Abstract
Diaphanous-related formins (DRFs) are key regulators of actin cytoskeletal dynamics whose in vitro actin assembly activities are thought to be regulated by autoinhibition. However, the in vivo consequences of autoinhibition and the involvement of DRFs in specific biological processes are not well understood. In this study, we show that in the DRFs FRLα (formin-related gene in leukocytes α) and mouse diaphanous 1, autoinhibition regulates a novel membrane localization activity in vivo as well as actin assembly activity in vitro. In FRLα, the Rho family guanosine triphosphatase Cdc42 relieves the autoinhibition of both membrane localization and biochemical actin assembly activities. FRLα is required for efficient Fc-γ receptor–mediated phagocytosis and is recruited to the phagocytic cup by Cdc42. These results suggest that mutual autoinhibition of biochemical activity and cellular localization may be a general regulatory principle for DRFs and demonstrate a novel role for formins in immune function.
Introduction
Dynamic control of the actin cytoskeleton is essential for cell polarization, migration, and division (Evangelista et al., 2003; Pollard and Borisy, 2003). Proper actin remodeling involves the activation of different eukaryotic nucleation factors that can generate specific types of new actin filaments. The actin-related protein 2/3 (Arp2/3) complex nucleates new actin filaments while remaining anchored to the sides of existing filaments, creating branched actin networks (Higgs and Pollard, 2001). Formin proteins nucleate unbranched filaments, creating actin cables, contractile rings, and stress fibers (Evangelista et al., 2002; Sagot et al., 2002; Sharpless and Harris, 2002; Zigmond, 2004). Although the Arp2/3 complex is known to play important roles in processes such as cell polarization, motility, and vesicle trafficking, much less is know about the biological functions of formins, especially in higher eukaryotes. Similarly, although molecular regulation of the Arp2/3 complex by upstream signals is understood in some detail (Higgs and Pollard, 2001), much less is known about the regulation and function of the much larger formin family.
The formins are defined by a conserved C-terminal formin homology (FH) 2 domain that mediates effects on actin (Wallar and Alberts, 2003; Watanabe and Higashida, 2004; Zigmond, 2004; Higgs, 2005; Higgs and Peterson, 2005). The FH2 domain functions as a dimer and has varying effects in different formins, including actin filament nucleation, filament severing, and barbed-end binding with elongation (anticapping effect) or without elongation (capping effect; Evangelista et al., 2002; Kovar et al., 2003; Li and Higgs, 2003; Pring et al., 2003; Harris et al., 2004; Romero et al., 2004; Xu et al., 2004).
The function of the conserved N-terminal region found in most formins is still unclear. In vitro and in vivo studies with mouse diaphanous 1 (mDia1) have demonstrated a role for its N terminus in mediating the autoinhibition of FH2 activity (see the following paragraph; Watanabe et al., 1999; Alberts, 2001; Ishizaki et al., 2001; Li and Higgs, 2003, 2005). Other studies with the fungal formins Bni1p, Fus1, and SepA have implicated their N termini in regulating localization in vivo (Petersen et al., 1998; Ozaki-Kuroda et al., 2001; Sharpless and Harris, 2002). For both Bni1p and Fus1, perturbation of cellular localization disrupts biological activity (Petersen et al., 1998; Ozaki-Kuroda et al., 2001). It is unclear whether the effects of the N terminus on FH2 activity and cellular localization are linked or whether the N terminus simply serves different roles in different formins. Elucidating the general regulation of formin localization and activity in higher eukaryotes is important for understanding the biological activities of these molecules.
