Abstract
Release of apoptogenic proteins such as cytochrome c from mitochondria is regulated by pro- and anti-apoptotic Bcl-2 family proteins, with pro-apoptotic BH3-only proteins activating Bax and Bak. Current models assume that apoptosis induction occurs via the binding and inactivation of anti-apoptotic Bcl-2 proteins by BH3-only proteins or by direct binding to Bax. Here, we analyze apoptosis induction by the BH3-only protein BimS. Regulated expression of BimS in epithelial cells was followed by its rapid mitochondrial translocation and mitochondrial membrane insertion in the absence of detectable binding to anti-apoptotic Bcl-2 proteins. This caused mitochondrial recruitment and activation of Bax and apoptosis. Mutational analysis of BimS showed that mitochondrial targeting, but not binding to Bcl-2 or Mcl-1, was required for apoptosis induction. In yeast, BimS enhanced the killing activity of Bax in the absence of anti-apoptotic Bcl-2 proteins. Thus, cell death induction by a BH3-only protein can occur through a process that is independent of anti-apoptotic Bcl-2 proteins but requires mitochondrial targeting.
Introduction
Sensitivity and resistance to apoptosis are to a large degree regulated by pro- and anti-apoptotic members of the Bcl-2 protein family. How this regulation is achieved is under intense investigation (Adams, 2003; Danial and Korsmeyer, 2004). Structural and functional similarities divide the Bcl-2 family into three groups. The pro-apoptotic multidomain proteins (containing the Bcl-2 homology [BH] domains 1–3) Bax and/or Bak are required for mitochondrial permeabilization during apoptosis (Lindsten et al., 2000; Zong et al., 2001; Adams, 2003; Danial and Korsmeyer, 2004). The activation of Bax/Bak is caused consecutively to the activation of the BH3-only group of Bcl-2 family proteins (which have in common only the short BH3 domain; Huang and Strasser, 2000; Puthalakath and Strasser, 2002). The anti-apoptotic group of Bcl-2 proteins contains Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1 (Cory and Adams, 2002).
The various interactions between these proteins are crucial for the life-death decision. However, many details of these interactions are still unclear. Direct binding of Bax to Bcl-2 was demonstrated early on (Oltvai et al., 1993), but the significance of this interaction is questionable because it depends on the presence of certain detergents (Hsu and Youle, 1997) and because Bcl-2 is localized on intracellular membranes, whereas Bax is largely soluble in the cytosol (Suzuki et al., 2000; Schinzel et al., 2004). Bak, on the other hand, is an integral protein of the outer mitochondrial membrane, and it has recently been demonstrated to be sequestered there and kept inactive by the two anti-apoptotic Bcl-2 proteins, Mcl-1 and Bcl-xL (Willis et al., 2005).
Eight BH3-only proteins are known. It is firmly established that their BH3 domains can bind to anti-apoptotic Bcl-2 proteins, which prevents their activating Bax/Bak (Petros et al., 2000; Liu et al., 2003; Chen et al., 2005). This has led to the proposition that BH3-only proteins induce apoptosis, at least in part, through the neutralization of Bcl-2-like proteins. This view has gained support by the recent demonstration of a selectivity in binding between BH3-only proteins and Bcl-2-like proteins (Chen et al., 2005), reproduced in another study using a different technical approach (Certo et al., 2006). Although both studies are potentially limited by the use of BH3 domain peptides rather than whole proteins, the results are intriguing. The selectivity of binding found in these studies could explain the varying apoptosis-inducing potency of BH3-only proteins, and the model has been elegantly confirmed by the demonstration that combining BH3-only proteins that can bind to Bcl-xL and Mcl-1 leads to the release and, presumably, the auto-activation of Bak (Willis et al., 2005).
The above studies have engendered two models of BH3-only protein action. One model (the direct binding model) proposes that the BH3-only proteins Bim and tBid (and perhaps Puma) can directly bind and activate Bax/Bak (Letai et al., 2002; Certo et al., 2006), whereas the remaining BH3-only proteins (Bik, Puma, Noxa, Bad, Bmf, and Hrk) can only sensitize, i.e., release Bim and tBid from their site of sequestration to Bcl-2-like proteins (and Bim/tBid would then go on to activate Bax/Bak). Although it is difficult to demonstrate the interaction of Bim/tBid with Bax/Bak in intact cells, this model has received support from studies with purified proteins and artificial membranes (Kuwana et al., 2002, 2005).
The second model (the displacement model) proposes that Bax/Bak can auto-activate once the inhibition imposed by Bcl-2-like proteins has been removed by BH3-only proteins, but that no direct interaction between BH3-only proteins and Bax/Bak is required (Chen et al., 2005; Willis and Adams, 2005). Although this model has been supported experimentally for Bak (see above), it is still difficult to see how this mechanism could work for Bax. One step in the activation of Bax has to be the mitochondrial translocation. Although this Bax translocation and insertion is seen during apoptosis induction (Gross et al., 1998; Eskes et al., 2000), there is no signal known that could initiate this, and no interaction partner is known that could determine Bax translocation.
BH3-only proteins are activated upon apoptotic stimuli from the outside. However, the study of BH3-only protein action in intact cells is not easy. One problem is that most BH3-only proteins are subject to post-translational regulation, and their activation and activity are very difficult to measure. For instance, Bim is expressed in normal cells and is activated upon a suitable stimulus, but is it unclear how Bim is activated on a molecular level. Furthermore, apoptotic stimuli may activate more than one BH3-only protein, and this may depend on the cell type used. Lastly, most apoptotic stimuli have massive side effects. UV irradiation, for instance, will activate BH3-only proteins in many cells but also activate other pathways, such as stress kinases and the DNA-damage response, confounding the observed results.
To escape these problems and to analyze the consequences of BH3-only protein activation in human cells, we used a strategy of inducible expression of Bim in HeLa epithelial cells. The inducible expression of BimS was sufficient to induce apoptosis. This enabled us to investigate the interaction of BimS with other Bcl-2 proteins and the events triggered by BimS at the cell's mitochondria.
Results
We first tested the major isoform of Bim, BimEL. In transient transfection experiments of T-REx-HeLa cells (a cell line stably carrying the tetracycline repressor, in which expression of the construct is induced by tetracycline [tet]) BimEL induced apoptosis of transfected cells (∼50% dead cells after 24 h in the presence of tet). Upon transfection and selection, nine clones were obtained that stably carried the tet-inducible BimEL construct. All of these clones expressed BimEL, and the expression was induced to varying degrees by tet addition. The expression exceeded in all clones by far the endogenous levels in T-REx-HeLa cells, which is easily detectable by Western blotting (unpublished data). However, tet-induced expression of BimEL completely failed to induce apoptosis in these cells (unpublished data).
Although this was unexpected, it might be explained by the fact that BimEL is in many cells expressed at relatively high levels and is probably activated by molecular events that have not been fully worked out. We thus turned to the expression of BimS (mouse BimS was used, as at the time the identity of human BimS was not unequivocal in the databases). BimS is the shortest of the regularly expressed Bim isoforms, is normally expressed at very low levels, and is not subject to any known post-translational activation (O'Connor et al., 1998; Puthalakath et al., 1999). When expressed transiently as above, BimS was more active than BimEL (∼80% dead cells at 24 h), confirming earlier results (O'Connor et al., 1998). Two stable cell lines carrying the inducible BimS construct were analyzed in detail. There was no major difference in the parameters analyzed between these two lines.
