Abstract
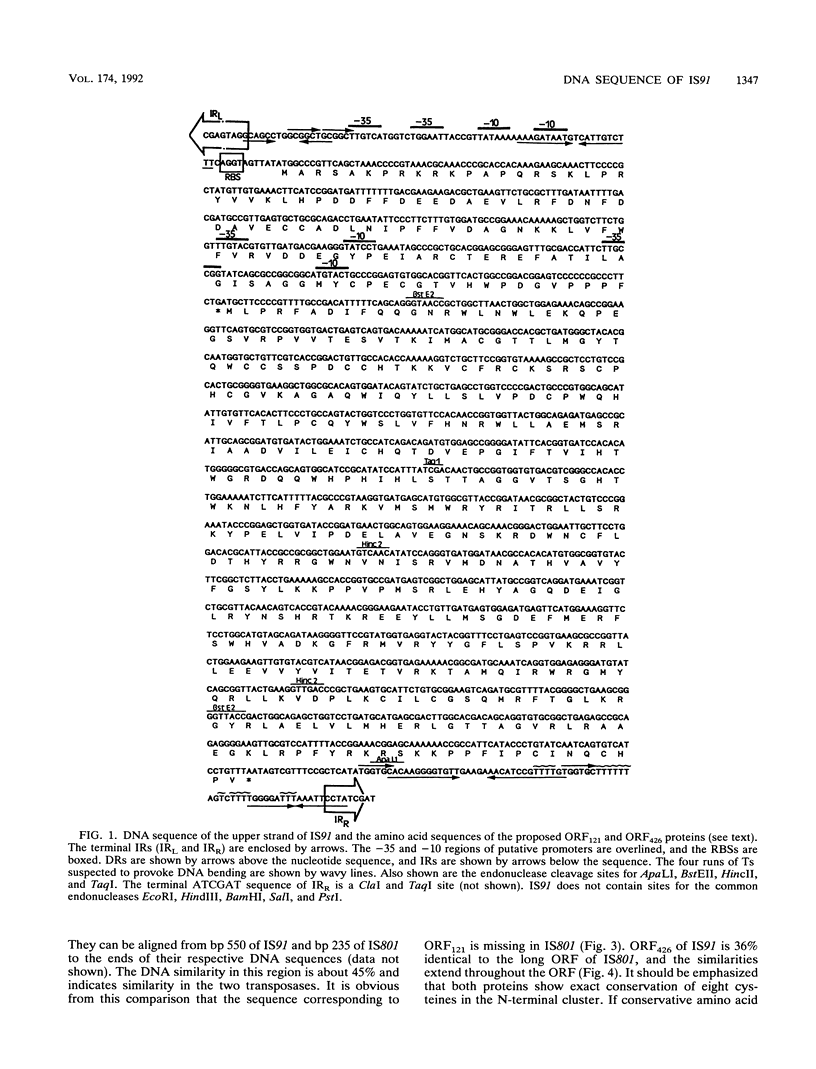
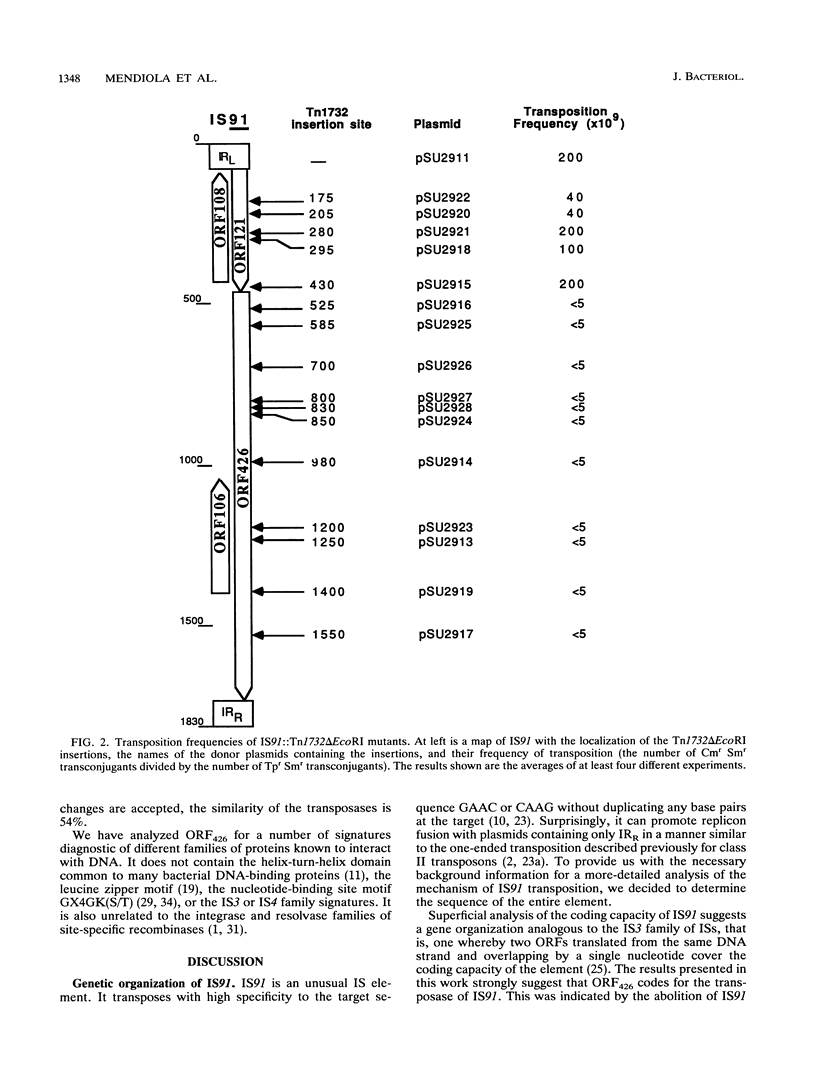
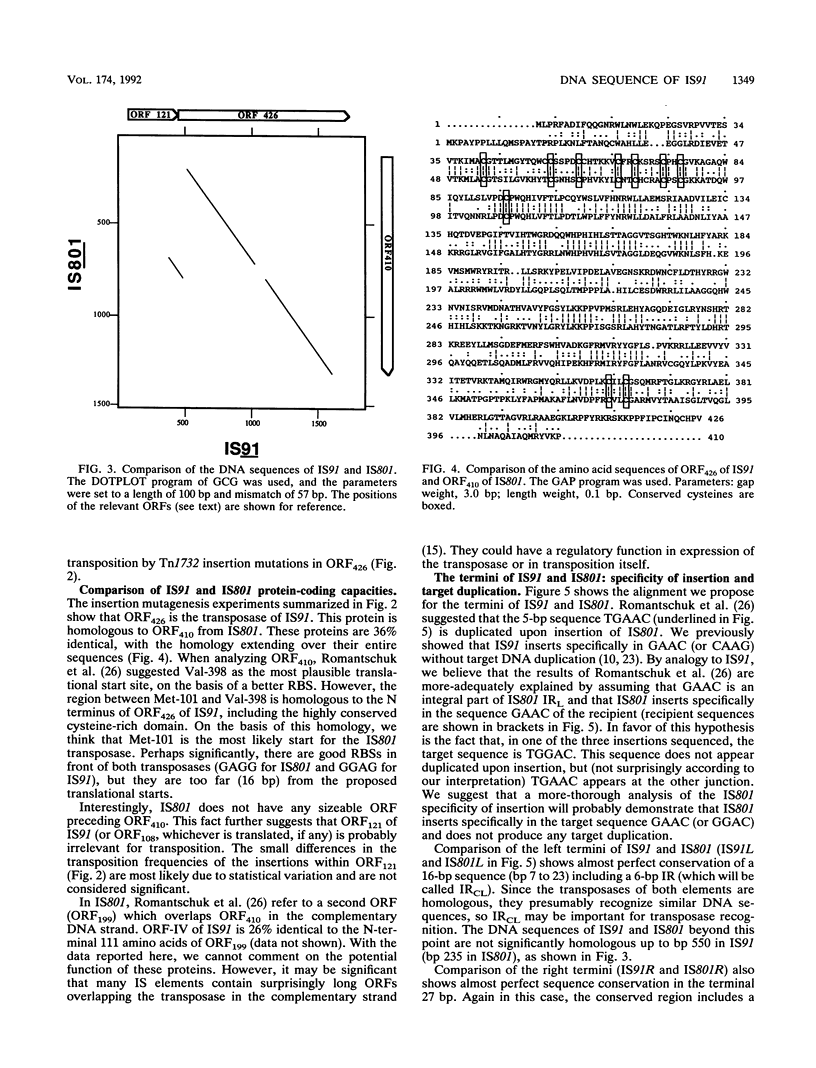
IS91 is a 1,830-bp insertion sequence that inserts specifically at the sequence CAAG or GAAC of the target and does not duplicate any sequence upon insertion (23). By transposon mutagenesis, we have identified open reading frame 426 (ORF426; bp 454 to 1731) as the putative ORF for the transposase. It displays a cysteine-rich, potential metal-binding domain in its N-terminal region. Adjacent to ORF426, there is an ORF (ORF121) which precedes and terminally overlaps ORF426 by one amino acid. Tn1732 insertions in ORF121 do not affect the transposition frequency. IS91 has sequence similarities to IS801 from Pseudomonas syringae. Their putative transposases are 36% identical, including conservation of the cysteine-rich cluster. The information concerning IS801 insertion specificity and target duplication has been reevaluated in the light of our results.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila P., de la Cruz F., Ward E., Grinsted J. Plasmids containing one inverted repeat of Tn21 can fuse with other plasmids in the presence of Tn21 transposase. Mol Gen Genet. 1984;195(1-2):288–293. doi: 10.1007/BF00332761. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Aroca E., Mendiola M. V., Zabala J. C., de la Cruz F. Transposition of IS91 does not generate a target duplication. J Bacteriol. 