Abstract
The limb- and heart-specific Tbx5 transcription factor coexpresses with and directly binds to the novel PDZ-LIM domain protein, LMP4. LMP4 is distributed in the cytoplasm associated with the actin cytoskeleton. In the presence of LMP4, Tbx5 shuttles dynamically between the nucleus and cytoplasm and, in a complex with LMP4, localizes to actin filaments. Nuclear and cytoplasmic Tbx5 distribution in developing chicken wings suggests the functional significance of the LMP4–Tbx5 interaction. In primary epicardial cells, we demonstrate that Tbx5 protein subcellular relocalization can be stimulated by external signals that induce cell differentiation. To test whether the relocalization from nuclear to cytoplasmic sites interferes with downstream gene expression, we used limb-specific Fgf10 and heart-specific Anf promoter-luciferase reporters and demonstrate that LMP4 acts as a repressor of Tbx5 activity. These studies reveal a previously unknown mechanism for Tbx transcription factor regulation in vertebrate limb and heart development and provide a better understanding of the molecular basis of hand/heart birth defects associated with Tbx5 mutations.
Introduction
Tbx5 and -4 belong to the family of T-box transcription factors that share a homologous DNA binding domain (T-domain) first described in the mouse brachyury (or T) gene product (Herrmann et al., 1990; Kispert and Herrmann, 1993). Studies in chicken (Logan and Tabin, 1998; Rodriguez-Esteban et al., 1999; Takeuchi et al., 1999), zebrafish (Ahn et al., 2002; Garrity et al., 2002), and mouse (Agarwal et al., 2003; Naiche and Papaioannou, 2003; Rallis et al., 2003) revealed that both Tbx5 and -4 play critical roles in the outgrowth and specification of vertebrate forelimbs and hindlimbs, respectively. In addition to the limbs, Tbx5 has been shown to be required for proper heart development in zebrafish (Garrity et al., 2002) and mouse (Bruneau et al., 2001). In the chicken, Tbx4 expression has also been described in the heart, complementing the asymmetrical Tbx5 expression in this organ and suggesting parallel pathways for these transcription factors in the limbs and heart (Krause et al., 2004). Although there is strong evidence that Tbx5 and -4 are critical for embryonic development, little is known about how the transcription factors are regulated and function at the cellular level.
In a protein–protein interaction screen, we recently identified from chicken a new protein called LMP4, by its ability to interact with the C-terminal transactivation domain of the Tbx5 and -4 transcription factors (Krause et al., 2004). In chicken embryos, LMP4 is expressed in the developing eye, heart, forelimbs, and hindlimbs, all organs that express either Tbx5 or -4 (Logan et al., 1998; Bruneau et al., 1999; Krause et al., 2004). LMP4 is a member of an emerging class of scaffolding proteins, denoted PDZ-LIM proteins, which appear to function in fundamental biological processes, including cytoskeletal organization, cell lineage specification, and organ development (Dawid et al., 1998; Fanning and Anderson, 1999). PDZ-LIM proteins contain cassettes of two different types of protein–protein interaction domains: a single N-terminal PDZ domain and one or three C-terminal LIM domains. The PDZ domain is an 85-amino-acid β-barrel protein interaction motif that binds to both C-terminal peptides and internal sequences of target proteins (Harris and Lim, 2001). The PDZ domains of the PDZ-LIM proteins Enigma homologue (ENH) 1 and CLP-36 both bind to α-actinin, and this interaction localizes the proteins to actin filaments (Nakagawa et al., 2000; Vallenius et al., 2000). The LIM domain is a 55-amino-acid sequence that contains two zinc finger–like motifs with conserved cysteine residues (Kadrmas and Beckerle, 2004). The LIM domains of PDZ-LIM proteins have been found to interact with protein kinases, such as Clik1 (Vallenius and Makela, 2002), PKC (Kuroda et al., 1996), and receptor tyrosine kinases (Wu and Gill, 1994; Wu et al., 1996). All of the described binding partners for PDZ-LIM proteins suggest a role for this protein family as mediators, regulating protein function and/or signaling.
PDZ-LIM family proteins can be subdivided into two subclasses depending on the number of LIM domains present. For example, CLP-36 contains a single C-terminal LIM domain, whereas ENH1 and LMP1 contain three. The ENH and LMP proteins share significant sequence homology between their PDZ and LIM domains. However, there still appears to be specificity within the binding motifs. The PDZ domains of rat ENH1 and human Enigma (an LMP protein) bind to α-actinin (Nakagawa et al., 2000) and β-tropomyosin (Guy et al., 1999), respectively, whereas the LIM domains of each protein bind different isoforms of PKC (Kuroda et al., 1996).
We have proposed that LMP4 interacts with Tbx5 and -4 and regulates their activities by localizing the transcription factors out of the nucleus (Krause et al., 2004). Building on our previous developmental studies, we focus on Tbx5 and use it as a model to understand the mechanism of LMP4–Tbx interactions. We have conducted a detailed cellular investigation using cell biology and biochemical techniques to test our hypothesis and uncover a novel mechanism that regulates Tbx protein subcellular localization and transcriptional activity.
Results
Tbx5 and LMP4 are localized to the cytoplasm in the developing chicken wing
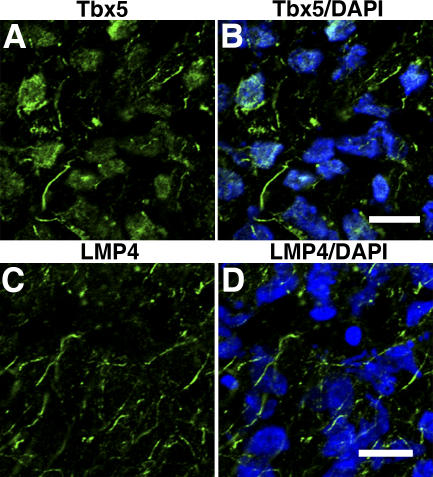
Previously, we showed coexpression of Tbx5 and LMP4 mRNA in chicken embryos during heart and forelimb development. Additionally, Tbx5 and LMP4 protein binding was shown in vitro, and initial experiments with transfected cells suggested that the Tbx5 and LMP4 proteins colocalized at cytoplasmic sites (Krause et al., 2004). To extend our earlier findings, we wished to determine whether such protein interactions occur within a developmental context in vivo. To gain insight into this question, we turned to the developing chicken wing. Wings from Hamburger-Hamilton (HH) stage 36 (Hamburger and Hamilton, 1951) chicken embryos were sectioned, and serial sections were stained with Tbx5- (Khan et al., 2002) and LMP4-specific antibodies (Fig. S1 and supplemental text, available at http://www.jcb.org/cgi/content/full/jcb.200511109/DC1). As expected, we detected Tbx5 protein in mesenchymal cells but not in the outer epidermal layer (Fig. 1, A and B; and not depicted). Interestingly, Tbx5 was localized both within the nucleus and at cytoplasmic sites; however, the ratio of nuclear to cytoplasmic distribution varied in different regions of the limb. This finding demonstrates that Tbx5 protein in the developing chicken wing is not strictly localized to the nucleus. Cytoplasmic localization of TBX5 has also been reported in the human lung (Collavoli et al., 2003). In adjacent tissue sections, the expression of LMP4 was predominantly cytoplasmic, revealing a punctate and filamentous pattern, reminiscent of Tbx5 cytoplasmic localization (Fig. 1, C and D). No obvious nuclear localization was detected for LMP4. The expression of Tbx5 and LMP4 at the RNA level in HH stage 36 wings was confirmed by RT-PCR, whereas the Tbx5-specific antiserum did not detect protein in respective hindlimb cryosections (unpublished data). Of note, in experiments with developing chicken hearts, we have observed similar subcellular localization patterns for Tbx5 and LMP4 (unpublished data). This would indicate a more general role for nuclear and cytoplasmic Tbx5 distribution and suggest functional significance of the LMP4–Tbx5 interaction in vivo.
Figure 1.
