Abstract
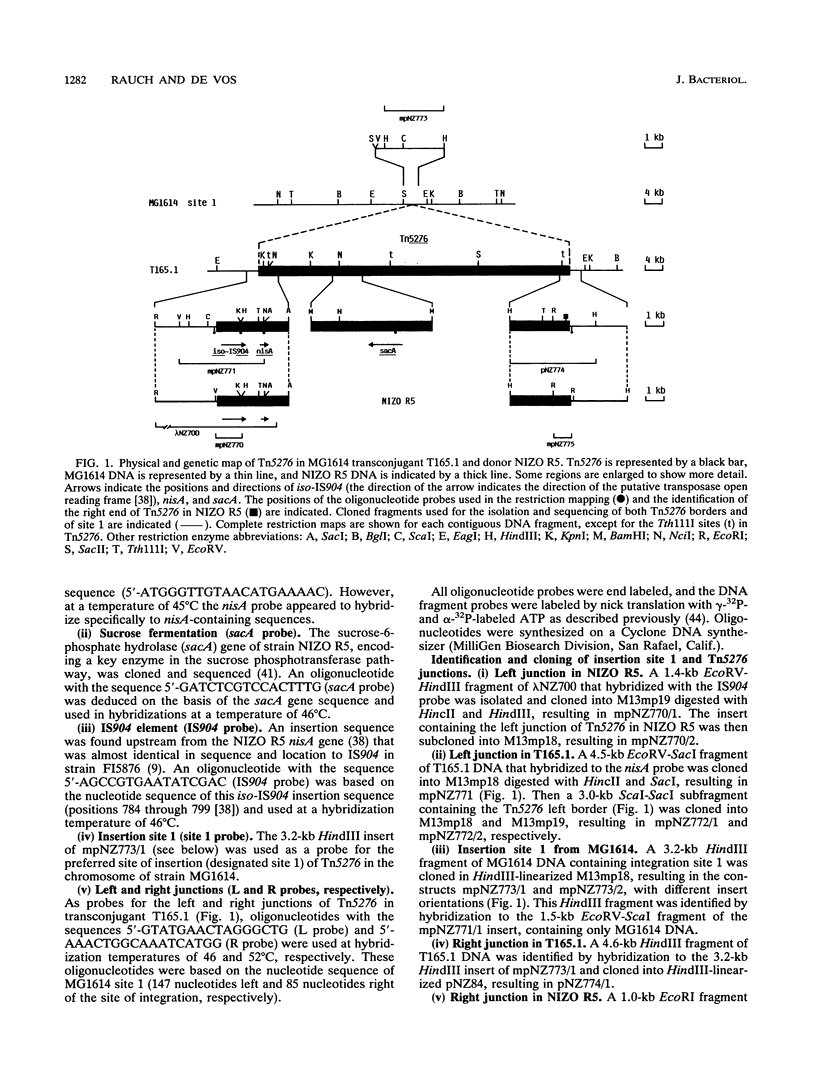
A novel, chromosomally located conjugative transposon in Lactococcus lactis, Tn5276, was identified and characterized. It encodes the production of and immunity to nisin, a lanthionine-containing peptide with antimicrobial activity, and the capacity to utilize sucrose via a phosphotransferase system. Conjugal transfer of Tn5276 was demonstrated from L. lactis NIZO R5 to different L. lactis strains and a recombination-deficient mutant. The integration of Tn5276 into the plasmid-free strain MG1614 was analyzed by using probes based on the gene for the nisin precursor (nisA) and the gene for sucrose-6-phosphate hydrolase (sacA). The transposon inserted at various locations in the MG1614 chromosome and showed a preference for orientation-specific insertion into a single target site (designated site 1). By using restriction mapping in combination with field inversion gel electrophoresis and DNA cloning of various parts of the element including its left and right ends, a physical map of the 70-kb Tn5276 was constructed, and the nisA and sacA genes were located. The nucleotide sequences of Tn5276 junctions in donor strain NIZO R5 and in site 1 of an MG1614-derived transconjugant were determined and compared with that of site 1 in recipient strain MG1614. The results show that the A + T-rich ends of Tn5276 are flanked by a direct hexanucleotide repeat in both the donor and the transconjugant but that the element does not contain a clear inverted repeat.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Isolation of a recombination-deficient mutant of Streptococcus lactis ML3. J Bacteriol. 1983 Aug;155(2):930–932. doi: 10.1128/jb.155.2.930-932.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman G. W., Banerjee S., Hansen J. N. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988 Nov 5;263(31):16260–16266. [PubMed] [Google Scholar]
- Caillaud F., Courvalin P. Nucleotide sequence of the ends of the conjugative shuttle transposon Tn1545. Mol Gen Genet. 1987 Aug;209(1):110–115. doi: 10.1007/BF00329844. [DOI] [PubMed] [Google Scholar]
- Campbell A., Berg D. E., Botstein D., Lederberg E. M., Novick R. P., Starlinger P., Szybalski W. Nomenclature of transposable elements in prokaryotes. Gene. 1979 Mar;5(3):197–206. doi: 10.1016/0378-1119(79)90078-7. [DOI] [PubMed] [Google Scholar]
- Caparon M. G., Scott J. R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989 Dec 22;59(6):1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- Dodd H. M., Horn N., Gasson M. J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990 Mar;136(3):555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- Donkersloot J. A., Thompson J. Simultaneous loss of N5-(carboxyethyl)ornithine synthase, nisin production, and sucrose-fermenting ability by Lactococcus lactis K1. J Bacteriol. 1990 Jul;172(7):4122–4126. doi: 10.1128/jb.172.7.4122-4126.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald G. F., Gasson M. J. In vivo gene transfer systems and transposons. Biochimie. 1988 Apr;70(4):489–502. doi: 10.1016/0300-9084(88)90085-5. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982 Nov 18;300(5889):281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- Gonzalez C. F., Kunka B. S. Transfer of Sucrose-Fermenting Ability and Nisin Production Phenotype among Lactic Streptococci. Appl Environ Microbiol. 1985 Mar;49(3):627–633. doi: 10.1128/aem.49.3.627-633.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hill C., Daly C., Fitzgerald G. F. Development of High-Frequency Delivery System for Transposon Tn919 in Lactic Streptococci: Random Insertion in Streptococcus lactis subsp. diacetylactis 18-16. Appl Environ Microbiol. 1987 Jan;53(1):74–78. doi: 10.1128/aem.53.1.74-78.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn N., Swindell S., Dodd H., Gasson M. Nisin biosynthesis genes are encoded by a novel conjugative transposon. Mol Gen Genet. 1991 Aug;228(1-2):129–135. doi: 10.1007/BF00282457. [DOI] [PubMed] [Google Scholar]