1987 Jan;169(1):442–443. doi: 10.1128/jb.169.1.442-443.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Aroca E., de la Cruz F., Zabala J. C., Ortiz J. M. Characterization of the new insertion sequence IS91 from an alpha-hemolysin plasmid of Escherichia coli. Mol Gen Genet. 1984;193(3):493–499. doi: 10.1007/BF00382089. [DOI] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Fayet O., Ramond P., Polard P., Prère M. F., Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990 Oct;4(10):1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Hanson I. M., Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991 Feb 8;64(3):483–484. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Korneluk R. G., Quan F., Gravel R. A. Rapid and reliable dideoxy sequencing of double-stranded DNA. Gene. 1985;40(2-3):317–323. doi: 10.1016/0378-1119(85)90055-1. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Martinez E., Bartolomé B., de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988 Aug 15;68(1):159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- Mendiola M. V., de la Cruz F. Specificity of insertion of IS91, an insertion sequence present in alpha-haemolysin plasmids of Escherichia coli. Mol Microbiol. 1989 Jul;3(7):979–984. doi: 10.1111/j.1365-2958.1989.tb00247.x. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prère M. F., Chandler M., Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990 Jul;172(7):4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantschuk M., Richter G. Y., Mukhopadhyay P., Mills D. IS801, an insertion sequence element isolated from Pseudomonas syringae pathovar phaseolicola. Mol Microbiol. 1991 Mar;5(3):617–622. doi: 10.1111/j.1365-2958.1991.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982 Oct 11;10(19):5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Schwartz E., Kröger M., Rak B. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 1988 Jul 25;16(14B):6789–6802. doi: 10.1093/nar/16.14.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubben D., Schmitt R. Tn1721 derivatives for transposon mutagenesis, restriction mapping and nucleotide sequence analysis. Gene. 1986;41(2-3):145–152. doi: 10.1016/0378-1119(86)90093-4. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala J. C., García-Lobo J. M., Diaz-Aroca E., de la Cruz F., Ortiz J. M. Escherichia coli alpha-haemolysin synthesis and export genes are flanked by a direct repetition of IS91-like elements. Mol Gen Genet. 1984;197(1):90–97. doi: 10.1007/BF00327927. [DOI] [PubMed] [Google Scholar]
- Zabala J. C., de la Cruz F., Ortiz J. M. Several copies of the same insertion sequence are present in alpha-hemolytic plasmids belonging to four different incompatibility groups. J Bacteriol. 1982 Jul;151(1):472–476. doi: 10.1128/jb.151.1.472-476.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