Tbx5 and LMP4 protein localization in chicken wings. Serial cryosections of HH stage 36 wings were stained for Tbx5 (A and B) or LMP4 (C and D). (A and B) Tbx5 colocalized with nuclear marker DAPI (B) but was also localized to filamentous structures in the cytoplasm. (C and D) LMP4 (C) was localized to filamentous structures in the cytoplasm with no colocalization with DAPI (D). Comparison of merged images (B and D) shows a very similar staining pattern in the cytoplasm for Tbx5 and LMP4 in the wing. Bars, 10 μm.
Differentiation of chicken epicardial cells changes Tbx5 subcellular localization
Finding that Tbx5 protein displayed cytoplasmic distribution during development supported our model that LMP4 may regulate Tbx5 on a cellular level. To elucidate in more detail the mechanism of Tbx5 and LMP4 interactions, we turned to cultured cells. During chicken heart development, epicardial cells arise from the proepicardial organ, migrate over the heart, and through a process of epithelial-to-mesenchymal transition (EMT) give rise to cardiac fibroblasts and coronary smooth muscle cells that contribute to the myocardial wall, atrioventricular cushion and valves, and coronary vasculature (Mikawa and Gourdie, 1996; Gittenberger-de Groot et al., 1998). Epicardial cells have been shown to natively express Tbx5 (Hatcher et al., 2004), and in vitro cultures of these cells have been used as a cell differentiation model during EMT (Dettman et al., 1998; Lu et al., 2001). We have used such a chicken primary epicardial cell culture system to determine if Tbx5 and LMP4 subcellular localization would change after stimulating the cells to differentiate. Cultured epicardial cells maintained in serum-free media remain in an undifferentiated state, characterized by predominantly cortical actin and absence of differentiation markers such as calponin (Lu et al., 2001). In contrast, cells cultured in chicken embryonic heart–conditioned media differentiate into epicardially derived cells (EPDCs), which lose their cortical actin organization and begin to express the smooth muscle marker calponin (Lu et al., 2001; Morabito et al., 2001).
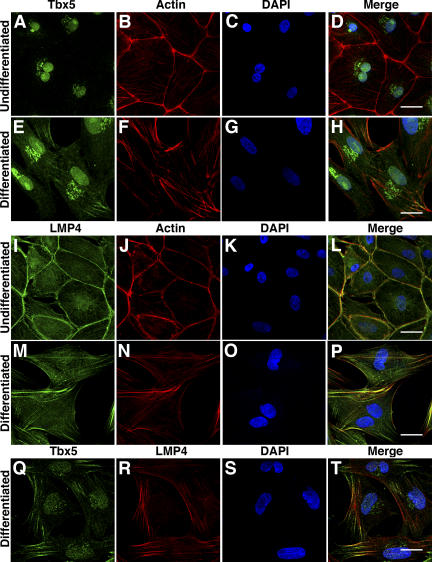
Monolayers of chicken epicardial cells were grown from HH stage 25 hearts in serum-free media. These epicardial cells were then processed for indirect confocal microscopy to detect endogenous Tbx5 and LMP4 using specific antibodies. Alternatively, cultures were induced to differentiate into EPDCs using embryonic heart–conditioned media followed by immunocytochemical analysis. Using the Tbx5-specific antiserum, the transcription factor was detected in cultured epicardial cells and EPDCs by confocal microscopy (Fig. 2, A–H) and confirmed by Western blot using cell lysates (not depicted). Interestingly, Tbx5 protein localization was found to change in the chicken primary heart cultures depending on cellular context. In epicardial cells cultured in serum-free media, the actin cytoskeleton was predominantly cortical, consistent with undifferentiated epicardial cells (Fig. 2, A–D). A further indication for the undifferentiated status of these cells is the absence of calponin expression (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200511109/DC1; Lu et al., 2001). In these epicardial cultures, Tbx5 was predominantly nuclear (Fig. 2 A), similar to previous observations in transfected cells (Collavoli et al., 2003; Krause et al., 2004; Zaragoza et al., 2004). However, when epicardial cells were shifted from serum-free to heart-conditioned culture media, they differentiated into EPDCs (Fig. 2, E–H). The altered differentiation status was indicated by the drastic change in cell morphology, as outlined by the reorganization of actin, from predominantly cortical to mostly filamentous stress fibers (Lu et al., 2001). In addition, the cells expressed the differentiation marker calponin (Fig. S2). In the EPDCs, Tbx5 changed its localization from predominantly nuclear to a combination of nuclear and cytoplasmic distribution (Fig. 2 E). Furthermore, a significant amount of the cytoplasmic Tbx5 protein colocalized with phalloidin-stained actin filaments (Fig. 2 H). The distribution of Tbx5 within the EPDCs demonstrates that the transcription factor is not strictly localized to the nucleus and that its localization is regulated depending on cell differentiation status, a finding that has not previously been described.
Figure 2.
Localization of Tbx5 and LMP4 during epicardial cell differentiation. HH stage 25 chicken primary epicardial cells were cultured and stained for endogenous Tbx5 (A–H) or LMP4 (I–P) using specific antibodies. (A–D) Undifferentiated epicardial cells stained for Tbx5 (A), Alexa 488 phalloidin pseudocolored red (B), and the nuclear marker DAPI (C). Merged image (D) shows Tbx5 colocalization with DAPI but not with actin-stained phalloidin. (E–H) Epicardial cells induced to differentiate into EPDCs. Tbx5 localization (E) was nuclear but also displayed a filamentous cytoplasmic distribution. Merged image (H) shows Tbx5 colocalization with the nucleus and actin filaments. (I–L) Epicardial cells stained for LMP4 (I), actin (J), and DAPI (K). Merged image (L) shows colocalization of LMP4 with actin. (M–P) Differentiated EPDCs displayed filamentous and cortical localization of LMP4 (M), consistent with the reorganization of actin into stress fibers (N). No significant localization of LMP4 was detected within the nucleus (P). LMP4 also displayed nonfilamentous cytoplasmic localization in undifferentiated and differentiated epicardial cells (I and M). (Q–T) Differentiated EPDCs stained with anti-Tbx5 (Q), anti–LMP4-rhodamine (R), and DAPI (S). Merged image (T) shows significant colocalization of Tbx5 and LMP4 in the cytoplasm along actin filaments. EPDC differentiation was confirmed by the detection of calponin (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200511109/DC1). Bars, 20 μm.
Our model suggests that Tbx5 cytoplasmic localization is due to direct interactions with LMP4, and our previous study showed that LMP4 localizes Tbx5 to actin in transfected COS-7 cells (Krause et al., 2004). To correlate the dynamic localization of Tbx5 to the LMP4 binding partner, we assayed for expression of LMP4 in the epicardial cells. As detected by indirect fluorescence (Fig. 2, I–P) and by Western blot of cell lysates (not depicted), using the LMP4-specific antiserum, chicken epicardial and EPDC cultures natively expressed LMP4. In the undifferentiated epicardial cells, LMP4 predominantly colocalized with cortical actin, outlining cell margins (Fig. 2, I–L). LMP4, as well as other PDZ-LIM protein family members, has been shown to associate with the actin cytoskeleton, supporting the localization in epicardial cells (Guy et al., 1999; Nakagawa et al., 2000; Krause et al., 2004; Torrado et al., 2004). Additional cytoplasmic localization was also observed in epicardial cultures for LMP4, along with some staining in or around the nucleus. When epicardial cells were shifted to differentiation medium, the cells began to express calponin (Fig. S2) and LMP4 remained predominantly localized to actin, now organized to stress fibers (Fig. 2, M–P). No distinctive nuclear localization could be detected in differentiating EPDCs. Thus, although Tbx5 displayed a dramatic localization shift, LMP4 remained cytoplasmic and associated with actin as the epicardial cells differentiated. Interestingly, the filamentous localization patterns for Tbx5 and LMP4 in the differentiating EPDCs were strikingly similar. Costaining EPDCs for both Tbx5 and LMP4 demonstrated that the two proteins colocalized along the actin cytoskeleton, supporting the notion that the proteins bind each other (Fig. 2, Q–T). The epicardial cell data suggest a potential new role for Tbx5 and/or LMP4 during EMT in the heart, and this possibility is currently under investigation (unpublished data). In addition, the shift of subcellular localization, concomitant with the change of culture conditions of the epicardial cells, suggests that Tbx5 localization may be regulated by external signals.