In the diaphanous-related formin (DRF) subfamily of formins, a short (∼20 residue) conserved region called the diaphanous autoregulatory domain (DAD) follows the FH2 domain in sequence (Alberts, 2001; Higgs and Peterson, 2005). In mDia1, the DAD binds to the N terminus to inhibit the actin assembly activity of the FH2 domain through an unknown mechanism (Alberts, 2001; Li and Higgs, 2003). The DAD-binding element in the N terminus is called the diaphanous inhibitory domain (DID; Li and Higgs, 2005). Rho binds to the mDia1 N terminus through a region that spans a portion of the DID and an adjacent sequence element termed the G region (for GTPase binding; Otomo et al., 2005; Rose et al., 2005). Rho binding destabilizes interactions between the N and C termini, leading to partial activation of the mDia1 FH2 domain (Watanabe et al., 1999; Li and Higgs, 2003). The mDia1 N terminus forms a dimer through its dimerization domain (DD) and coiled coil (CC) region (Otomo et al., 2005; Rose et al., 2005). Based on sequence similarity, the G-DID-DD-CC architecture of the mDia1 N terminus is likely to hold for other DRF family members (Higgs, 2005). The formin FHOD1 is also likely to be autoinhibited based on cytoskeletal effects of mutant proteins studied in vivo (Koka et al., 2003). However, the generality of FH2 domain autoinhibition through a DAD–DID interaction and activation by Rho GTPases has yet to be established directly for DRFs other than mDia1.
FRLα (formin-related gene in leukocytes α) is a macrophage-enriched DRF whose FH2 domain can nucleate new actin filaments and sever existing filaments in vitro (Yayoshi-Yamamoto et al., 2000; Harris et al., 2004). FRLα has been reported to bind to Rac and modulate cell adhesion, migration, and survival, but its molecular function in macrophages has not been explored in detail (Yayoshi-Yamamoto et al., 2000). Because of its unique expression profile, we hypothesized that understanding FRLα may reveal new aspects of formin biology in higher eukaryotes as well as elucidate general principles governing the biological and biochemical regulation of formin proteins.
We find that the FRLα N terminus binds to the C terminus in a DAD-dependent manner and inhibits the actin assembly activity of the FH2 domain. For both FRLα and mDia1, autoinhibition also regulates a previously unrecognized plasma membrane localization activity of the N-terminal domains. This activity has both GTPase-dependent and -independent components. For FRLα, active Cdc42 relieves the autoinhibition of FH2 activity in vitro and membrane localization activity in macrophages. Knockdown experiments reveal that FRLα is required for efficient Fc-γ receptor–mediated phagocytosis, which is consistent with its role as a macrophage-enriched Cdc42 effector. Live cell imaging shows that FRLα is transiently recruited to the phagocytic cup in a Cdc42-dependent manner. These studies reveal a general mechanism of DRF regulation in which autoinhibition controls actin assembly activity and cellular localization. In addition, we identify a new biological function for DRFs in the immune system of higher eukaryotes.
Results
Autoinhibition regulates the actin assembly activity and intracellular localization of FRLα
Most previous in vivo studies of DRF N-terminus function have used fragments based on GTPase-binding domain (GBD)–FH3 sequence elements (Yayoshi-Yamamoto et al., 2000; Kato et al., 2001). However, recent structural studies of the mDia1 N terminus have demonstrated that these sequence elements do not demarcate structural elements of the protein. Therefore, the results from previous studies using constructs representing divided structural domains are difficult to interpret (Yayoshi-Yamamoto et al., 2000; Kato et al., 2001). In this study, we have used predicted structural domains of FRLα based on sequence alignments with mDia1 to examine the function of its N- and C-terminal domains (Otomo et al., 2005). Our N-terminal construct (residues 1–450) contains the regions of FRLα that align with the G, DID, and DD elements of mDia1 (FRLα appears to lack a CC region), whereas our C-terminal construct (residues 612–1,094) contains the FH2 and DAD domains (Fig. 1 A). Pull-down assays show that the immobilized GST-tagged N terminus can interact directly with the untagged C terminus (Fig. 1 B). Binding is also observed when immobilized maltose-binding protein (MBP)–tagged C terminus is used to pull down GST-tagged N terminus (Fig. 1 C). The DAD motif is required for binding because a C-terminal construct mutated at a conserved leucine residue within the DAD motif (L1062D) cannot interact with the N-terminal fragment (Fig. 1, B and C). Mutation of the same residue in mDia1 (unpublished data) or mDia2 (Alberts, 2001) also prevents interactions between N- and C-terminal fragments.
Figure 1.