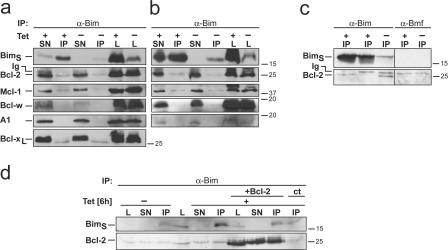
In these clones, tet addition induced apoptosis in a dose-dependent manner (Fig. S1 a, available at http://www.jcb.org/cgi/content/full/jcb.200610148/DC1). Cell death was rapid, with ∼50% of cells containing active caspase-3 after 2 h (Fig. S1 b) and ∼90% of cells containing condensed nuclei after 4–6 h (not depicted). We then proceeded to analyze the molecular events during apoptosis induction by BimS. Both models of BH3-only protein action, especially the displacement model, predict that BimS will induce apoptosis through binding to anti-apoptotic Bcl-2 proteins. We therefore tested the association of BimS with Bcl-2 by immunoprecipitation (IP). IP with antibodies specific for Bim precipitated BimS from both clones (Fig. 1 a and Fig. S3 a). A fraction of Bcl-2, Mcl-1, and Bcl-xL was coprecipitated with Bim from uninduced cells (Fig. 1, a and c), suggesting that the “leaky” BimS that was expressed in these cells was associated with Bcl-2, Mcl-1, and Bcl-xL (see Fig. 1 c for a specificity control). Induction with tet led to the increase in the amount of BimS isolated (Fig. 1 a). However, the amount of Bcl-2, Mcl-1, and Bcl-xL coprecipitated with BimS appeared to be essentially the same as in uninduced cells, suggesting that the induced, apoptosis-inducing fraction of BimS is not bound to Bcl-2, Mcl-1, or Bcl-xL. Although Bcl-w and A1 were detectable in the cells by Western blotting, they were not seen in IP products, suggesting that none or very little of these proteins was bound to BimS (Fig. 1 a). These results indicate that, in order to induce apoptosis, BimS does not need to bind to detectable amounts of anti-apoptotic Bcl-2 proteins in HeLa cells. To test this in a different cellular system, 293 human fibroblasts carrying the tet repressor (T-REx-293 cells) were transiently transfected with the BimS construct, and binding of BimS to Bcl-2 family proteins was tested. As in the case of HeLa cells, small amounts of Bcl-2 and Mcl-1 could be coprecipitated with BimS, and a very faint band was seen when probing for Bcl-w (Bcl-xL was undetectable in these cells). However, as in the HeLa clones, the amounts precipitated appeared essentially the same when BimS expression was induced by tet (Fig. 1 b). Notably, we could detect some Bax by co-IP in these experiments, when higher amounts of mitochondria were used or when mitochondria or whole-cell lysates were used from the T-REx-293 cells transiently transfected with the BimS construct (see Fig. S3, b–d). Weak binding of BimS (unlike BimEL or BimL) to Bax in detergent lysates has been observed before but was found not to be required for apoptosis induction, as a BimS mutant that had lost this binding capacity still induced apoptosis equally well (Marani et al., 2002; Willis et al., 2007).
Figure 1.
Interaction between BimS and prosurvival proteins as tested by immunoprecipitation. BimS-expressing clone A2 (a, c, and d) or T-Rex-293 cells transiently transfected with the BimS construct (b) were left untreated or treated with tetracycline for 6 h (A2 clone) or 16 h (T-Rex-293 cells). Mitochondrial lysates (a–c) or whole-cell lysates (d) were immunoprecipitated with anti-Bim antibodies. Aliquots of mitochondrial (L, 45 μg of protein) or total cell lysates (L, 22 μg), post-IP supernatant (SN), and IP pellets (IP) were loaded and membranes were probed for Bim, Bcl-2, Mcl-1, Bcl-w, A1, and Bcl-xL. The migration of immunoglobulin light chain (Ig) is indicated but was not observed in all experiments. (c) Left, pellet fractions after Bim-IP from isolated mitochondria (first lane), cytosolic fraction (middle lane) after 4 h of tet and from mitochondria without tet (third lane). Right, specificity control with antibodies against an unrelated protein (mBmf). (d) Binding of tet-induced BimS to transiently expressed Bcl-2. Cells were transfected with control vector or an expression vector for human Bcl-2, lysed, and Bim was immunoprecipitated as above. Bim and Bcl-2 were detected by Western blotting. All Western blots are representative of three independent experiments (the experiment shown in panel d was done twice).
As an additional control, BimS cells were transiently transfected with an expression construct for Bcl-2. As expected, the transfected cells were protected against BimS-induced apoptosis (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200610148/DC1). However, in cells overexpressing Bcl-2, tet-induced BimS clearly associated with Bcl-2 as judged by the increased amount of Bcl-2 coprecipitated with BimS (Fig. 1 d, compare lanes where the IP pellet was loaded). This indicates that in the situation where Bcl-2 acts to block BimS-induced apoptosis, it does so via direct binding of BimS. Conversely, when the apoptosis-inducing activity of BimS is not blocked, no binding between BimS and anti-apoptotic Bcl-2 proteins can be detected.
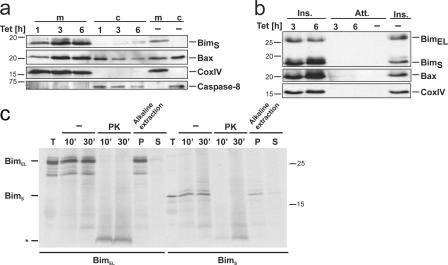
In the attempt to understand the molecular function of BimS we next analyzed the subcellular localization of induced BimS. In uninduced cells, a small amount of BimS was detectable in the fraction containing mitochondria. Tet-induced BimS was initially also found in mitochondria, and only when higher levels were reached (especially in the 3C4 clone) part of it was seen in the cytosol (Fig. 2 a and Fig. S4, respectively, for the two clones tested; there was some difference between the clones as to localization, which may be linked to different amounts of BimS induced or possibly to different mitochondrial import capacity [see below]). Intriguingly, mitochondrial BimS was integrally inserted into mitochondrial membranes, as measured by alkaline treatment of mitochondria (which allows separation of integral membrane proteins from soluble/peripherally attached proteins; Fig. 2 b). Some (endogenous) BimEL was also found expressed in mitochondria, possibly a fraction that had been activated but blocked by anti-apoptotic proteins. BimEL was also inserted into mitochondrial membranes, as was Bax (Fig. 2 b). In vitro import experiments using radiolabeled Bim proteins confirmed these results. Both in vitro–translated BimEL and BimS proteins became associated with isolated HeLa mitochondria upon incubation in a typical mitochondrial import reaction (Fig. 2 c). When mitochondria were then treated with protease the Bim proteins were readily digested, suggesting an exposed localization at the outer membrane of mitochondria. Alkaline extraction experiments following the import experiments indicate that Bim was integrally inserted into mitochondrial membranes (Fig. 2 c). Similar results were obtained when yeast mitochondria (from Saccharomyces cerevisiae) were used (not depicted).
Figure 2.
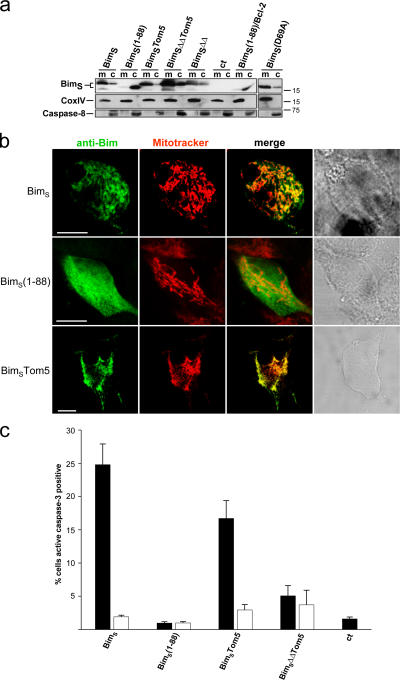
BimS localizes to and inserts into mitochondrial membranes in HeLa cells and in isolated mitochondria. (a) Mitochondria (m) from T-REx-HeLa cells (A2 clone) uninduced or tet-induced as indicated were isolated and separated alongside the soluble cytosolic fractions (c) on 12.5% SDS-PAA gels. CoxIV is a marker for mitochondria, caspase-8 for cytosolic proteins. (b) Isolated mitochondria were treated with 0.1 M Na2CO3 to separate membrane inserted (Ins.) from soluble (Att., membrane attached, intermembrane space and matrix) fractions. Both fractions were analyzed by Western blotting for Bim, CoxIV as a marker for mitochondrial membranes and Bax. (c) Import of in vitro–translated BimEL or BimS into isolated HeLa mitochondria. Radiolabeled precursors of BimEL and BimS were incubated for 10 and 30 min at 30°C with mitochondria isolated from HeLa cells. Subsequently, the samples were split. One third was treated with 50 μg/ml proteinase K (PK) for 20 min on ice, another one was left untreated. The last third was subjected to alkaline extraction resulting in a pellet fraction (P; containing integral membrane proteins) and a supernatant fraction (S; containing soluble and peripherally attached proteins). For comparison, 10% of the total input of radiolabeled precursors was included (T). Mitochondria were reisolated and import was analyzed by SDS-PAGE and autoradiography. *, very likely proteolysis products.