Individual expression of Tbx5 or LMP4 results in localization to separate cellular compartments
The cytoplasmic localization of Tbx5 in vivo and in cultured epicardial cells in the developing chicken wing supports our model of LMP4 regulating Tbx5 localization and activity. However, the epicardial cell cultures also demonstrate that Tbx5–LMP4 interactions are more complex than simply being expressed within the same cell. To further elucidate the mechanism of Tbx5–LMP4 interactions and its impact on Tbx5 activity, we used COS-7 cells. COS-7 cells provide a more amenable system than the primary chicken epicardial cells. The cells do not express either Tbx5 or LMP4 (unpublished data) and, therefore, allowed us to dissect the localization and function of each protein separately as well as in combination.
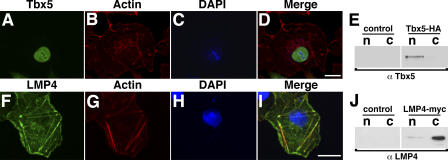
Our group, as well as others, has shown nuclear localization of Tbx5 in transfected cells (Collavoli et al., 2003; Krause et al., 2004; Zaragoza et al., 2004). However, many of these experiments used large fusion proteins such as EGFP for detection, which we have found to cause Tbx5 to function at suboptimal levels (see Fig. 6). To reduce the risk for functional interference, we have constructed nontagged and small C-terminal epitope–tagged Tbx5 expression plasmids. Nontagged Tbx5 or Tbx5-HA expression constructs were transfected into COS-7 cells, and protein localization was detected by indirect fluorescence using anti-HA or Tbx5-specific antibodies. Identical results were obtained with tagged or nontagged Tbx5; however, for consistency with other experiments, data for Tbx5-HA are shown (Fig. 3, A–D). Using confocal immunofluorescence detection, Tbx5-HA displayed a clear nuclear localization in COS-7 cells. This microscopic examination was further verified by Western blot analysis of separated cytoplasmic and nuclear fractions (Fig. 3 E). Empty vector controls displayed no specific localization by Western blot or immunofluorescence (Fig. 3 E and not depicted). For detection, we used the specific Tbx5 and anti-HA antibodies interchangeably with identical results (unpublished data).
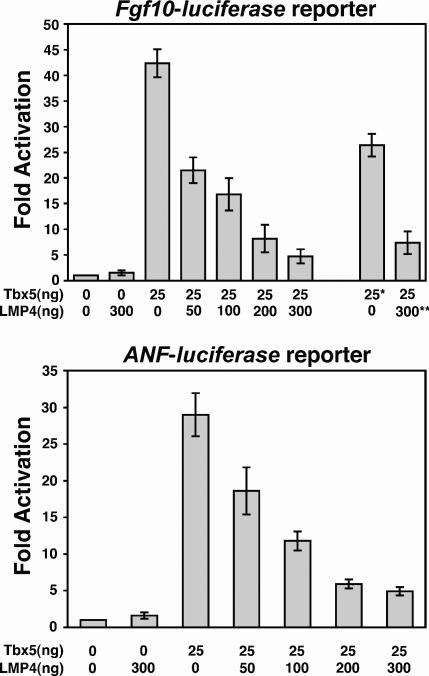
Figure 6.
LMP4 represses Tbx5 transcriptional activity on the Fgf10 and ANF target promoters. COS-7 cells were transfected with constant amounts of each respective reporter, Tbx5, and increasing amounts of LMP4 expression plasmid. Changes in luciferase reporter expression are indicated as fold activation. Asterisk indicates EGFP-Tbx5 fusion protein, revealing compromised activation of the Fgf10-luciferase reporter construct. Double asterisks indicate LMP4-EYFP fusion protein, revealing compromised repression of Tbx5 activity. Data shown are from two independent experiments performed in triplicate. Data are normalized to Renilla luciferase to control for differences in transfection efficiency.
Figure 3.
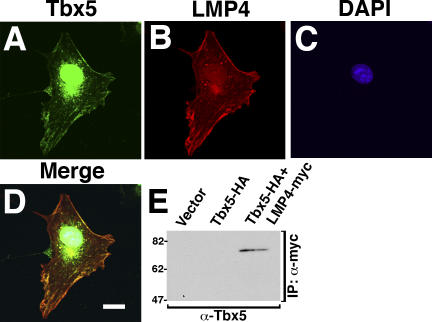
In single transfections, chicken Tbx5 and LMP4 localize to separate cellular compartments. (A–E) COS-7 cells transfected with Tbx5-HA and its expression detected using anti-Tbx5 antibodies (A). Cells were counterstained for actin using Alexa Fluor 633 phalloidin (B) and the nucleus using DAPI (C). The merged image (D) shows Tbx5 exclusively localized to the nucleus. Western blot of fractionated protein lysates from COS-7 cells transfected with Tbx5-HA, revealing exclusive nuclear localization of the Tbx5 protein (E). (F–J) COS-7 cells transfected with LMP4-myc and its expression detected using LMP4 antibodies (F). Cells were counterstained for actin (G) and the nucleus (H) as in B and C, respectively. The merged image (I) shows colocalization of LMP4 to actin stress fibers with no obvious nuclear localization. Western blot of fractionated protein lysates from COS-7 cells transfected with LMP4-myc, indicating localization of LMP4 to the cytoplasm (J). Empty vector transfections were used as controls for cellular fractionation. c, cytoplasmic fraction; n, nuclear fraction. Bars, 20 μm.
Previously, we showed that LMP4 localizes to cytoplasmic sites using an HcRed-LMP4 fusion expression construct (Krause et al., 2004). To verify our earlier findings, similar to Tbx5, we used expression plasmids containing LMP4 without a tag or with a small C-terminal myc-epitope tag for transfections. LMP4 or LMP4-myc transfected into COS-7 cells was detected by indirect fluorescence using the LMP4-specific antiserum. Fig. 3 (F–I) represents the data with LMP4-myc; however, identical results were obtained with nontagged LMP4 (not depicted). LMP4 protein displayed a filamentous distribution that overlapped with phalloidin-stained actin. This pattern is comparable to our previous data with HcRed-LMP4 and those obtained using an anti-myc antibody for detection (Krause et al., 2004). Cellular fractionation confirmed LMP4 cytoplasmic localization (Fig. 3 J). We note that we could not detect with either method conclusive nuclear localization for LMP4 in these single-transfection experiments. Empty vector controls displayed no specific localization by Western blot or immunofluorescence (Fig. 3 J and not depicted). Thus, using cell biology and biochemical methods, individually expressed Tbx5 and LMP4 localize to separate subcellular compartments in COS-7 cells—the nucleus and actin cytoskeleton, respectively.
Coexpression of Tbx5 and LMP4 leads to Tbx5–LMP4 interactions at cytoplasmic sites
We next cotransfected COS-7 cells with LMP4-myc and Tbx5-HA, and protein localization was determined by indirect fluorescence using anti-HA and anti-myc antibodies (Fig. 4, A–D). The anti-HA antibody detected Tbx5-HA within the nucleus but also at cytoplasmic structures (Fig. 4 A). LMP4-myc, as detected by anti-myc, produced a localization pattern comparable to single transfections, indicating association with actin filaments (Fig. 4 B). However, we also observed a higher level of nonfilamentous cytoplasmic staining for this protein. Comparing Tbx5 and LMP4 localization in the merged image revealed colocalization of Tbx5 and LMP4 within the cytoplasm (Fig. 4 D), along polymerized actin (phalloidin stain not depicted; Krause et al., 2004), and at additional unidentified cytoplasmic sites. We note that in transfected COS-7 cells, we were only able to detect cytoplasmic localized Tbx5 in the presence of LMP4.
Figure 4.
