCTTAAAAATGACAAGTTCTACAAGGATTCTCTTGAACTAATTGTAGAACTTGTCACGG synthesized by IDT were annealed and cloned into BglII and HindIII sites of the pSuperior.puro vector from OligoEngine to generate a single-unit shRNA construct. The shRNA unit, containing an H1 promoter and San-shRNA flanked by XhoI and SalI sites, was amplified by PCR and cloned into the SalI site of the single-unit shRNA construct to generate the two-unit shRNA construct. This procedure was repeated again to generate the three-unit shRNA construct that was used in this study. The inducible San-shRNA construct and pcDNA6/TR (Invitrogen) were cotransfected into HeLa cells using Lipofectamine (Invitrogen) according to the manufacturer's protocol. The next day, cells were selected with 1 μg/ml puromycin and 3 μg/ml blasticidin. 3 wk later, individual clones were picked up, and the expression of shRNA was induced with 3 μg/ml doxycycline for 4 d. A clone showing the best knockdown effect was used for the rescue experiment. In the rescue experiment, the rescuing plasmids were transfected into the inducible San-shRNA cell line right before the induction. The second round of DNA transfection was performed the next day to boost the transfection efficiency, and cells were harvested 4 d later for analysis. All the rescue constructs were based on pCS2 and tagged with His6 tag. To make their transcripts resistant to San-shRNA, the DNA sequence “AATGACAAGTTCTACAAGGAT” in San cDNA was scrambled to “AACGATAAATTTTATAAAGAC,” which does not change the resulting amino acid sequence (the substitutions are underlined).
Construction of the SMC1-GFP stable cell line
The SMC1 gene, with GFP fused in-frame at its C-terminal end, was cloned into the pIRESneo3 vector (CLONTECH Laboratories, Inc.) for eukaryotic gene expression. The construct was transfected into HeLa cells, and a G418-resistant stable cell line expressing SMC1-GFP was generated and used for further experiments.
In vitro acetylation assay
The assay was performed as previously described (Hou and Zou, 2005). The final concentration for 14C-labeled acetyl-CoA is ∼1 mM. The chromosome pellet was prepared as previously described (Mendez and Stillman, 2000).
Chromosome spreads and immunofluorescent staining
The procedure to prepare mitotic chromosome spreads has been previously described (Hou and Zou, 2005). To prepare the spreads for immunofluorescent staining, the cells were incubated in hypertonic buffer (60–70 mM KCl) for 10 min and cytospun onto the slides. For statistical comparison, the percentage of the mitotic spreads exhibiting various status of sister chromatid cohesion was calculated. After repeating at least three trials, the statistic significance was determined by a paired two-sided t test and indicated as the P value.
For immunofluorescent staining of regular HeLa cells, chromosome spreads or cells growing on the coverslip were fixed with cold methanol (−20°C) for 2 min and rinsed briefly with PBS. The cells were permeabilized with PBS containing 0.2% Triton X-100 for 10 min, followed by blocking with 3% BSA in PBS for 30 min. After that, the cells were incubated with the primary antibody in 3% BSA for 1 h, followed by the incubation with the secondary antibody in donkey serum with three washes in between. After three washes with PBS, the coverslips were mounted onto the slides and sealed. To stain Sgo1, the cells were fixed with 4% paraformaldehyde (in PBS) instead of methanol.
For immunofluorescent staining of the SMC1-GFP HeLa cells, chromosome spreads or cells growing on the coverslip were extracted with PBS + 0.2% Triton X-100 before being fixed with 4% formaldehyde in PBS for 30 min. Blocking was performed in SNBP buffer (0.02% saponin, 0.05% NaN3, and 1% BSA in PBS) for 30 min. Cells were washed three times with PBS after incubation with anti-GFP (Invitrogen) for 1 h. After the incubation with secondary antibody for 1 h, cells were washed three times. After a brief incubation with DAPI in PBS for 5 min, the coverslip was mounted and sealed.
Epifluorescence and confocal microscopy were performed at room temperature on an Axioplan 2 microscope and a LSM510 META Laser Scanning Confocal Microscope, respectively (both Carl Zeiss MicroImaging, Inc.). A Plan-APOCHROMAT 100×/1.4 NA oil objective or a Plan NEOFLUAR 10×/0.30 NA was used on the Axioplan2 and the images were acquired with a 1,388 × 1,040 AxioCam HRm charge-coupled device camera controlled by OpenLab (Improvision). A Plan-APOCHROMAT 100×/1.4 NA oil objective was used on the LSM510, and the images were acquired with accompanied software. Gamma adjustments and necessary cropping were performed using Photoshop 7.0 (Adobe).