The exposed localization and the membrane integration of Bim imply that Bim is integrally inserted into the mitochondrial outer membrane, suggesting that it translocates to the surface of the mitochondria (most likely directed by an intrinsic mitochondrial targeting signal), where it is inserted into the lipid bilayer of the outer membrane through the activity of the TOM complex (the general translocase of the mitochondrial outer membrane). Bim thus seems to follow the main import pathway for mitochondrial outer membrane proteins (Paschen and Neupert, 2001).
We noticed in these experiments that the expression of BimS caused an early shift of Bax to mitochondria. This was detectable as both an increase in mitochondrial Bax and, perhaps more distinctly, a cytosolic depletion of Bax that was clearly visible a few hours after BimS induction (Fig. 2 a and Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200610148/DC1). Similar results were obtained when T-REx-293 cells were transiently transfected with wild-type BimS construct and induced with tet. No translocation of Bax to the mitochondria was seen when the empty expression vector was used (unpublished data).
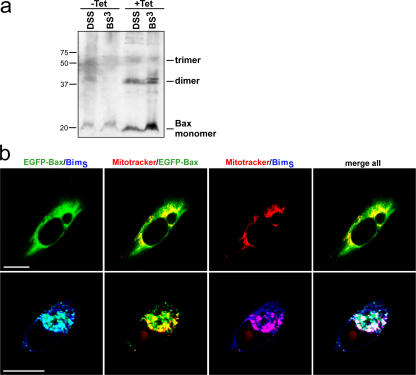
The activation of Bax has been measured by a conformational change at the N terminus (Hsu and Youle, 1997) or the C-terminal part (Dewson et al., 2003), or as oligomerization at mitochondria (Eskes et al., 2000). Bands corresponding to the described Bax dimers and trimers were found when mitochondrial proteins were cross-linked upon BimS induction (Fig. 3 a). Bax activation as measured by staining for N- or C-terminal conformational change was also detected (not depicted).
Figure 3.
BimS-expression causes Bax oligomerization and mitochondrial recruitment of Bax. (a) BimS was induced in HeLa cells (A2 clone) as indicated for 6 h. Mitochondria were isolated and treated with two different cross-linkers as described in Materials and Methods. After cross-linking, the reaction was analyzed by Western blotting with an anti-Bax monoclonal antibody. (b) BimS cells (A2 clone) were transiently transfected with the EGFP-Bax fusion construct and were analyzed by confocal microscopy untreated (top) or upon BimS-induction (bottom, 5 h). Localization of EGFP-Bax (green), mitochondria (red), BimS (blue), and various overlays are shown. Bars, 10 μm.
In order better to visualize mitochondrial translocation of Bax, we used an EGFP-Bax fusion protein that has been described before to translocate to mitochondria during apoptosis (Schinzel et al., 2004). Laser-scanning microscopy confirmed that tet-induced BimS was predominantly located on the cell's mitochondria (Fig. 3 b). Upon transient transfection EGFP-Bax was localized in the cytosol, but upon induction of BimS translocated to the mitochondria where it colocalized with BimS (Fig. 3 b). It is worth noting that mitochondrial Bax at all times appeared to be inserted into mitochondrial membranes rather than be loosely attached (Fig. 2 b). Thus, the sole manipulation of tet-dependent induction of BimS causes the translocation of Bax to mitochondria, accompanied by Bax activation.
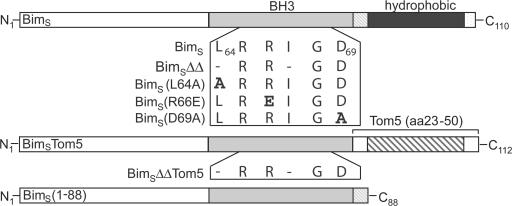
These results suggested that the mitochondrial translocation, but not the interaction with anti-apoptotic Bcl-2 proteins, was the key event in apoptosis induction by BimS. We therefore pursued the two-pronged strategy of analyzing BimS mutants that were engineered to differ either in translocation or in their capacity to interact with Bcl-2. Fig. 4 gives a schematic representation of the mutants used.
Figure 4.
Schematic representation of BimS and BimS mutant proteins used in this study. Mutants were generated that lack the hydrophobic C terminus (1–88, bottom) or where the C terminus was replaced with the mitochondrial membrane anchor of yeast Tom5 (BimSTom5, middle) or that contain various mutations in the BH3 domain as indicated in the box.
It has been noted before that the C terminus of Bim (which is part of the protein in all major splice forms) contains a potential membrane insertion sequence, and it has been shown early on that overexpressed Bim is targeted to intracellular membranes (O'Connor et al., 1998). Further, our above data suggest that active Bim is a tail-anchored protein, which is targeted to mitochondria through this hydrophobic C-terminal domain (see Fig. 2). To analyze the role of mitochondrial translocation of BimS, three mutants were generated. The first one, BimS(1–88), was generated by introducing a stop codon after amino acid 88, deleting the hydrophobic C terminus. The second mutant (BimSTom5) was engineered to consist of the amino acids 1–84 of BimS followed by the mitochondrial tail-anchor sequence of yeast Tom5 (which is imported into the outer mitochondrial membrane), a well-investigated member of the mitochondrial outer membrane protein translocation complex (Horie et al., 2003). The third mutant (BimSΔΔTom5) differed from the second by the deletion of two amino acid residues in the BH3 domain predicted to abolish its pro-apoptotic activity (Fig. 4).
Localization of the mutants in transient transfection of T-REx-293 cells was as predicted (Fig. 5 a): BimS(1–88) was detected almost exclusively in the cytosol, whereas BimSTom5 was mostly found on mitochondria. The majority of BimSΔΔTom5 was mitochondrial with a cytosolic fraction, possibly an overflow from mitochondria as this mutant was expressed at higher levels. BimSΔΔ (full-length BimS containing the BH3 domain double deletion, Fig. 4) was also targeted to mitochondria, as was BimS(D69A). Co-expression of Bcl-2 failed to direct BimS(1–88) to mitochondria (Fig. 5 a). Laser-scanning microscopy was used to confirm these results with respect to BimS, BimS(1–88), and BimSTom5 (Fig. 5 b). The apoptosis-inducing activity of the mutants was tested by transient transfection of T-REx-293 cells, where the expression was regulated by tet. As shown in Fig. 5 c, BimS(1–88) had completely lost its apoptosis inducing activity, while retargeting of this mutant by a mitochondrial targeting sequence (BimSTom5) restored this activity not completely but to a substantial degree. The BH3 domain deletion (BimSΔΔTom5) again almost abolished it (Fig. 5 c shows the results for caspase-3 activation; similar results were seen when apoptotic morphology was assessed [Fig. S5 a, available at http://www.jcb.org/cgi/content/full/jcb.200610148/DC1]). Therefore, mitochondrial targeting and an intact BH3 domain are required for apoptosis induction by BimS in this system.
Figure 5.