In cotransfected cells, chicken Tbx5 and LMP4 interact at cytoplasmic sites. (A–D) COS-7 cells cotransfected with Tbx5-HA and LMP4-myc. Cells were stained with anti-HA for Tbx5 (A), anti-myc for LMP4 (B), and the nuclear stain DAPI (C). The merged image (D) shows colocalization of Tbx5 and LMP4 outside the nucleus, predominantly along actin fibers. Coimmunoprecipitation of Tbx5-HA and LMP4-myc from COS-7 protein lysates (E). LMP4-myc was immunoprecipitated with myc antibodies, and the Western blot was processed with Tbx5-specific antibodies. Protein molecular mass markers in kD are indicated on the left of the Western blot. Bar, 20 μm.
The colocalization of Tbx5 and LMP4 within the cytoplasm supports our previous in vitro binding studies and suggests that the two proteins interact in cells (Krause et al., 2004). To confirm the interactions by independent means, we performed protein coimmunoprecipitations. Lysates of COS-7 cells cotransfected with LMP4-myc and Tbx5-HA were subjected to immunoprecipitation with anti-myc antibodies and Western blot analysis (Fig. 4 E). Probing the Western blot with the Tbx5-specific antibody demonstrated that the transcription factor coprecipitated with LMP4 (Fig. 4 E, third lane). The vector controls and individual Tbx5-HA transfections did not result in immunoprecipitates with the anti-myc antibody (Fig. 4 E, first and second lanes). Nuclear/cytoplasmic fractionation experiments of cells coexpressing Tbx5 and LMP4 did not reveal any evidence for nuclear localization of LMP4, despite the appearance of some weak staining in the nuclear area of cotransfected cells (Fig. 4 B). The fractionation experiments also did not detect Tbx5, LMP4, or the Tbx5–LMP4 complex within the soluble fraction, indicating that they are part of additional protein complexes (unpublished data). The coimmunoprecipitation of LMP4 and Tbx5 supports the cellular colocalization and confirms the binding of the two proteins in the cell. In reciprocal coimmunoprecipitation experiments, Tbx5-HA was also able to coprecipitate LMP4-myc (unpublished data). Thus, Tbx5 localization outside the nucleus is a result of its interaction with LMP4.
Actin destabilization does not change Tbx5–LMP4 binding or colocalization
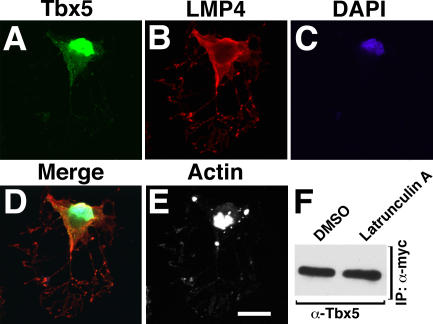
Because the Tbx5–LMP4 complex forms at actin filaments, it was important to investigate what role an intact actin cytoskeleton would have in mediating the interaction. To determine this, Tbx5 and LMP4 localization was observed in COS-7 cells with destabilized actin. 24 h after transfection of Tbx5-HA and LMP4-myc, cells were treated with 2 μM latrunculin A for 1 h to disrupt filamentous actin and then processed for confocal microscopy using indirect fluorescence (Fig. 5). Using the anti-HA antibody, Tbx5 was detected both in the nucleus and cytoplasm of actin-disrupted cells (Fig. 5 A). LMP4 was detected with the anti-myc antibody and found only in the cytoplasm of actin-disrupted cells (Fig. 5 B). As in nontreated cells, both Tbx5 and LMP4 appear to colocalize within the cytoplasm of latrunculin A–treated cells (Fig. 5 D); however, the proteins displayed no clear subcellular localization (Fig. 5 E). Despite the lack of specific localization, Tbx5 and LMP4 were still able to interact as demonstrated by imaging and coimmunoprecipitation (Fig. 5 F). Similar results were also obtained when actin was disrupted using 5 μM cytochalasin B for 1 h on cotransfected COS-7 cells (unpublished data). It remains to be determined whether an intact actin cytoskeleton is needed for initial Tbx5–LMP4 binding or whether it has a predominant role in the proper subcellular localization of the protein complex. However, it appears that the interaction of both proteins is maintained despite the lack of a complete actin cytoskeleton.
Figure 5.
Filamentous actin is required for cytoplasmic Tbx5–LMP4 complex localization. (A–E) COS-7 cells cotransfected with Tbx5-HA and LMP4-myc. 24 h after transfection, cells were treated with 2 μM latrunculin A for 60 min to sequester actin monomers. Cells were processed with anti-HA for Tbx5 (A), anti-myc for LMP4 (B), the nuclear stain DAPI (C), and Alexa Fluor 633 phalloidin to detect actin (E). The merged image (D) shows that the Tbx5–LMP4 complex no longer displays a filamentous pattern. For comparison, actin distribution of the cell in A–D is shown (E). (F) Coimmunoprecipitation of Tbx5 and LMP4 after actin disruption. Tbx5-HA was coprecipitated along with LMP4-myc in lysates from latrunculin A and DMSO control-treated COS-7 cells. Bar, 20 μm.
LMP4 represses Tbx5 transcriptional activity
Tbx5 has been shown to function as a transcription factor, activating target genes in the developing limb and heart (Bruneau et al., 2001; Hiroi et al., 2001). In the mouse, the limb-specific Fgf10 and the heart-specific atrial natriuretic factor (ANF) genes have been shown to be immediate downstream targets of Tbx5 (Bruneau et al., 2001; Agarwal et al., 2003). DNA fragments containing the respective promoters were ligated to the luciferase gene, and the resulting reporter constructs were used to determine Tbx5 transcriptional activity as a function of subcellular relocalization. COS-7 cells were transfected with constant amounts of the luciferase reporter and Tbx5 plasmids and increasing amounts of LMP4 expression plasmids. To achieve optimal protein activities in this assay, nontagged and small-tagged constructs were used. Tbx5 alone revealed a robust activation of both the Fgf10 and ANF reporters (Fig. 6), verifying that the chicken Tbx5 transcription factor can activate the respective mouse gene promoters at levels comparable to those of mouse Tbx5 (Bruneau et al., 2001; Agarwal et al., 2003). Cotransfecting LMP4 along with Tbx5, however, resulted in decreased Tbx5 activity on both of the reporters. The ability of LMP4 to repress Tbx5 transcriptional activity was dose dependent, as increasing amounts of LMP4 caused a linear reduction in Tbx5 activity (Fig. 6). The maximum amount of LMP4 tested (300 ng) led to a repression of Tbx5 (25 ng) of 90 and 83% using the Fgf10 and ANF promoters, respectively. Of note, our work with EGFP-Tbx5 and LMP4-EYFP revealed that such large fusion proteins have a significant reduction in activity. For example, transfection of COS-7 cells with 25 ng EGFP-Tbx5 resulted in an ∼30% reduction of transcriptional activity on the Fgf10 promoter as compared with an equivalent amount of Tbx5-HA or nontagged Tbx5 (Fig. 6). All luciferase reporter data were normalized to Renilla luciferase to account for variability in transfection efficiency and expression. Luciferase assays were performed in triplicate, and data were collected from two independent experiments. Thus, LMP4 modulates Tbx5 transcriptional activity by relocalizing the transcription factor out of the nucleus.