Online supplemental material
Fig. S1 shows the cloning of the human homologue of San. Fig. S2 shows that San-siRNA cells were arrested in metaphase. Fig. S3 shows that the endogenous San was depleted by siRNA in HeLa cells stably expressing SMC1-GFP, and that SMC1-GFP faithfully reported the dynamics of the cohesin complex. Fig. S4 shows that Sgo1 localizes next to the crest signals on the chromosome spreads with both paired and unpaired chromatids. The online version of this article is available at http://www.jcb.org/cgi/content/full/jcb.200701043/DC1.
Supplementary Material
Acknowledgments
We thank Yu Hong for her excellent technical support. We also thank Dr. Hongtao Yu for kindly providing the antibody to human Sgo1, and Dr. Johan R. Lillehaug for generously providing the antibodies to NatH and Ard1.
This work is supported by a research grant (1-FY02-207) from the March of Dimes Birth Defects Foundation to K. Yokomuri. H. Zou is the Kenneth G. and Elaine A. Langone Scholar supported by the Damon Runyon Cancer Foundation (DRS-#35-03). This work is also supported by a Research Scholar Grant from the American Cancer Society (RSG-04-171-01-CCG) and a research grant from the Welch Foundation (I-1594) to H. Zou.
Abbreviations used in this paper: ACC6, aminoglycoside 6′-N-acetyltransferase; Plk, pololike kinase; SNAT, serotonin N-acetyltransferase.
References
- Angus-Hill, M.L., R.N. Dutnall, S.T. Tafrov, R. Sternglanz, and V. Ramakrishnan. 1999. Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol. 294:1311–1325. [DOI] [PubMed] [Google Scholar]
- Arnesen, T., D. Anderson, C. Baldersheim, M. Lanotte, J.E. Varhaug, and J.R. Lillehaug. 2005. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem. J. 386:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen, T., D. Anderson, J. Torsvik, H.B. Halseth, J.E. Varhaug, and J.R. Lillehaug. 2006. Cloning and characterization of hNAT5/hSAN: an evolutionarily conserved component of the NatA protein N-alpha-acetyltransferase complex. Gene. 371:291–295. [DOI] [PubMed] [Google Scholar]
- Bellows, A.M., M.A. Kenna, L. Cassimeris, and R.V. Skibbens. 2003. Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res. 31:6334–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands, A., and R.V. Skibbens. 2005. Ctf7p/Eco1p exhibits acetyltransferase activity–but does it matter? Curr. Biol. 15:R50–R51. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth, and K. Nasmyth. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 5:243–254. [DOI] [PubMed] [Google Scholar]
- Dai, J., B.A. Sullivan, and J.M. Higgins. 2006. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell. 11:741–750. [DOI] [PubMed] [Google Scholar]
- Edwards, S., C.M. Li, D.L. Levy, J. Brown, P.M. Snow, and J.L. Campbell. 2003. Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol. Cell. Biol. 23:2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., H. Yu, and M.W. Kirschner. 1998. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell. 2:163–171. [DOI] [PubMed] [Google Scholar]
- Fluge, O., O. Bruland, L.A. Akslen, J.E. Varhaug, and J.R. Lillehaug. 2002. NATH, a novel gene overexpressed in papillary thyroid carcinomas. Oncogene. 21:5056–5068. [DOI] [PubMed] [Google Scholar]
- Gandhi, R., P.J. Gillespie, and T. Hirano. 2006. Human Wapl Is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 16:2406–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi, M., S. Just, A. Mun, S. Ross, P. Rucknagel, Y. Dubaquie, A. Ehrenhofer-Murray, and S. Rospert. 2003. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23:7403–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, P.J., and T. Hirano. 2004. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 14:1598–1603. [DOI] [PubMed] [Google Scholar]
- Hauf, S., E. Roitinger, B. Koch, C.M. Dittrich, K. Mechtler, and J.M. Peters. 2005. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, A.B., M.A. Namboodiri, D.C. Klein, and F. Dyda. 1999. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell. 97:361–369. [DOI] [PubMed] [Google Scholar]
- Hou, F., and H. Zou. 2005. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol. Biol. Cell. 16:3908–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, D., A. Schleiffer, F. Eisenhaber, K. Mechtler, C.H. Haering, and K. Nasmyth. 2002. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 12:323–328. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., R.M. MacCallum, and M.J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499–520. [DOI] [PubMed] [Google Scholar]