The N terminus of FRLα negatively regulates actin assembly activity of the C-terminal FH2 domain. (A) Schematic diagrams of FRLα and mDia1 constructs used in this study. (B) Interaction between the N and C termini of FRLα. 3 μM GST-tagged N-terminal proteins were immobilized on glutathione–Sepharose beads and used to pull down 3 μM C-terminal constructs. Purified proteins are shown in lanes 2, 3, 6, and 9. Unbound proteins in the flow through (FT) and proteins remaining on the beads after five washes (B) are shown for each experiment. GST–N terminus was used to pull down wild-type C terminus (lanes 4 and 5) or C terminus L1062D containing a point mutation in the DAD domain (lanes 7 and 8). GST–N terminus T126D (Cdc42-binding mutant) was used to pull down wild-type C terminus (lanes 10 and 11). For each experiment, the volumes loaded on the SDS-PAGE gel were adjusted to equalize protein concentrations in the flow through and bead lanes. Gels are stained with Coomassie blue. (C) Similar to B except that the MBP-tagged wild-type C terminus (lane 2) or mutant C terminus (lane 6) was immobilized on amylose resin and used to pull down GST–N terminus (lane 3). Flow through and bead samples from the wild-type C-terminal pull down (lanes 4 and 5) or the mutant C-terminal pull down (lanes 7 and 8) are shown. (D) Dose-dependent inhibition of the actin assembly activity of 200 nM FRLα C terminus in the presence of the N terminus monitored by the increase in fluorescence of 4 μM pyrene-actin (5% labeled) upon incorporation into filaments. (E) Maximum actin assembly rate versus the concentration of N terminus. (F) Effect of the L1062D DAD motif mutation on the ability of 400 nM N terminus to inhibit 4 μM actin assembly (5% pyrene labeled) by 400 nM wild-type or mutant C terminus. Note that mutant C terminus alone or mutant C terminus + N terminus curves overlap significantly.
As previously described, the FH2 domain of FRLα can stimulate actin filament assembly, although it is ∼50-fold less potent than the FH2 domain of mDia1 (Fig. 1 D; Li and Higgs, 2003; Harris et al., 2004). The N terminus inhibits the activity of the C terminus in a dose-dependent manner with an IC50 of ∼100 nM (Fig. 1 E) but has no effect on the L1062D DAD mutant or FH2-independent actin assembly (Fig. 1, D and F). Thus, the biochemical activity of the FRLα FH2 domain is regulated by high affinity DAD-mediated autoinhibitory interactions with the N terminus.
We transfected RAW 264.7 cells, a mouse macrophage cell line, with GFP-tagged FRLα. Full-length FRLα is cytoplasmic and is excluded from the nucleus, as shown by confocal images of cells coexpressing FRLα-GFP and monomeric RFP (mRFP), a uniformly distributed fluorescent control (Fig. 2 A). In contrast, the N-terminal fragment of FRLα is located primarily at the plasma membrane, suggesting that in the full-length protein, the cellular localization of the N terminus may be controlled by binding to the C terminus (Fig. 2 B). To test this hypothesis, we determined the cellular localization of FRLα-GFP proteins with mutations that would impair the N + C interaction. Introduction of the L1062D DAD mutation results in plasma membrane localization of full-length FRLα (Fig. 2 C). In mDia1, an L260E mutation in the N-terminal DID region blocks DAD binding without affecting Rho interactions (Otomo et al., 2005). The analogous mutation in full-length FRLα (V281E) causes the protein to localize at the plasma membrane (Fig. 2 D). Quantitation of the ratio of GFP fluorescence intensity at the plasma membrane to the intensity in the cytosol shows that all membrane-localized FRLα constructs exhibit 2–4.5-fold enrichment at the plasma membrane, whereas cytosolic FRLα constructs have a ratio near 1 (Fig. 2 F). The degree of enrichment is largely independent of the overall expression level for all constructs shown here (Fig. 2 F).
Figure 2.