In the presence of LMP4, Tbx5 shuttles dynamically between the nucleus and cytoplasm
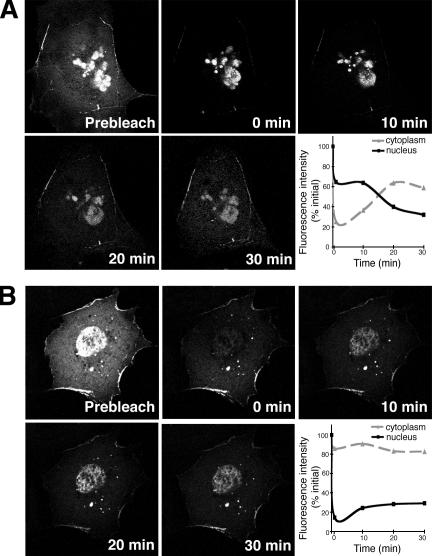
To determine if Tbx5 relocalization and transcriptional modulation by LMP4 was due to shuttling of the transcription factor out of the nucleus, FRAP experiments in COS-7 cells were performed. For this series of experiments, it was essential to use an EGFP-Tbx5 fusion construct to visualize protein in living cells. EGFP-Tbx5 revealed a reduced level of transcriptional activity compared with nontagged or small HA-tagged Tbx5 (Fig. 7); however, the fusion protein retained the ability to colocalize with LMP4 within the cytoplasm of cotransfected cells (not depicted; Krause et al., 2004). COS-7 cells were cotransfected with EGFP-Tbx5 and LMP4-myc or HcRed-LMP4 and grown on glass-bottomed culture dishes for live cell confocal microscopy. As expected, in the background of LMP4, EGFP-Tbx5 was detected in both nuclear and cytoplasmic compartments (Fig. 7). To observe shuttling of the transcription factor, the EGFP fluorescence signal in either the cytoplasm or nucleus was bleached using the 488-nm laser at maximum intensity, and its recovery within the bleached compartment was observed over time. After photobleaching the cytoplasm, the EGFP fluorescent signal showed significant recovery over a 30-min time window (Fig. 7 A and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200511109/DC1). It is important to note that the fluorescence recovery in the cytoplasm occurred with concomitant decrease of fluorescence in the nucleus, indicative of active shuttling of EGFP-Tbx5 out of the nucleus to cytoplasmic sites. Quantitative data analysis for a representative cell is shown in Fig. 7 A. Likewise, reciprocal experiments involving photobleaching the nucleus (Fig. 7 B and Video 2) displayed EGFP fluorescence recovery of the nuclear compartment at the expense of the cytoplasmic signal over a similar 30-min time frame. This result indicates movement of EGFP-Tbx5 from cytoplasmic sites into the nucleus. As a control, whole cell FRAP of cotransfected cells was performed. No EGFP recovery was observed in control cells within the same time period in this setup, indicating that the experimental fluorescence recovery is due to protein shuttling and not to translation of additional EGFP-Tbx5 or maturation of EGFP (Fig. S3). Therefore, in the presence of LMP4, Tbx5 subcellular localization is dynamic and the transcription factor can shuttle between the cytoplasm and nucleus.
Figure 7.
Dynamic shuttling of Tbx5 between nuclear and cytoplasmic compartments. COS-7 cells cotransfected with EGFP-Tbx5 and LMP4. (A) Photobleaching of cytoplasmic EGFP-Tbx5. Graph displays quantitative fluorescent data for a representative cell. (B) Photobleaching of nuclear EGFP-Tbx5. Graph displays representative quantitative fluorescent data. Bleaching of whole cell displayed no fluorescence recovery within the displayed time frame (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200511109/DC1). All FRAP assays were performed with a minimum of three cells.
Discussion
Tbx5 subcellular localization is dynamic and related to cell differentiation
We previously hypothesized that Tbx5 transcriptional activity may be modulated by dynamic interactions between Tbx5 and LMP4 proteins and localization of the complex to the actin cytoskeleton (Krause et al., 2004). Here, we provide evidence in support of the Tbx5–LMP4 regulatory model. Individually, Tbx5 and LMP4 localize to separate cellular compartments— the nucleus and cytoplasm, respectively. However, when expressed within the same cell, Tbx5 is no longer strictly nuclear. The Tbx5 and LMP4 proteins bind and colocalize in the cytoplasm, predominantly in association with actin. The change in Tbx5 localization caused by LMP4 represses its ability to activate target promoters, as shown by in vitro luciferase reporter assays. The regulatory model would also imply a dynamic Tbx5–LMP4 complex assembly/disassembly, responding to external stimuli or signal transduction pathways and ultimately modulating Tbx5 protein activity. This notion is supported by relocalization of Tbx5 in differentiating epicardial cells and by dynamic shuttling of Tbx5 between the cytoplasmic and nuclear compartments in transfected COS-7 cells, as observed by FRAP. In addition, the coexpression of both Tbx5 and LMP4 proteins in developing chicken wings and, importantly, the localization of native Tbx5 outside of the nucleus in these limb cells indicate in vivo relevance. In chicken epicardial cells, we observed coexpression of Tbx5 and LMP4; however, in contrast to COS-7 cells, Tbx5 localization was predominantly nuclear in these cells. Cytoplasmic and actin-associated Tbx5 was not observed until cells were induced to differentiate. Cultured epicardial cells are well known to undergo such EMTs when stimulated with embryonic heart–conditioned medium or TGFs such as TGFβ (Dettman et al., 1998; Lu et al., 2001; Morabito et al., 2001; Compton et al., 2006). For the first time, we have demonstrated a concomitant relocalization of Tbx5 proteins from the nucleus to the cytoplasm during this process, strongly suggesting that specific signaling pathways, potentially involving TGFβ-like factors, are involved in regulating Tbx5–LMP4 interactions and Tbx5 activity. The data presented are in agreement with the Tbx5–LMP4 regulatory model and point toward a complex pathway regulating Tbx5 activity by altering its localization depending on the developmental context of the cell.
An emerging role for LMP4 as a signal mediator in Tbx5 regulation
The nuclear concentration of many transcription factors is a dynamic balance that is determined by competing processes of nuclear import and export and by the presence of anchor proteins in both the nucleus and the cytoplasm. For example, NF-κB/Rel proteins, which are involved in diverse biological processes, were initially identified as constitutive nuclear transcription factors. Subsequent analysis, however, revealed that in most cells, NF-κB is sequestered in the cytoplasm via its interaction with IκB family proteins and only released into the nucleus in response to specific stimuli (Karin and Ben-Neriah, 2000). Additionally, the GLI-1 transcription factor has been shown to be not strictly nuclear but also cytoplasmic (Dahmane et al., 1997; Ruiz i Altaba, 1999). GLI-1 cytoplasmic localization has been shown to be due to interactions with Suppressor-of-Fused, and the export of GLI-1 from the nucleus regulates its transcriptional activity (Kogerman et al., 1999). A similar novel mechanism may be emerging with LMP4 and Tbx5. When complexed with LMP4, a pool of Tbx5 is localized outside the nucleus in association with the actin cytoskeleton, thereby limiting the transcription factor's availability and activity in the nucleus. However, the specific signaling cascades that regulate the expression, localization, and function of Tbx5 have yet to be identified. Based on our initial studies with primary chicken epicardial cultures, it appears that specific stimuli are involved in the relocalization of Tbx5 during differentiation. Recently, TGFβ has been shown to induce differentiation of chicken epicardial cells into EPDCs (Compton et al., 2006). It will be of interest to determine whether TGFβ in concert with LMP4 is required for Tbx5 relocalization in differentiating epicardial cells or to identify the nature of other specific upstream signals.
In addition to external stimuli or signaling pathways that may modulate Tbx5 protein activity, the mechanism by which Tbx5 is shuttled out of the nucleus into the cytoplasm for interaction with LMP4 is not yet understood. One hypothesis is that a small amount of LMP4 is at least temporarily present in the nucleus and, in response to a given stimulus/signal, acts as a shuttling vector for Tbx5. Alternatively, it is possible that an as-yet-unidentified transport protein shuttles Tbx5 out of the nucleus, where it is then able to interact with LMP4. Finally, Tbx5 itself may be using an intrinsic shuttling signal to translocate to the cytoplasm, where LMP4 is waiting to localize it to actin sites. These options can be experimentally tested, and studies are under way to investigate which cellular mechanism is responsible for altering Tbx5 subcellular localization.