- Kenna, M.A., and R.V. Skibbens. 2003. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 23:2999–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima, T.S., S. Hauf, M. Ohsugi, T. Yamamoto, and Y. Watanabe. 2005. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting shugoshin localization. Curr. Biol. 15:353–359. [DOI] [PubMed] [Google Scholar]
- Kitajima, T.S., T. Sakuno, K. Ishiguro, S. Iemura, T. Natsume, S.A. Kawashima, and Y. Watanabe. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 441:46–52. [DOI] [PubMed] [Google Scholar]
- Kraft, C., F. Herzog, C. Gieffers, K. Mechtler, A. Hagting, J. Pines, and J.M. Peters. 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22:6598–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng, S., B. Hegemann, B.H. Peters, J.J. Lipp, A. Schleiffer, K. Mechtler, and J.M. Peters. 2006. Wapl controls the dynamic association of cohesin with chromatin. Cell. 127:955–967. [DOI] [PubMed] [Google Scholar]
- Madril, A.C., R.E. Johnson, M.T. Washington, L. Prakash, and S. Prakash. 2001. Fidelity and damage bypass ability of Schizosaccharomyces pombe Eso1 protein, comprised of DNA polymerase eta and sister chromatid cohesion protein Ctf7. J. Biol. Chem. 276:42857–42862. [DOI] [PubMed] [Google Scholar]
- Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan, G.L., B. Pfander, and S. Jentsch. 2006. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 23:723–732. [DOI] [PubMed] [Google Scholar]
- Polevoda, B., and F. Sherman. 2003. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325:595–622. [DOI] [PubMed] [Google Scholar]
- Riedel, C.G., V.L. Katis, Y. Katou, S. Mori, T. Itoh, W. Helmhart, M. Galova, M. Petronczki, J. Gregan, B. Cetin, et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 441:53–61. [DOI] [PubMed] [Google Scholar]
- Rollins, R.A., M. Korom, N. Aulner, A. Martens, and D. Dorsett. 2004. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 24:3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic, A., J.C. Waters, and T.J. Mitchison. 2004. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 118:567–578. [DOI] [PubMed] [Google Scholar]
- Skibbens, R.V. 2004. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics. 166:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R.V., L.B. Corson, D. Koshland, and P. Hieter. 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T.S., P. Yiu, M.F. Chou, S. Gygi, and J.C. Walter. 2004. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 6:991–996. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., T. Yonekawa, Y. Kawasaki, M. Kai, K. Furuya, M. Iwasaki, H. Murakami, M. Yanagida, and H. Okayama. 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20:3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Z. Hao, M. Kai, and H. Okayama. 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20:5779–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., Y. Sun, S.E. Harley, H. Zou, and H. Yu. 2004. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. USA. 101:18012–18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., H. Shu, W. Qi, N.A. Mahmood, M.C. Mumby, and H. Yu. 2006. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell. 10:575–585. [DOI] [PubMed] [Google Scholar]
- Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda, Y., K. Furuya, G. Goshima, K. Nagao, K. Takahashi, and M. Yanagida. 2002. Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12:347–358. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I.C., S. Hauf, A. Meinke, and J.M. Peters. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 103:399–410. [DOI] [PubMed] [Google Scholar]
- Watrin, E., A. Schleiffer, K. Tanaka, F. Eisenhaber, K. Nasmyth, and J.M. Peters. 2006. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 16:863–874. [DOI] [PubMed] [Google Scholar]
- Williams, B.C., C.M. Garrett-Engele, Z. Li, E.V. Williams, E.D. Rosenman, and M.L. Goldberg. 2003. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr. Biol. 13:2025–2036. [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot, L.E., K. Draker, G.D. Wright, and A.M. Berghuis. 1999. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure. 7:497–507. [DOI] [PubMed] [Google Scholar]
- Zou, H., T.J. McGarry, T. Bernal, and M.W. Kirschner. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 285:418–422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.