Autoinhibition regulates FRLα localization. Confocal images of RAW cells coexpressing GFP fusion proteins and mRFP. The mRFP serves as an evenly distributed control fluorophore. For these and all subsequent confocal images, the intensities for each channel have been normalized against the maximum value in the cell depicted in each image. As illustrated in F, the degree of membrane localization is not dependent on the total expression level of FRLα proteins. GFP fusion proteins are as follows: (A) full-length FRLα-GFP; (B) FRLα N terminus–GFP; (C) full-length FRLα-GFP containing an L1062D mutation in the DAD; (D) full-length FRLα-GFP containing a V281E mutation in the DID; and (E) mini-FRLα–GFP consisting of the N terminus of FRLα fused directly to the DAD domain with an artificial (Gly-Gly-Ser)2 linker. (F) Membrane enrichment versus expression level for the indicated constructs. Each data point represents one transfected cell. As described in Materials and methods, for each cell expressing a given GFP fusion protein, ratios were obtained for the GFP intensity at the plasma membrane against the GFP intensity in the cytosol. The ratios are plotted against the total GFP expression level in each cell (expressed as the fluorescence intensity per unit area). (G) Membrane enrichment versus expression level for the indicated constructs. Each data point represents one transfected cell. (H) Confocal images of cells coexpressing mRFP and the indicated mutant mini-FRLα–GFP fusion protein. The schematic protein diagram refers to the mini-FRLα construct depicted in Fig. 1 A and contains an asterisk signifying that the point mutations are all contained within the DAD region.
To test whether interactions between the N terminus and DAD are sufficient to mediate the control of FRLα cellular localization, we constructed a truncated FRLα protein called mini-FRLα, which lacks the FH2 domain. Mini-FRLα contains the G, DID, and DD regions tethered by a (Gly-Gly-Ser)2 linker to the DAD-containing C-terminal 101 residues (Fig. 1 A). When purified from overexpressing bacteria, this protein is dimeric as assessed by multiwavelength static light scattering (measured mol wt = 148.2 ± 0.2 kD compared with 71.9 kD for the monomer; unpublished data), which is the expected molecular organization of full-length FRLα. When expressed in macrophages, mini-FRLα–GFP is located in the cytoplasm of transfected cells (Fig. 2, E and F).
In the crystal structure of the DAD–DID complex from mDia1, the DAD forms an amphipathic helix with its hydrophobic face contacting the DID. Mutation of conserved residues on the hydrophobic face significantly decreases DAD–DID affinity, and the mutation of hydrophilic residues on the solvent exposed face has little effect on this interaction (Lammers et al., 2005; Otomo et al., 2005; Rose et al., 2005). With mini-FRLα, the mutation of any one of the conserved hydrophobic residues I1058, I1059, or L1062 on the hydrophobic face of the analogous predicted helix results in plasma membrane localization of the protein (Fig. 2, G and H). In contrast, the mutation of residues G1053 or E1056 on the hydrophilic face has no effect on localization (Fig. 2, G and H). Furthermore, the FRLα N terminus is cytoplasmic in cells coexpressing the DAD-containing C-terminal 100 residues of the protein but is membrane localized in cells coexpressing an L1062D mutant C terminus (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200605006/DC1). These results suggest that binding of the hydrophobic face of the amphipathic DAD helix is necessary and sufficient to block membrane localization mediated by the FRLα N terminus. Our combined in vitro and in vivo data show that the interactions between the N and C termini are mutually autoinhibitory, with the N terminus blocking the actin assembly activity of the FH2 domain and the C terminus blocking the plasma membrane localization activity of the N terminus.
Cdc42 relieves FRLα autoinhibition in vitro and in vivo
We next sought to determine whether Rho GTPases could relieve the autoinhibition of FRLα. Previously reported pull-down assays indicated that an N-terminal fragment of FRLα can specifically associate with Rac1 in a nucleotide-independent manner (Yayoshi-Yamamoto et al., 2000). However, we could not detect an interaction between recombinant Cdc42 or Rac1 loaded with a GTP analogue, β,γ-imidoguanosine 5′-triphosphate (GMPPNP), and GST-tagged FRLα N terminus using similar assays (unpublished data). Unexpectedly, high concentrations of Cdc42-GMPPNP can relieve N-terminal inhibition of the FRLα FH2 domain (Fig. 3 A). The Cdc42 effect is dose dependent and saturates at ∼400 μM GTPase, a concentration that slightly decreases the activity of the isolated C terminus (Fig. 3, A and D). Under these conditions, although Cdc42-GMPPNP exhibits low potency, it essentially fully relieves inhibition by the N terminus. Cdc42-GDP is a much weaker activator compared with Cdc42-GMPPNP (Fig. 3, B and D). Rac1-GMPPNP is unable to relieve the autoinhibition of FRLα actin assembly activity (Fig. 3 C) even at concentrations (∼400 μM) at which Cdc42-GMPPNP is maximally effective (Fig. 3 C). A V161D mutation in the mDia1 DID decreases Rho binding without affecting DAD binding (Otomo et al., 2005). The analogous T126D mutation in the FRLα N terminus does not affect binding to the C terminus (Fig. 1 B) or inhibition of FH2-mediated actin assembly (Fig. 3 E) but blocks the ability of Cdc42-GMPPNP to relieve inhibition (Fig. 3 E).