In this context, it is noteworthy that PDZ-LIM proteins are thought to have diverse roles as regulators of cytoarchitecture, cell motility, signal transduction, and gene expression (Bach, 2000; Kadrmas and Beckerle, 2004). Several family member proteins such as Enigma, CLP-36, and the Cypher/ZASP proteins interact via their PDZ domains with the cytoskeleton (Guy et al., 1999; Zhou et al., 1999; Vallenius et al., 2000). Consistent with these findings, it is not surprising that in our studies, LMP4 is also colocalizing with the actin cytoskeleton. Of note, CLP-36's C-terminal LIM domain binds to and relocalizes the nuclear Clik1 kinase to actin stress fibers (Vallenius and Makela, 2002). LIM domains in general are known to mediate protein interactions, and the close LMP4 family member Enigma binds to the insulin receptor (Wu and Gill, 1994), receptor tyrosine kinases (Wu et al., 1996), and PKC (Kuroda et al., 1996). Although the exact roles of PDZ-LIM proteins such as Enigma in signaling cascades are currently speculative, the association with signal receptors and/or transducers provides an attractive link and points to an involvement in regulated signaling events, eliciting a change in binding partners. The presence of LMP4 in epicardial cultures, which display a differentiation response to external signals along with a significant relocalization of Tbx5, also suggests involvement of this PDZ-LIM protein in a signaling cascade.
Nuclear versus cytoplasmic Tbx5 localization and its relation to development and disease
A cytoplasmic distribution for TBX5 in human lung during development has been indicated (Collavoli et al., 2003). Our observations with developing wings, primary epicardial cells, and transfected cells reveal a previously unidentified localization of Tbx5 in both the nucleus and the cytoplasm. The actin-associated distribution of the Tbx5 transcription factor is particularly striking in the primary chicken EPDCs and would suggest that an equilibrium of Tbx5 in the nucleus and cytoplasm is important for the proper maintenance of its functions within the cell and, in turn, the organism. Although the system may compensate for some changes in protein levels, acting as a capacitor, significant over- or underexpression would be expected to result in deleterious consequences. Few reports are available on Tbx5 gain-of-function/overexpression phenotypes in higher vertebrates, but those available support our findings. For instance, retroviral overexpression of Tbx5/4 in the respective chicken wing/leg bud resulted in limb truncations, similar to misexpression of the respective dominant-negative constructs (Rodriguez-Esteban et al., 1999). In addition, skeletal and cardiac malformations known as Holt-Oram syndrome (HOS) in humans are caused by mutations in TBX5. The majority of TBX5 mutations critical for disease manifestation are thought to result in early protein terminations and haploinsufficiency; however, increased TBX5 dosage, such as chromosome 12q2 duplication, has been reported to also result in HOS (Vaughan and Basson, 2000; Hatcher and Basson, 2001). These data would suggest a more generalized effect of Tbx5 gene dosage, both under- and overexpression, in causing fairly similar, if not identical, phenotypes. The data presented here would also imply that a change in Tbx5 level would have direct consequences on Tbx protein distribution in the cell. This in turn may interfere with the differentiation program such as EMT of epicardial cells. In this context, it may be of significance that epicardial cells contribute to the myocardial wall, atrioventricular cushions, and valves, all cardiac structures that are predominantly affected in HOS. Therefore, balanced cellular Tbx5 levels and appropriate localization appear to be critical, and LMP4 may play a central role in this regulation.
Experimental data coming from many animal models have provided clear evidence for the importance of Tbx5 in eye, limb, and heart development, and gene and RNA studies have provided some clues for the roles Tbx5 has in the cell. However, in addition to its role as a transcription factor, our new data may also point to unknown functions of Tbx5 outside the nucleus when associated with actin. An attractive possibility would be a direct role in regulating actin dynamics, a notion supported by the finding that Tbx5 function is involved in cell migration in zebrafish fin development (Ahn et al., 2002). A role in migratory behavior would be quite plausible also in light of the complex phenotypes of Tbx5 misexpression that have been observed in humans and animal models. This hypothesis can be tested, and future experiments mislocalizing Tbx5 to distinct cellular compartments and examining the resulting functional consequences will provide new insights into this question.
Materials and methods
Cell culture, transfection, and reporter assays
COS-7 cells were grown in DME supplemented with 10% FBS, 1% l-glutamine, and penicillin/streptomycin. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For reporter assays, COS-7 cells were grown in 12-well culture dishes and transfected with 300 ng reporter plasmid, 10 ng Renilla luciferase plasmid, and expression plasmids as described. The total amount of DNA transfected was held constant at 1 μg. Transfected cells were cultured for 36 h before lysis. Luciferase activity was measured using the Dual-Luciferase assay system (Promega), and samples were read on a Lumat LB 9501 (Berthold). All reporter assays were performed in triplicate, and the collected data from two independent experiments were normalized to the Renilla luciferase activity. Tbx5-HA expression plasmids were provided by M. Logan (Medical Research Council, London, UK), and luciferase reporter constructs were provided by B. Bruneau (Hospital for Sick Kids, Toronto, Canada).
To obtain chicken epicardial cell cultures, HH stage 25 (Hamburger and Hamilton, 1951) hearts were dissected and placed on fibronectin-coated coverslips in MEM without l-glutamine (Dettman et al., 1998). After 24 h, the hearts were removed and the cells were either fixed for immunocytochemistry or induced to differentiate into EPDCs. Differentiation was induced by culturing cells with whole heart–conditioned media (Morabito et al., 2001), and after 3 d, cells were fixed and processed for immunocytochemistry.
Immunofluorescence and imaging
COS-7 cells were fixed in 4% PFA followed by 1% Triton X-100 extraction and sequential incubation with primary and secondary antibodies in 1% BSA. Affinity-purified rabbit polyclonal anti-LMP4 (Fig. S1) and anti-Tbx5 (Khan et al., 2002) were used at a 1:500 dilution. Anti-HA (HA-7; Sigma-Aldrich), anti-myc (9E10; Sigma-Aldrich), and anti-calponin (CP-93; Sigma-Aldrich) were diluted 1:500. Primary antibodies were detected using Alexa 488– and Alexa 546–conjugated secondary antibodies at 1:500 dilutions (Invitrogen). Filamentous actin was detected using Alexa Fluor 488 or 633 phalloidin (Invitrogen). Nuclei were stained using DAPI (Roche). For double-staining experiments, LMP4 antibodies were directly coupled to rhodamine using the EZ-Label protein labeling kit (Pierce Biotechnology). Confocal microscopy was performed using a 510 META system (Carl Zeiss MicroImaging, Inc.) equipped with a Plan Apochromat 63×/1.4 oil differential interference contrast lens. Images were processed in Photoshop CS2 (Adobe).
Limb sectioning
Chicken wings from HH stage 36 were dissected in cold PBS, embedded in Tissue Tek OCT (Sakura Finetek), and frozen over dry ice. 10-μm sections were cut on a cryostat (CM3050S; Leica), fixed in 4% PFA, and processed for immunohistochemistry as described.
FRAP
Cells were transfected with EGFP-Tbx5 and either LMP4-myc or HcRed-LMP4. Cells were grown on uncoated glass-bottomed 35-mm culture dishes (No. 1.0; MatTek) containing DME/10% FBS and equilibrated on a 37°C heated stage fitted on a laser-scanning microscope (LSM 510; Carl Zeiss MicroImaging, Inc.). EGFP photobleaching was performed using the 488-nm laser line at 100% intensity. Changes in pixel intensity were analyzed using OpenLab 4.0 (Improvision). Whole cell bleaching of a cotransfected cell was performed to determine the onset of EGFP protein synthesis and maturation.