Subcellular localization of BimS mutants and their pro-apoptotic activity. (a) Localization as determined by cell fractionation. T-REx-293 cells were transiently transfected with wild-type or mutant constructs and induced 7 h later with tet for ∼15 h. Mitochondria (m) and cytosolic fractions (c) were prepared and tested for expression of BimS proteins. As a control (ct) the cells were transfected with the empty expression vector. BimS(1–88)/Bcl-2: cells were cotransfected with BimS(1–88) and an expression construct for hBcl-2. The mutants are described in Fig. 4. (b) Localization of C-terminal mutants as determined by confocal microscopy. Cells were transfected as above, induced 24 h later for 5 h with tet, and BimS was detected by staining with Bim-specific antibodies. Mitochondria were identified by MitoTracker staining. Bars, 10 μm. (c) T-Rex-293 cells were transiently transfected with the constructs indicated (ct, empty vector control). 7 h later, expression of BimS was induced by tet addition as shown. 15 h later, cells were harvested and percentage of active caspase-3–positive cells was determined by flow cytometry. Black bars indicate induction with tet; white bars without tet. The values give the mean of at least three independent experiments/SEM. Expression of transfected proteins was confirmed by Western blotting (not depicted).
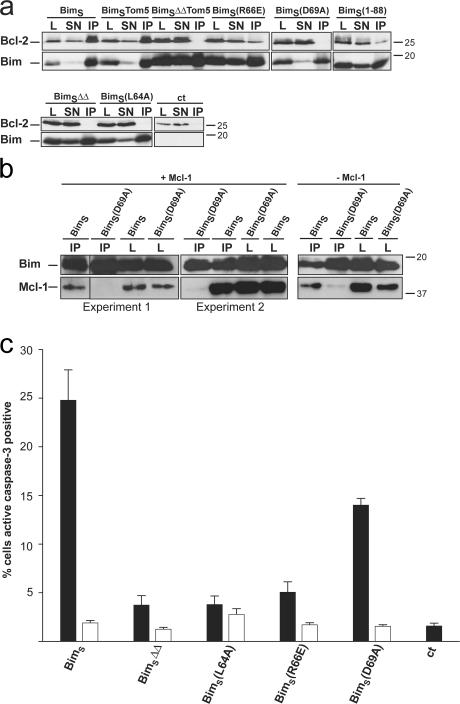
In the second line of experiments, mutations were made to the BH3 domain of BimS and their effect on binding to Bcl-2 and on apoptosis induction was tested. The double deletion mentioned above and three point mutations were introduced in BimS (Fig. 4). When binding to Bcl-2 was tested by cotransfection of Bcl-2 and the respective BimS mutant into T-REx-293 cells, interaction with Bcl-2 (measured by co-IP) was seen for BimS wild type, BimSTom5 and, albeit to a lesser extent, BimS(1–88) and BimS(R66E). It has to be pointed out that this assay only tests for interaction capacity and not actual interaction. BimS(1–88) is almost exclusively located in the cytosol, while Bcl-2 is membrane associated (compare Fig. 1 c with Fig. 5 a), so the co-IP very likely relies on an interaction enabled only upon detergent solubilization. No binding to Bcl-2 was detected for the mutants BimSΔΔ, BimSΔΔTom5, BimS(D69A), and BimS(L64A) (Fig. 6 a). BimSΔΔ and the point mutants showed almost no apoptosis inducing activity in this assay except for BimS(D69A), which conserved over half of the apoptosis inducing activity (Fig. 6 c and Fig. S5 b). This group of mutants therefore contains two examples that did not show correlation between Bcl-2 binding and apoptosis induction predicted by the displacement model. BimS(R66E) shows considerable binding to overexpressed Bcl-2 (Fig. 6 a) but fails to induce apoptosis, while BimS(D69A) fails to bind to Bcl-2 but has considerable apoptosis-inducing activity. When the mutant was expressed in T-Rex-293 cells, BimS(D69A)-IP brought down a small amount of endogenous Mcl-1. However, this amount was essentially the same when Mcl-1 was overexpressed (Fig. 6 b). This suggests that the small amount of coprecipitated Mcl-1 was bound to endogenous BimEL, which is also found in mitochondria and cannot be separated in this assay (see Fig. 2 b). The binding of BimS(D69A) to Mcl-1 is thus either abolished or at least very strongly reduced. As predicted by all models from the lack of binding, Bcl-2 was unable to protect TRex HeLa cells against the overexpression of BimS(D69A) in transient transfection assays, although it protected against BimS-induced apoptosis (not depicted). Notably, although BimS was found to bind to Bax, BimS(D69A) failed to do so when whole-cell lysates of T-REx-293 cells transiently transfected with the BimS mutant construct were used for Bim-IP (see Fig. S3 d). A summary of the results is given in Table I.
Figure 6.
Bcl-2/Mcl-1 binding and pro-apoptotic activity of BH3 domain mutants of BimS. T-REx-293 cells were transfected with the mutants indicated (see Fig. 4) together with an expression construct for hBcl-2 (a) or alone (c). After 7 h, BimS was induced with tet. (a) 18 h later Bim was immunoprecipitated from whole-cell extracts (200 μg) of T-REx-293 cells as above. Control (ct), cells were transfected with hBcl-2 vector and the empty expression vector instead of a BimS expression vector. (b) T-REx-293 cells were cotransfected with hMcl-1 vector and the wild-type BimS or the mutant construct BimS(D69A). 18 h later Bim was immunoprecipitated from whole-cell extracts (200 μg left, 300 μg middle and right) as above. Two experiments are shown. Right: same experiment showing binding of endogenous Mcl-1 to Bim where cells were solely transfected with wt BimS or the mutant construct BimS(D69A). (c) 15 h later cells positive for active caspase-3 were scored by flow cytometry after staining with a monoclonal antibody against active caspase-3 and a Cy3-conjugated secondary antibody. Values give mean of at least three independent experiments/SEM. Expression of transfected proteins was confirmed by Western blotting (not depicted).
Table I.
Summary of Bcl-2 binding, localization, and apoptosis-inducing activity of BimS mutants
| Proteins | Localization | Bcl-2 binding | Apoptosis |
|---|---|---|---|
| BimS | m | yes | yes |
| BimSΔΔ | m | no | no |
| BimS(L64A) | n.d. | no | no |
| BimS(R66E) | m* | yes | no |
| BimS(D69A) | m | no | yes |
| BimSTom5 | m | yes | yes |
| BimSΔΔTom5 | m | no | no |
| BimS(1-88) | c | yes | no |
n.d., not determined; m, mitochondria; c, cytosol. *, data not shown.
These results suggested that mitochondrial targeting rather than binding to anti-apoptotic Bcl-2 proteins was the determinant of apoptosis induction through BimS. To test this hypothesis in an independent system, we turned to the yeast S. cerevisiae. Although it is a matter of dispute whether yeast possesses a system for apoptotic response (Frohlich and Madeo, 2000; Wysocki and Kron, 2004), it is clear that the expression of Bax can kill yeast cells (Sato et al., 1994), which can be detected by growth arrest and which is accompanied by mitochondrial hyperpolarization (Gross et al., 2000). Notably, yeast has no recognized Bcl-2 family proteins. We therefore examined whether BimS would enhance Bax-dependent killing in yeast (which would have to occur in the absence of anti-apoptotic Bcl-2 proteins). BimS was expressed from a constitutive promoter and Bax was placed under the control of the tet-off system (i.e., Bax was expressed in the absence of tet).