- Inamine J. M., Burdett V. Structural organization of a 67-kilobase streptococcal conjugative element mediating multiple antibiotic resistance. J Bacteriol. 1985 Feb;161(2):620–626. doi: 10.1128/jb.161.2.620-626.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol. 1989 Mar;171(3):1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski J. J., Murphy E., Novick R. P., Rush M. G. Site-specificity of the chromosomal insertion of Staphylococcus aureus transposon Tn554. J Mol Biol. 1981 Oct 15;152(1):19–33. doi: 10.1016/0022-2836(81)90093-0. [DOI] [PubMed] [Google Scholar]
- Le Bouguénec C., de Cespédès G., Horaud T. Molecular analysis of a composite chromosomal conjugative element (Tn3701) of Streptococcus pyogenes. J Bacteriol. 1988 Sep;170(9):3930–3936. doi: 10.1128/jb.170.9.3930-3936.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. M. Plasmid Reference Center Registry of transposon(Tn) and insertion sequence (IS) allocations through December 1986. Gene. 1987;51(2-3):115–118. doi: 10.1016/0378-1119(87)90299-x. [DOI] [PubMed] [Google Scholar]
- Mulders J. W., Boerrigter I. J., Rollema H. S., Siezen R. J., de Vos W. M. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur J Biochem. 1991 Nov 1;201(3):581–584. doi: 10.1111/j.1432-1033.1991.tb16317.x. [DOI] [PubMed] [Google Scholar]
- Murphy E., Huwyler L., de Freire Bastos M. do C. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 1985 Dec 1;4(12):3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. C., Steele J. L., Daly C., McKay L. L. Concomitant conjugal transfer of reduced-bacteriophage-sensitivity mechanisms with lactose- and sucrose-fermenting ability in lactic streptococci. Appl Environ Microbiol. 1988 Aug;54(8):1951–1956. doi: 10.1128/aem.54.8.1951-1956.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgren M., Scott J. R. The presence of conjugative transposon Tn916 in the recipient strain does not impede transfer of a second copy of the element. J Bacteriol. 1991 Jan;173(1):319–324. doi: 10.1128/jb.173.1.319-324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch P. J., Beerthuyzen M. M., de Vos W. M. Nucleotide sequence of IS904 from Lactococcus lactis subsp. lactis strain NIZO R5. Nucleic Acids Res. 1990 Jul 25;18(14):4253–4254. doi: 10.1093/nar/18.14.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J. L., McKay L. L. Conjugal transfer of genetic material by Lactococcus lactis subsp. lactis 11007. Plasmid. 1989 Jul;22(1):32–43. doi: 10.1016/0147-619x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Steele J. L., McKay L. L. Partial characterization of the genetic basis for sucrose metabolism and nisin production in Streptococcus lactis. Appl Environ Microbiol. 1986 Jan;51(1):57–64. doi: 10.1128/aem.51.1.57-64.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen M. T., Chung Y. J., Hansen J. N. Characterization of the nisin gene as part of a polycistronic operon in the chromosome of Lactococcus lactis ATCC 11454. Appl Environ Microbiol. 1991 Apr;57(4):1181–1188. doi: 10.1128/aem.57.4.1181-1188.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from gram-positive bacteria. Gene. 1991 Sep 30;106(1):21–27. doi: 10.1016/0378-1119(91)90561-o. [DOI] [PubMed] [Google Scholar]
- Vijayakumar M. N., Priebe S. D., Guild W. R. Structure of a conjugative element in Streptococcus pneumoniae. J Bacteriol. 1986 Jun;166(3):978–984. doi: 10.1128/jb.166.3.978-984.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., van Asseldonk M., van Jeveren F., Siezen R., Simons G., de Vos W. M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989 May;171(5):2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley R. C., Pennock A., Ashton R. J., Davies A., Young M. Transfer of Tn1545 and Tn916 to Clostridium acetobutylicum. Plasmid. 1989 Sep;22(2):169–174. doi: 10.1016/0147-619x(89)90027-9. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Alen-Boerrigter I. J., Baankreis R., de Vos W. M. Characterization and overexpression of the Lactococcus lactis pepN gene and localization of its product, aminopeptidase N. Appl Environ Microbiol. 1991 Sep;57(9):2555–2561. doi: 10.1128/aem.57.9.2555-2561.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]