Coimmunoprecipitation and cellular fractionation
COS-7 cells were grown to 80–90% confluency in 10-cm culture dishes and transfected with the described plasmids. A modification of the subcellular fractionation protocol from Sun et al. (2003) was used. 24 h after transfection, the cells were rinsed with cold PBS, trypsinized, and pelleted at 1,500 rpm for 15 min at 4°C. The cells were lysed in homogenization buffer (10 mM Tris, pH 7.4, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.1 mM EGTA, 0.5% Nonidet-P40, and 5% sucrose) containing protease inhibitors (P8340; Sigma-Aldrich) for 10 min on ice, followed by further disruption with 15 strokes in a tightly fitting Dounce homogenizer. The homogenate was centrifuged at 6,000 rpm for 1 min at 4°C to pellet the nuclei. The supernatant was further centrifuged at 10,000 rpm for 10 min at 4°C, and this supernatant was saved as the cytosolic fraction. The nuclear pellet was passed through 5 ml sucrose buffer (10 mM Tris, pH 7.4, 15 mM NaCl, 60 mM KCl, and 10% sucrose) at 3,000 rpm for 5 min at 4°C, washed three times with wash buffer (10 mM Tris, pH 7.4, 15 mM NaCl, and 60 mM KCl), and resuspended in homogenization buffer that had the NaCl adjusted to 0.5 M. After incubation for 30 min at 4°C with rocking to extract the nuclear proteins, the extract was centrifuged at 10,000 rpm for 10 min at 4°C and the supernatant was saved as the nuclear fraction. Protein concentrations were determined for each of the fractions by BCA assay (Pierce Biotechnology) for subsequent SDS-PAGE (Laemmli, 1970) and immunoblot analysis (Towbin et al., 1979) with the indicated antibodies.
For coimmunoprecipitation, COS-7 cells were grown to 80–90% confluency in 10-cm culture dishes and transfected with 10 μg Tbx5-HA and 14 μg LMP4-myc. After 24 h, cells were lysed in lysis buffer (25 mM Tris-HCl, 100 mM NaF, 10 mM EGTA, 5 mM EDTA, 250 mM NaCl, 1% NP-40, 50 mM Na4P2O7·H2O, 0.5% DOC, and 10 mM ATP) containing protease inhibitors. Lysates were incubated on ice for 20 min followed by centrifugation at 52,000 rpm for 10 min. The supernatant was incubated with anti-myc–conjugated protein A–Sepharose beads (GE Healthcare) overnight at 4°C. The Sepharose beads were washed in lysis buffer, and the bound protein was eluted with SDS buffer, boiled, and analyzed by immunoblotting with the indicated antibodies.
Actin disruption
COS-7 cells were treated with 2 μM latrunculin A (Sigma-Aldrich) or 5 μM cytochalasin D (Sigma-Aldrich) for 60 min at 37°C. Parallel cultures were treated with the vehicle DMSO as a control. After treatment, the cells were immediately prepared for cell imaging or biochemical analysis.
Expression constructs
Full-length chicken Tbx5 was cloned into a pcDNA3.1 expression vector containing a HA tetramer tag. Tbx5 was additionally placed as an N-terminal fusion into a modified pEGFP-C1 vector suitable for the Gateway recombination system (Invitrogen). Full-length chicken LMP4 was cloned into pcDNA3.1 containing a myc C-terminal tag. Chicken LMP4 was also recombined as an N-terminal fusion into a modified HcRed-C1 expression vector suitable for the Gateway recombination system. Mouse ENH1 (available from GenBank/EMBL/DDBJ under accession no. DQ177283) fragments were cloned from mouse brain cDNA into the pGEX-6P-2 prokaryotic expression vector to create an N-terminal GST fusion protein (GE Healthcare). The PDZ/proline-rich fragment covers amino acids 1–414 and was amplified using forward primer 5′-ACGCGTCGACCATGAGCAACTACAGTGTGTCATTG-3′ and reverse primer 5′-ATAGTTTAGCGGCCGCTCACATGGGGGTCCGCTTGCCCG-3′. The ENH1 LIM 1/2/3 fragment covers amino acids 412–593 and was amplified using forward primer 5′-ACGCGTCGACCATGTGTGCCCACTGCAACCA-3′ and reverse primer 5′-ATAGTTTAGCGGCCGCTGATTTTCAAAAATTCACAGAATGAG-3′. Recombinant mouse ENH1 peptides were expressed in BL21 Escherichia coli as described previously (Krause et al., 2004).
LMP4 antibody design
To identify a region in chicken LMP4 suitable for specific antibody production, a multiprotein sequence alignment (MacVector 7.0; Accelrys) was conducted to compare two closely related subclasses of PDZ-LIM proteins: LMP and ENH proteins. LMP protein sequences from human LMP1 (Wu and Gill, 1994), rat LMP1 (Boden et al., 1998), and chicken LMP4 (Krause et al., 2004) were compared with ENH sequences from human ENH1 (Ueki et al., 1999), mouse ENH1 (Nakagawa et al., 2000), and rat ENH1 (Kuroda et al., 1996). From this alignment, a 17-amino-acid peptide (DPAFAERYAPDKTSTVL) was identified that was conserved in LMP proteins but not in ENH proteins. In addition, based on its predicted antigenicity and hydrophobicity, the peptide was suitable to elicit a good immune response. Peptide synthesis and rabbit immunization was performed by Invitrogen custom antibody services. The final rabbit antiserum was affinity purified on antigen peptide–conjugated columns and tested for specificity (Fig. S1 and supplemental text).
Online supplemental material
Fig. S1 and the accompanying supplemental text describe the production and testing of the LMP4-specific antisera. Fig. S2 shows the calponin control staining in epicardial cells for Fig. 2. Fig. S3 describes the negative control whole cell FRAP. Representative time-lapse videos of cytoplasmic FRAP (Fig. 7 A) and nuclear FRAP (Fig. 7 B) are provided online in Videos 1 and 2, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200511109/DC1.
Supplementary Material
Acknowledgments
We are grateful to Drs. B. Dettman and J. Topczewski for critical reading of the manuscript and to members of the Developmental Biology Core for stimulating discussions.
This work is supported by an American Heart Association Predoctoral Fellowship (to T. Camarata) and National Institutes of Health grant HL085834-01 (to H.-G. Simon).
Abbreviations used in this paper: ANF, atrial natriuretic factor; ENH, Enigma homologue; EPDC, epicardially derived cell; HH, Hamburger-Hamilton; HOS, Holt-Oram syndrome.