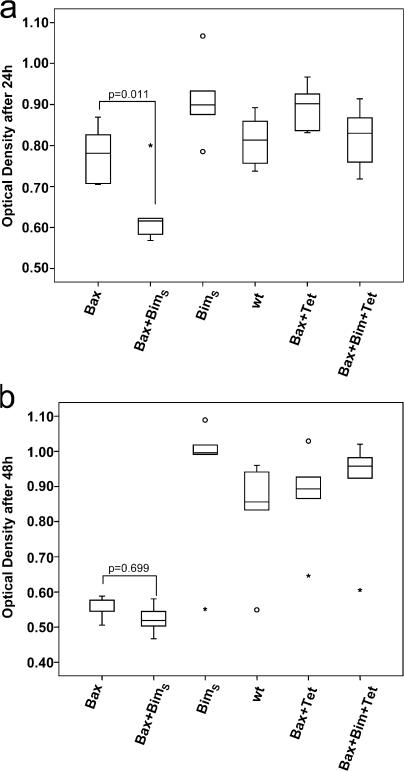
Yeast cells tolerated the expression of BimS without any growth delay or other detectable alteration, and no effect was seen in cells that carried the (switched-off) inducible Bax plasmid in the presence of tet (Fig. 7). At 48 h after removal of tet from the liquid culture, there was a clear reduction in growth that was the same in cells that expressed Bax and in cells that expressed BimS and Bax (Fig. 7 b). No further growth was observed in these cells at later time points (not depicted). When cell density was measured at earlier times, we noted that although Bax expression on its own had no significant growth-delaying effect at 24–30 h (P = 0.511; Bax vs. wt), there was a clear and significant reduction of cell growth at this point when BimS and Bax were coexpressed (Fig. 7 a; (P = 0.002, BimS + Bax vs. wt). This suggested that the expression of BimS, although not affecting growth on its own, accelerated the growth inhibition conferred by Bax. Notably, the expression of BimS(1–88) or BimSΔΔ together with Bax failed to enhance the Bax effect (unpublished data).
Figure 7.
BimS accelerates the growth inhibiting effect of Bax in yeast. Yeast strains containing BimS, Bax, or Bax/BimS were grown in the presence or absence of tetracycline (to induce Bax expression) as indicated. An untransformed yeast strain was used as control. After periods of 24–30 h (a) or 48 h (b), OD600 was measured. Boxes indicate median and range of individual values (25 and 75% are the lower and upper box limits). Vertical bars on boxes indicate range of values, asterisks and circles indicate individual outliers. Statistical significance was calculated using ANOVA and least square difference post-hoc tests. P values give levels of significance.
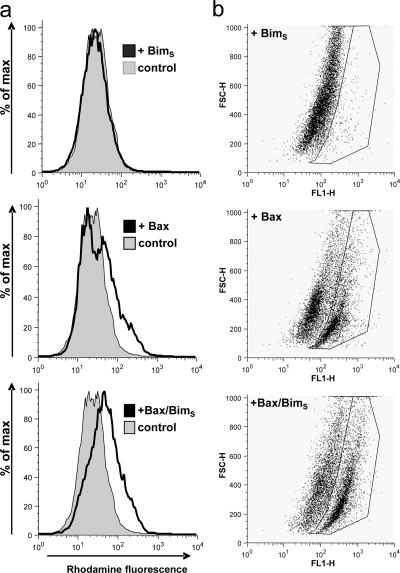
A similar acceleration/enhancement was seen when yeast were assayed for the second criterion, i.e., mitochondrial membrane potential. No change in uptake of the potential-sensitive dye rhodamine123 was seen due to the expression of BimS or in yeast carrying the uninduced Bax construct (Fig. 8, a and b, and unpublished data). Induction of Bax caused a partial shift of the population to higher fluorescence, as described previously (Gross et al., 2000), indicative of mitochondrial hyperpolarization. When Bax was induced in BimS-expressing cells this shift was more pronounced, suggesting that BimS either increased the population of yeast cells with hyperpolarized mitochondria or increased mitochondrial membrane potential in the individual cells. Dot blot analysis of the data showed that the observed shift in rhodamine123 uptake was largely due to the appearance of a distinct population of yeast cells, which was increased in size in cells expressing both BimS and Bax as compared with Bax alone (Fig. 8 b). When BimS(1–88) or BimSΔΔ were coexpressed with Bax, no enhancement of the Bax effect on mitochondrial membrane potential was seen (unpublished data). These data thus indicate a death-inducing effect of BimS in yeast cells, which depends on Bax but not on anti-apoptotic Bcl-2 proteins.
Figure 8.
Bax expression causes mitochondrial hyperpolarization in yeast, and this effect is enhanced by BimS. Yeast strains containing BimS, Bax, or Bax/BimS were grown in the absence of tetracycline (to induce Bax expression) for 24 h. An untransformed yeast strain was used as control. Yeast cells were grown to an OD600 of 0.2 and incubated with 5 μM rhodamine123 for 30 min at 30°C. Cells were analyzed by flow cytometry. (a) Histograms showing rhodamine123 fluorescence. (b) Two-dimensional dot-plot of rhodamine123 versus forward scatter. Data are representative of six individual experiments.
Discussion
In this study we analyzed molecular events during apoptosis induction by BimS. Although no evidence was found for binding of BimS to anti-apoptotic Bcl-2 proteins, we describe the mitochondrial translocation and insertion of BimS, which was sufficient to recruit and activate cytosolic Bax. Analysis of BimS mutants showed that apoptosis induction correlated with mitochondrial localization but not the ability to bind to Bcl-2. In yeast, BimS enhanced the death-inducing activity of Bax.
Surprisingly, high-level expression of BimEL failed to induce apoptosis when expressed in cells stably carrying the inducible construct. This may be a result of some experimental selection process (although the clones were established and selected alongside the BimS clones), or expression of some post-translational regulation mechanism that controls BimEL but not BimS. Only low levels of BimS are normally found in any cell that has been investigated. This is presumably linked to the fact that BimS is not subject to any post-translational regulation and is immediately active when expressed. Although it is conceivable that the apoptosis-inducing activity of BimS is substantially different from that one of BimEL, there is little evidence to suggest that. BimS contains the same BH3 domain and the same hydrophobic C terminus as the other major splice forms, whereas it lacks the DLC1-binding site and other parts of the longer forms. BimS was found to be, unlike the other forms, able to bind to Bax in certain detergents; however, a BH3 domain point mutant that had lost the ability to bind Bax was unaltered in its capacity to induce apoptosis, indicating that this binding is not physiologically relevant (Marani et al., 2002; Willis et al., 2007). We also observed some binding of BimS to Bax in co-IP experiments. However, BimS(D69A) showed no Bax binding, confirming the view that this interaction is not required for apoptosis induction by BimS.
When we investigated the association of BimS with mitochondria, some BimEL was also detected on mitochondria, and this fraction of BimEL was also inserted in the mitochondrial outer membrane (and presumably kept inactive by Bcl-2-like proteins). Furthermore, both BimEL and BimS were imported into isolated mitochondria. There seems no reason to assume that, once both are active, BimS acts in any way different from BimEL.
Bim can bind Bcl-2 and Bcl-2-like proteins very well, and it was therefore surprising to see that tet-induced BimS failed to do so in any detectable way. Perhaps there is the possibility that the amount of anti-apoptotic Bcl-2-like proteins that needs to be neutralized is so small as not to be detectable. However, there is no evidence to support this, and all models of interactions between Bcl-2 family members are based on solidly detectable results. Importantly, when Bcl-2 was overexpressed to inhibit BimS-induced apoptosis, a direct interaction was seen. The data thus confirm the view that Bcl-2 blocks BH3-only protein-induced apoptosis by direct binding. However, BimS does not appear to induce apoptosis by binding to Bcl-2-like anti-apoptotic proteins. This conclusion is also supported by the analysis of mutants in either mitochondrial localization or BH3 domain, where apoptosis induction was strictly correlated with mitochondrial localization but not with the ability to bind Bcl-2 (Table I; a different BimS mutant that had lost the ability to bind Bcl-2 but still induced apoptosis has been published previously [Marani et al., 2002]). Finally, in yeast, BimS enhanced the death-inducing activity of Bax, in the bona fide absence of Bcl-2-like proteins. Although each of these findings on its own may have their limitations, in their sum they strongly suggest that Bax activation and apoptosis by BimS occurs in the absence of binding to anti-apoptotic Bcl-2 proteins.
Although Bax can be demonstrated to be bound to Bcl-2-like proteins in some detergents (Hsu and Youle, 1997), it is difficult to know how much of this interaction is physiological. The displacement model of BH3-only protein function predicts that Bax is bound to Bcl-2 until this interaction is broken up by BH3-only proteins (leaving aside for the moment the question of different localization; [Willis et al., 2007]). We tested the interaction between Bax and Bcl-2-like proteins by Bax-IP in lysates of mitochondria containing 1% Triton X-100 (which allows co-IP of Bax and Bcl-2). Some Bcl-2 was coprecipitated; the amount of Bcl-xL was greater (as reported [Hsu and Youle, 1997]), and some Mcl-1 was co-IPed as well (Fig. S3 b). There was no clear evidence of a BimS-dependent reduction of these interactions. However, the evidence is probably not conclusive because much more Bax is found on mitochondria upon BimS induction.
Importantly, Bak has been found to be sequestered by Mcl-1 and Bcl-xL, and an elegant model has been proposed how Bcl-xL and Mcl-1 could restrain Bak and stop it from auto-activating (Willis et al., 2005). Very recently, predictions of both models of BH3-only function have been put to the test, and neither model has come away unscathed. The direct activation model has it that no Bax/Bak dependent apoptosis can occur in the absence of activator BH3-only proteins, but taking away these proteins led either only to a reduction of apoptosis (Kim et al., 2006) or had no effect (Willis et al., 2007). The Bak auto-activation proposed by the replacement model has also been queried because mutants of Bax and Bak that have lost detectable binding to anti-apoptotic Bcl-2-like proteins were functionally unaltered (Kim et al., 2006). In any case, although there is the possibility that Bax is also sequestered and kept inactive by anti-apoptotic Bcl-2 proteins, it seems clear that the activation of Bax has to involve the additional step of mitochondrial translocation. Conflicting results have been reported regarding the question which domain of Bax is important for this process (Priault et al., 2003; Schinzel et al., 2004; Cartron et al., 2005), and Bax can permeabilize artificial membranes in the absence of other proteins (Kuwana et al., 2002), suggesting that Bax is not regularly imported into mitochondria but uses an alternative way of translocation. However, some signal must trigger this translocation. Although a peptide representing the BH3 domain of Bim (or Bid) can activate Bax (Letai et al., 2002), high concentrations of peptide are required, and recent work shows that tethering Bid peptide to membranes strongly enhances its Bax-activating activity (Oh et al., 2006). Similarly, we detected no killing activity in BimS(1–88) (lacking the mitochondrial targeting domain), but substantial activity in BimSTom5 (targeted to the outer mitochondrial membrane); the differences seen between studies using peptides and BH3-only protein may therefore simply be a matter of abundance.
The essential activity of BimS thus appears to be the activation of Bax. Although some interaction between the proteins can be demonstrated, mutant analysis shows that this detectable interaction is not required for apoptosis induction. Furthermore, experiments by many investigators with chemical cross-linkers typically showed only dimeric and trimeric Bax on mitochondrial membranes, arguing against a complex containing Bax and BH3-only proteins (although large complexes perhaps would go unnoticed). It is completely unclear how BimS conveys an activation signal to cytosolic Bax. Perhaps BimS causes changes to composition and topology of the outer mitochondrial membrane that will initiate an alteration in Bax. At least in most cells, some Bax is found on mitochondria, possibly in exchange and balance with cytosolic Bax. It seems therefore conceivable that the insertion of BimS into mitochondria somehow affects mitochondrial Bax and removes it from the reaction, causing recruitment of Bax from the cytosol until a critical concentration is reached in mitochondria and Bax auto-activates.
A final point is the question whether the behavior we saw for Bim also holds true for other BH3-only proteins. Published studies, including analyses of gene-deficient mice, indicate that Bim, Bid, and Puma are the strongest apoptosis inducers within this group. This may be because, as suggested, the capacity of directly activating Bax (Letai et al., 2002) or their affinity for all anti-apoptotic Bcl-2 proteins rather than only a subset (Chen et al., 2005; Certo et al., 2006). However, another point can now be made for mitochondrial insertion. It has been noted before that active Bid localizes to mitochondrial membranes (Grinberg et al., 2002; Esposti et al., 2003), and mitochondrial import of Bim was directly shown in this study. Of the other BH3-only proteins known, only Puma has a consensus mitochondrial tail-anchor sequence. A third possibility for the differing activity thus appears to be the ability to localize to mitochondrial membranes.
Materials and methods
Cell lines and protein induction
T-REx-293 cells transiently transfected with different mBimS mutants and T-REx-HeLa cell lines (Invitrogen) that stably express the tetracycline repressor from the pCDNA6/TR vector were cultured in DME supplemented with 10% fetal calf serum (Tet negative; PAA Laboratories), 5 μg/ml blasticidin, 50 μg/ml gentamycin and 20 μg/ml vancomycin at 37°C/5% CO2. T-REx-HeLa cell lines stably expressing human hBimEL or murine mBimS were additionally supplemented with zeocin (125 μg/ml). Induction of mBimS mutant proteins was performed 7–24 h after electroporation of T-REx-293 cells. For immunoprecipitation of BimS mutants, T-REx-293 cells were cotransfected with a pEF-hBcl-2 construct (a gift from Dr. David Huang, Walter and Eliza Hall Institute for Medical Research, Victoria, Australia) or with pCDNA-hMcl-1 (a gift from Dr. Joseph Opferman, St. Jude Children's Research Hospital, Memphis, TN).
Generation of stable T-REx-HeLa cell lines
Stable T-REx-HeLa cell lines expressing mBimS protein were established using pcDNA4 vectors coding for mBimS (see below) according to the manufacturer's instructions (Invitrogen).
Construction of mBimS expression vectors
The genes coding for mBimS were amplified by PCR and subcloned into the pcDNA4/TO/myc-HisA vector (Invitrogen) with a stop codon at the 3-prime end to avoid transcription of the myc-His tag. For generation of the BimS-BH3 mutant proteins, the QuickChange II Site-Directed Mutagenesis kit (Stratagene) was used. C-terminal truncated BimS protein (BimS(1–88)) was obtained by PCR. Replacement of the C-terminal domain (aa 85–110) including the hydrophobic part of mBimS with the hydrophobic domain of the yeast Tom5 protein (aa 23–50) was performed by two-step PCR. mBimSΔΔTom5 was generated using the same mutation strategy as above.
Subcellular fractionation
Cells were harvested in isotonic mitochondrial buffer (MB) (210 mM mannitol, 70 mM sucrose, 1 mM EDTA, and 10 mM Hepes [pH 7.5]) supplemented with 1× complete protease inhibitor cocktail (Roche) and the mitochondria were isolated as described earlier (Eskes et al., 2000) All fractions (membrane and soluble fractions) were stored in MB-EGTA buffer at −80°C until further analyses. Protein concentrations were determined by the Bradford assay in MB-EGTA-1% Triton buffer. Intracellular localization of Bim and Bax during induction of BimS in T-REx-HeLa cells or of BimS mutants in T-REx-293 cells was analyzed by loading equal protein amounts of the membrane fraction and the soluble fraction on 12.5% SDS-PAA gels. Bim and Bax were detected with anti-Bim (Sigma-Aldrich) or anti-Bax antibodies (clone 6A7; Upstate Biotechnology). A monoclonal 20E8 antibody against cytochrome c oxidase subunit IV (CoxIV; MoBiTec) and a monoclonal antibody directed against caspase-8 (1C12 clone; Cell Signaling Technology) were used to detect marker proteins.
Insertion of Bax and mBimS into mitochondrial membranes
The insertion of Bax and BimS into the mitochondrial membrane during BimS induction was tested by alkali extraction of the mitochondrial pellets as described earlier (Eskes et al., 2000). The membranes were pelleted by centrifugation (260,000 g for 1 h at 4°C); proteins (the alkali-sensitive fraction) were precipitated by 12% TCA and the corresponding volume of supernatants and mitochondrial pellets were separated by SDS-PAGE.