References
- Agarwal, P., J.N. Wylie, J. Galceran, O. Arkhitko, C. Li, C. Deng, R. Grosschedl, and B.G. Bruneau. 2003. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 130:623–633. [DOI] [PubMed] [Google Scholar]
- Ahn, D., M.J. Kourakis, L.A. Rohde, L.M. Silver, and R.K. Ho. 2002. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 417:754–758. [DOI] [PubMed] [Google Scholar]
- Bach, I. 2000. The LIM domain: regulation by association. Mech. Dev. 91:5–17. [DOI] [PubMed] [Google Scholar]
- Boden, S.D., Y. Liu, G.A. Hair, J.A. Helms, D. Hu, M. Racine, M.S. Nanes, and L. Titus. 1998. LMP-1, a LIM-domain protein, mediates BMP-6 effects on bone formation. Endocrinology. 139:5125–5134. [DOI] [PubMed] [Google Scholar]
- Bruneau, B.G., M. Logan, N. Davis, T. Levi, C.J. Tabin, J.G. Seidman, and C.E. Seidman. 1999. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 211:100–108. [DOI] [PubMed] [Google Scholar]
- Bruneau, B.G., G. Nemer, J.P. Schmitt, F. Charron, L. Robitaille, S. Caron, D.A. Conner, M. Gessler, M. Nemer, C.E. Seidman, and J.G. Seidman. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 106:709–721. [DOI] [PubMed] [Google Scholar]
- Collavoli, A., C.J. Hatcher, J. He, D. Okin, R. Deo, and C.T. Basson. 2003. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J. Mol. Cell. Cardiol. 35:1191–1195. [DOI] [PubMed] [Google Scholar]
- Compton, L.A., D.A. Potash, N.A. Mundell, and J.V. Barnett. 2006. Transforming growth factor-beta induces loss of epithelial character and smooth muscle cell differentiation in epicardial cells. Dev. Dyn. 235:82–93. [DOI] [PubMed] [Google Scholar]
- Dahmane, N., J. Lee, P. Robins, P. Heller, and A. Ruiz i Altaba. 1997. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 389:876–881. [DOI] [PubMed] [Google Scholar]
- Dawid, I.B., J.J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14:156–162. [DOI] [PubMed] [Google Scholar]
- Dettman, R.W., W. Denetclaw Jr., C.P. Ordahl, and J. Bristow. 1998. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev. Biol. 193:169–181. [DOI] [PubMed] [Google Scholar]
- Fanning, A.S., and J.M. Anderson. 1999. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J. Clin. Invest. 103:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity, D.M., S. Childs, and M.C. Fishman. 2002. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 129:4635–4645. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot, A.C., M.P. Vrancken Peeters, M.M. Mentink, R.G. Gourdie, and R.E. Poelmann. 1998. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ. Res. 82:1043–1052. [DOI] [PubMed] [Google Scholar]
- Guy, P.M., D.A. Kenny, and G.N. Gill. 1999. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol. Biol. Cell. 10:1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger, V., and H.L. Hamilton. 1951. A series of normal stages in the development of the chick embryo. J. Morphol. 88:49–92. [PubMed] [Google Scholar]
- Harris, B.Z., and W.A. Lim. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114:3219–3231. [DOI] [PubMed] [Google Scholar]
- Hatcher, C.J., and C.T. Basson. 2001. Getting the T-box dose right. Nat. Med. 7:1185–1186. [DOI] [PubMed] [Google Scholar]
- Hatcher, C.J., N.Y. Diman, M.S. Kim, D. Pennisi, Y. Song, M.M. Goldstein, T. Mikawa, and C.T. Basson. 2004. A role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol. Genomics. 18:129–140. [DOI] [PubMed] [Google Scholar]
- Herrmann, B.G., S. Labeit, A. Poustka, T.R. King, and H. Lehrach. 1990. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 343:617–622. [DOI] [PubMed] [Google Scholar]
- Hiroi, Y., S. Kudoh, K. Monzen, Y. Ikeda, Y. Yazaki, R. Nagai, and I. Komuro. 2001. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 28:276–280. [DOI] [PubMed] [Google Scholar]
- Kadrmas, J.L., and M.C. Beckerle. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5:920–931. [DOI] [PubMed] [Google Scholar]
- Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621–663. [DOI] [PubMed] [Google Scholar]
- Khan, P., B. Linkhart, and H.G. Simon. 2002. Different regulation of T-box genes Tbx4 and Tbx5 during limb development and limb regeneration. Dev. Biol. 250:383–392. [PubMed] [Google Scholar]
- Kispert, A., and B.G. Herrmann. 1993. The Brachyury gene encodes a novel DNA binding protein. EMBO J. 12:4898–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogerman, P., T. Grimm, L. Kogerman, D. Krause, A.B. Unden, B. Sandstedt, R. Toftgard, P.G. Zaphiropouls. 1999. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1:312–319. [DOI] [PubMed] [Google Scholar]
- Krause, A., W. Zacharias, T. Camarata, B. Linkhart, E. Law, A. Lischke, E. Miljan, and H.G. Simon. 2004. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev. Biol. 273:106–120. [DOI] [PubMed] [Google Scholar]
- Kuroda, S., C. Tokunaga, Y. Kiyohara, O. Higuchi, H. Konishi, K. Mizuno, G.N. Gill, and U. Kikkawa. 1996. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J. Biol. Chem. 271:31029–31032. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Logan, M., and C. Tabin. 1998. Targeted gene misexpression in chick limb buds using avian replication-competent retroviruses. Methods. 14:407–420. [DOI] [PubMed] [Google Scholar]
- Logan, M., H.G. Simon, and C. Tabin. 1998. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development. 125:2825–2835. [DOI] [PubMed] [Google Scholar]
- Lu, J., T.E. Landerholm, J.S. Wei, X.R. Dong, S.P. Wu, X. Liu, K. Nagata, M. Inagaki, and M.W. Majesky. 2001. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev. Biol. 240:404–418. [DOI] [PubMed] [Google Scholar]
- Mikawa, T., and R.G. Gourdie. 1996. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev. Biol. 174:221–232. [DOI] [PubMed] [Google Scholar]
- Morabito, C.J., R.W. Dettman, J. Kattan, J.M. Collier, and J. Bristow. 2001. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev. Biol. 234:204–215. [DOI] [PubMed] [Google Scholar]
- Naiche, L.A., and V.E. Papaioannou. 2003. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 130:2681–2693. [DOI] [PubMed] [Google Scholar]
- Nakagawa, N., M. Hoshijima, M. Oyasu, N. Saito, K. Tanizawa, and S. Kuroda. 2000. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem. Biophys. Res. Commun. 272:505–512. [DOI] [PubMed] [Google Scholar]
- Rallis, C., B.G. Bruneau, J.D. Buono, C.E. Seidman, J.G. Seidman, S. Nissim, C.J. Tabin, and M.P.O. Logan. 2003. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 130:2741–2751. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Esteban, C., T. Tsukui, S. Yonei, J. Magallon, K. Tamura, and J.C. Izpisua-Belmonte. 1999. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature. 398:814–818. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba, A. 1999. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 126:3205–3216. [DOI] [PubMed] [Google Scholar]
- Sun, H., X. Tu, M. Prisco, A. Wu, I. Casiburi, and R. Baserga. 2003. Insulin-like growth factor I receptor signaling and nuclear translocation of insulin receptor substrates 1 and 2. Mol. Endocrinol. 17:472–486. [DOI] [PubMed] [Google Scholar]
- Takeuchi, J.K., K. Koshiba-Takeuchi, K. Matsumoto, A. Vogel-Hopker, M. Naitoh-Matsuo, K. Ogura, N. Takahashi, K. Yasuda, and T. Ogura. 1999. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature. 398:810–814. [DOI] [PubMed] [Google Scholar]
- Torrado, M., V.V. Senatorov, R. Trivedi, R.N. Fariss, and S.I. Tomarev. 2004. Pdlim2, a novel PDZ-LIM domain protein, interacts with alpha-actinins and filamin A. Invest. Ophthalmol. Vis. Sci. 45:3955–3963. [DOI] [PubMed] [Google Scholar]
- Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki, N., N. Seki, K. Yano, Y. Masuho, T. Saito, and M. Muramatsu. 1999. Isolation, tissue expression, and chromosomal assignment of a human LIM protein gene, showing homology to rat enigma homologue (ENH). J. Hum. Genet. 44:256–260. [DOI] [PubMed] [Google Scholar]
- Vallenius, T., and T.P. Makela. 2002. Clik1: a novel kinase targeted to actin stress fibers by the CLP-36 PDZ-LIM protein. J. Cell Sci. 115:2067–2073. [DOI] [PubMed] [Google Scholar]
- Vallenius, T., K. Luukko, and T.P. Makela. 2000. CLP-36 PDZ-LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. J. Biol. Chem. 275:11100–11105. [DOI] [PubMed] [Google Scholar]
- Vaughan, C.J., and C.T. Basson. 2000. Molecular determinants of atrial and ventricular septal defects and patent ductus arteriosus. Am. J. Med. Genet. 97:304–309. [DOI] [PubMed] [Google Scholar]
- Wu, R.Y., and G.N. Gill. 1994. LIM domain recognition of a tyrosine-containing tight turn. J. Biol. Chem. 269:25085–25090. [PubMed] [Google Scholar]
- Wu, R., K. Durick, Z. Songyang, L.C. Cantley, S.S. Taylor, and G.N. Gill. 1996. Specificity of LIM domain interactions with receptor tyrosine kinases. J. Biol. Chem. 271:15934–15941. [DOI] [PubMed] [Google Scholar]
- Zaragoza, M.V., L.E. Lewis, G. Sun, E. Wang, L. Li, I. Said-Salman, L. Feucht, and T. Huang. 2004. Identification of the TBX5 transactivating domain and the nuclear localization signal. Gene. 330:9–18. [DOI] [PubMed] [Google Scholar]
- Zhou, Q., P. Ruiz-Lozano, M.E. Martone, and J. Chen. 1999. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 274:19807–19813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.