Immunoprecipitation
Cells or isolated mitochondria were extracted in lysis buffer (50 mM Hepes [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% [vol/vol] glycerol, and 1% [vol/vol] Triton X-100) supplemented with a protease inhibitor cocktail for 30 min on ice. The lysate was spun at 13.000 g for 10 min. For immunoprecipitation of Bim, protein G–Sepharose beads were washed twice with lysis buffer followed by preincubation with monoclonal rat anti-Bim antibody (3C5 clone, a gift from Dr. Andreas Strasser, Walter and Eliza Hall Institute for Medical Research, Victoria, Australia) for 1 h at 4°C. As a control an antibody directed against mBmf (provided by Dr. Strasser) was used. The beads were incubated in the presence of 45–200 μg of proteins overnight with constant agitation at 4°C. After washing four times with lysis/1% (vol/vol) Triton X-100 buffer (see above), bound protein was eluted off the beads by boiling in 2× Laemmli sample buffer and resolved by SDS-PAGE. Bax-IP was performed with 130 μg of mitochondria as described by the manufacturer with a human-specific polyclonal Bax antibody (1:100; Cell Signaling Technology) using the same buffers as decribed above. Detection of Bim, Bcl-2, Mcl-1, Bcl-w, A1, Bcl-xL, and Bax was done by Western blotting using antibodies directed against Bim and Bax (see above). Bcl-2, Mcl-1 (clone 22), and Bcl-xL(clone 2H12) antibodies were from BD Biosciences; Bcl-w (clone 31H4) and A1 antibodies were from Cell Signaling Technology.
Assays for apoptosis
After induction of BimS or BimS mutants, cells were washed in PBS before fixation in 4% formaldehyde for 20 min at room temperature. Cells were washed in PBS and incubated in the presence of monoclonal anti-active-caspase-3 antibody (BD Biosciences) in permeabilization buffer (0.5% [wt/vol] BSA and 0.5% [wt/vol] saponin in PBS). Cells were washed in permeabilization buffer and incubated with species-specific Cy3-conjugated secondary antibody (Dianova). Flow cytometry was performed using a FACSCalibur (Becton Dickinson).
Detection of Bax activation by cross-linking
Mitochondria were isolated from clone A2 6 h after tetracycline induction (control: untreated) and were incubated either with disuccinimidyl subernate (DSS, Pierce Chemical Co.) or bis(sulfosuccinimidyl) subernate (BS3; Pierce Chemical Co.) as decribed earlier (Eskes et al., 2000). Samples were directly dissolved in Laemmli sample buffer and then analyzed by Western blotting with an anti-Bax antibody (Clone 3; BD Biosciences).
Confocal microscopy
For localization of EGFP-Bax during BimS induction, BimS clone A2 was transiently transfected with an expression plasmid for EGFP-Bax (a gift from Dr. Christoph Borner, University of Freiburg, Freiburg, Germany). Cells were then seeded onto coverslips for overnight culture, and mitochondria were stained with MitoTracker Orange CMTMRos (Invitrogen) 30 min before washing with fresh media and tetracycline induction of BimS. To prevent detachment of cells, 25 μM of zVAD-fmk was added to block caspase-dependent apoptosis. Cells were fixed in 3.7% formaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 4 min, and blocked with 5% heat-inactivated donkey serum for 10 min. Primary antibody incubations with rabbit anti-Bim (StressGen Biotechnologies) were performed for 40 min, followed by incubation with secondary donkey anti–rabbit-Cy5 antibody (1:150; Dianova) for 30 min. 0.5% BSA-PBS was used for blocking, antibody incubations, and wash steps, and coverslips were mounted in Mowiol.
For localization of BimS and BimS mutants, T-REx-293 cells were transiently transfected with the respective expression plasmid, seeded onto coverslips and induced 7 h later for further 15 h with tet, stained for mitochondria, and processed as described. Cells were stained with rat anti-Bim (1:100, clone 3C5), followed by donkey anti–rat Alexa-488 (1:200; Invitrogen).
Cells were examined at room temperature with either a laser-scanning microscope (LSM 510 Axiovert 100M, 100×/1.3 oil immersion lens; Carl Zeiss MicroImaging, Inc.) or a confocal microscope (TCS SP5, 63×/1.2 water immersion lens; Leica) using the respective imaging medium. Pinholes were set to scan layers < 1μm, at a resolution of 1024 × 1024 pixels. Acquired images were exported using the in-built Zeiss or Leica Image Browser, respectively. Adobe Photoshop was used to adjust image size and resolution, and to enhance contrast of the whole image for better visibility in some pictures.
In vitro mitochondrial import
Radiolabeled (35S-Met) BimEL and BimS proteins were generated using the TNT reticulocyte transcription/translation system (Promega). Mitochondrial import experiments were performed essentially as described previously (Okamoto et al., 2002).
Analysis of cell death in yeast
The coding sequence of mouse Bax was amplified by PCR and cloned into the tet-off plasmid pCM189 (Gari et al., 1997). Mouse BimS and BimS mutants were inserted into the constitutive-expression vector p415-ADH, respectively (Mumberg et al., 1995). The plasmids were transformed into the S. cerevisiae strain EGY-48. Yeast cells were grown in SD medium containing 2% lactate and 1 μg/ml tetracycline. For induction of Bax, cells were shifted to medium without tetracycline (start at OD600 = 0.1). After ∼8 h, cultures were diluted to OD600 = 0.1 (no differences of growth were seen at this point in time between cultures), and culture was continued. OD600 was measured at indicated time points. Significance of the observed differences was tested by ANOVA followed by the least square difference post-hoc test on data from independent experiments. For FACS analysis of the mitochondrial membrane potential, yeast cells were incubated with 5 μM rhodamine123 for 30 min at 30°C in the dark. Subsequently, cells were isolated by centrifugation, washed in PBS, and resuspended in PBS. Stained cells were immediately analyzed in a FACSCalibur cytometer (Becton Dickinson).
Online supplemental materials
Fig. S1 shows induction of apoptosis by tet-dependent BimS induction. Fig. S2 shows how Bcl-2 overexpression blocks BimS-induced apoptosis in the A2 BimS clone. Fig. S3 describes interaction of Bax, BimS/BimS(D69A), and prosurvival Bcl-2-like proteins by IP. Fig. S4 shows subcellular localization of Bax and BimS in the 3C4 BimS clone. Fig. S5 shows pro-apoptotic activity of BimS and BimS mutants by Hoechst staining. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200610148/DC1.
Supplementary Material
Acknowledgments
We thank Tibor Schuster (Institute for Medical Statistics, Technische Universität München) for help with statistics, Stephie Potthoff and Juliane Vier for technical assistance, and Dr. Andreas Strasser (Walter and Eliza Hall Institute, Melbourne, Australia) for reagents.
This work was supported by the Wilhelm-Sander Stiftung.
Abbreviations used in this paper: BH, Bcl-2 homology; IP, immunoprecipitation; tet, tetracycline.
References
- Adams, J.M. 2003. Ways of dying: multiple pathways to apoptosis. Genes Dev. 17:2481–2495. [DOI] [PubMed] [Google Scholar]
- Cartron, P.F., H. Arokium, L. Oliver, K. Meflah, S. Manon, and F.M. Vallette. 2005. Distinct domains control the addressing and the insertion of Bax into mitochondria. J. Biol. Chem. 280:10587–10598. [DOI] [PubMed] [Google Scholar]
- Certo, M., G. Moore Vdel, M. Nishino, G. Wei, S. Korsmeyer, S.A. Armstrong, and A. Letai. 2006. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 9:351–365. [DOI] [PubMed] [Google Scholar]
- Chen, L., S.N. Willis, A. Wei, B.J. Smith, J.I. Fletcher, M.G. Hinds, P.M. Colman, C.L. Day, J.M. Adams, and D.C. Huang. 2005. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 17:393–403. [DOI] [PubMed] [Google Scholar]
- Cory, S., and J.M. Adams. 2002. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer. 2:647–656. [DOI] [PubMed] [Google Scholar]
- Danial, N.N., and S.J. Korsmeyer. 2004. Cell death: critical control points. Cell. 116:205–219. [DOI] [PubMed] [Google Scholar]
- Dewson, G., R.T. Snowden, J.B. Almond, M.J. Dyer, and G.M. Cohen. 2003. Conformational change and mitochondrial translocation of Bax accompany proteasome inhibitor-induced apoptosis of chronic lymphocytic leukemic cells. Oncogene. 22:2643–2654. [DOI] [PubMed] [Google Scholar]
- Eskes, R., S. Desagher, B. Antonsson, and J.C. Martinou. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20:929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposti, M.D., I.M. Cristea, S.J. Gaskell, Y. Nakao, and C. Dive. 2003. Proapoptotic Bid binds to monolysocardiolipin, a new molecular connection between mitochondrial membranes and cell death. Cell Death Differ. 10:1300–1309. [DOI] [PubMed] [Google Scholar]
- Frohlich, K.U., and F. Madeo. 2000. Apoptosis in yeast–a monocellular organism exhibits altruistic behaviour. FEBS Lett. 473:6–9. [DOI] [PubMed] [Google Scholar]
- Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 13:837–848. [DOI] [PubMed] [Google Scholar]
- Grinberg, M., R. Sarig, Y. Zaltsman, D. Frumkin, N. Grammatikakis, E. Reuveny, and A. Gross. 2002. tBID Homooligomerizes in the mitochondrial membrane to induce apoptosis. J. Biol. Chem. 277:12237–12245. [DOI] [PubMed] [Google Scholar]
- Gross, A., J. Jockel, M.C. Wei, and S.J. Korsmeyer. 1998. Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. EMBO J. 17:3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, A., K. Pilcher, E. Blachly-Dyson, E. Basso, J. Jockel, M.C. Bassik, S.J. Korsmeyer, and M. Forte. 2000. Biochemical and genetic analysis of the mitochondrial response of yeast to BAX and BCL-X(L). Mol. Cell. Biol. 20:3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie, C., H. Suzuki, M. Sakaguchi, and K. Mihara. 2003. Targeting and assembly of mitochondrial tail-anchored protein Tom5 to the TOM complex depend on a signal distinct from that of tail-anchored proteins dispersed in the membrane. J. Biol. Chem. 278:41462–41471. [DOI] [PubMed] [Google Scholar]
- Hsu, Y.T., and R.J. Youle. 1997. Nonionic detergents induce dimerization among members of the Bcl-2 family. J. Biol. Chem. 272:13829–13834. [DOI] [PubMed] [Google Scholar]
- Huang, D.C., and A. Strasser. 2000. BH3-only proteins-essential initiators of apoptotic cell death. Cell. 103:839–842. [DOI] [PubMed] [Google Scholar]
- Kim, H., M. Rafiuddin-Shah, H.C. Tu, J.R. Jeffers, G.P. Zambetti, J.J. Hsieh, and E.H. Cheng. 2006. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8:1348–1358. [DOI] [PubMed] [Google Scholar]
- Kuwana, T., M.R. Mackey, G. Perkins, M.H. Ellisman, M. Latterich, R. Schneiter, D.R. Green, and D.D. Newmeyer. 2002. Bid, bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 111:331–342. [DOI] [PubMed] [Google Scholar]
- Kuwana, T., L. Bouchier-Hayes, J.E. Chipuk, C. Bonzon, B.A. Sullivan, D.R. Green, and D.D. Newmeyer. 2005. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 17:525–535. [DOI] [PubMed] [Google Scholar]
- Letai, A., M.C. Bassik, L.D. Walensky, M.D. Sorcinelli, S. Weiler, and S.J. Korsmeyer. 2002. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2:183–192. [DOI] [PubMed] [Google Scholar]
- Lindsten, T., A.J. Ross, A. King, W.X. Zong, J.C. Rathmell, H.A. Shiels, E. Ulrich, K.G. Waymire, P. Mahar, K. Frauwirth, et al. 2000. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell. 6:1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., S. Dai, Y. Zhu, P. Marrack, and J.W. Kappler. 2003. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 19:341–352. [DOI] [PubMed] [Google Scholar]
- Marani, M., T. Tenev, D. Hancock, J. Downward, and N.R. Lemoine. 2002. Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol. Cell. Biol. 22:3577–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 156:119–122. [DOI] [PubMed] [Google Scholar]
- O'Connor, L., A. Strasser, L.A. O'Reilly, G. Hausmann, J.M. Adams, S. Cory, and D.C. Huang. 1998. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17:384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, K.J., S. Barbuto, K. Pitter, J. Morash, L.D. Walensky, and S.J. Korsmeyer. 2006. A membrane-targeted BID BCL-2 homology 3 peptide is sufficient for high potency activation of BAX in vitro. J. Biol. Chem. 281:36999–37008. [DOI] [PubMed] [Google Scholar]
- Okamoto, K., A. Brinker, S.A. Paschen, I. Moarefi, M. Hayer-Hartl, W. Neupert, and M. Brunner. 2002. The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 21:3659–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltvai, Z.N., C.L. Milliman, and S.J. Korsmeyer. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 74:609–619. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., and W. Neupert. 2001. Protein import into mitochondria. IUBMB Life. 52:101–112. [DOI] [PubMed] [Google Scholar]
- Petros, A.M., D.G. Nettesheim, Y. Wang, E.T. Olejniczak, R.P. Meadows, J. Mack, K. Swift, E.D. Matayoshi, H. Zhang, C.B. Thompson, and S.W. Fesik. 2000. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 9:2528–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault, M., P.F. Cartron, N. Camougrand, B. Antonsson, F.M. Vallette, and S. Manon. 2003. Investigation of the role of the C-terminus of Bax and of tc-Bid on Bax interaction with yeast mitochondria. Cell Death Differ. 10:1068–1077. [DOI] [PubMed] [Google Scholar]
- Puthalakath, H., and A. Strasser. 2002. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 9:505–512. [DOI] [PubMed] [Google Scholar]
- Puthalakath, H., D.C. Huang, L.A. O'Reilly, S.M. King, and A. Strasser. 1999. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell. 3:287–296. [DOI] [PubMed] [Google Scholar]
- Sato, T., M. Hanada, S. Bodrug, S. Irie, N. Iwama, L.H. Boise, C.B. Thompson, E. Golemis, L. Fong, H.G. Wang, et al. 1994. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc. Natl. Acad. Sci. USA. 91:9238–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel, A., T. Kaufmann, M. Schuler, J. Martinalbo, D. Grubb, and C. Borner. 2004. Conformational control of Bax localization and apoptotic activity by Pro168. J. Cell Biol. 164:1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., R.J. Youle, and N. Tjandra. 2000. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 103:645–654. [DOI] [PubMed] [Google Scholar]
- Willis, S.N., and J.M. Adams. 2005. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17:617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, S.N., L. Chen, G. Dewson, A. Wei, E. Naik, J.I. Fletcher, J.M. Adams, and D.C. Huang. 2005. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 19:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, S.N., J.I. Fletcher, T. Kaufmann, M.F. van Delft, L. Chen, P.E. Czabotar, H. Ierino, E.F. Lee, W.D. Fairlie, P. Bouillet, et al. 2007. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 315:856–859. [DOI] [PubMed] [Google Scholar]
- Wysocki, R., and S.J. Kron. 2004. Yeast cell death during DNA damage arrest is independent of caspase or reactive oxygen species. J. Cell Biol. 166:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, W.X., T. Lindsten, A.J. Ross, G.R. MacGregor, and C.B. Thompson. 2001. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 15:1481–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.