Figure 3.
Active Cdc42 relieves the autoinhibition of FRLα actin assembly activity. Actin assembly assays were performed with 4 μM actin (5% pyrene-labeled). (A) Effect on the inhibition of actin assembly by 200 nM C terminus in the presence of 400 nM N terminus by the addition of the indicated amount of Cdc42-GMPPNP. (B) Similar to A except using Cdc42-GDP. (C) Similar to A except using Rac1-GMPPNP. (D) Actin assembly rate versus the concentration of Cdc42 based on curves in A and B. (E) Actin assembly by 200 nM C terminus in the presence of 1 μM wild-type N terminus or 1 μM T126D N terminus with or without 200 μM Cdc42-GMPPNP.
When cotransfected with constitutively active Cdc42, both FRLα-GFP and mini-FRLα–GFP localize at the plasma membrane (Fig. 4, A, B, and I). The localization of FRLα is nucleotide and GTPase specific, as the protein remains primarily cytosolic in cells coexpressing dominant-negative Cdc42 or constitutively active Rac1 or RhoA (Fig. 4, C–E and I). In all cases, expression of the different GTPase mutants was confirmed by immunostaining (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200605006/DC1). Like our GFP fusions of FRLα, endogenous FRLα is primarily cytosolic in the absence of Cdc42. In cells transfected with constitutively active Cdc42, endogenous FRLα becomes enriched at the plasma membrane (Fig. S2).
Figure 4.
Active Cdc42 relieves the autoinhibition of FRLα localization. (A and B) Confocal images of RAW cells coexpressing mRFP, constitutively active Cdc42, and full-length FRLα-GFP (A) or mini-FRLα–GFP (B). (C–E) Confocal images of RAW cells expressing mRFP, full-length FRLα-GFP, and dominant-negative Cdc42 (C), constitutively active Rac1 (D), or constitutively active RhoA (E). (F) Confocal images of a cell expressing mRFP and the FRLα N terminus T126D-GFP mutant. (G and H) Confocal images of cells coexpressing N terminus–GFP and an mRFP fusion of WASP-GBD (G) or a mutant WASP-GBD (H) that does not bind Cdc42. For each transfection, 1 μg FRL construct was cotransfected with 4 μg GBD construct. (I) Membrane enrichment versus expression level for the indicated constructs. Each data point represents one transfected cell. (J) Coimmunoprecipitation experiments using lysates from 293T cells coexpressing myc-tagged Rho GTPase mutants and different FRLα-GFP constructs. Lysates were immunoprecipitated using anti-GFP or control IgG antibodies and blotted with anti-GFP or anti-myc antibodies. For each experiment (lanes a–c, d–f, g–i, j–l, m–o, and p–r), the lysate (starting material) is shown in the first lane followed by the material bound to control IgG beads or anti-GFP beads in the next two lanes. MW, molecular weight; SM, starting material.
Because Cdc42 is membrane anchored through prenylation of its C-terminal CAAX motif, we tested whether Cdc42 was directly recruiting FRLα to the plasma membrane of RAW cells. The T126D mutant FRLα N terminus, which is unresponsive to Cdc42 in actin assembly assays, is enriched at the plasma membrane but to a lesser degree than the analogous wild-type fragment (Fig. 4, F and I). As with our other FRLα constructs, the degree of enrichment does not vary systematically with varying expression levels (Fig. 4 I). Thus, Cdc42 binding likely contributes to but is not necessary for membrane enrichment of the FRLα N terminus. However, signaling through Cdc42 does appear to be required for this membrane localization activity because coexpression of the FRLα N terminus with the GBD of the Wiskott-Aldrich syndrome protein (WASP; a reagent that at high levels should sequester the active GTPase) causes the FRLα fragment to be cytoplasmic (Figs. 4 G and S1). This relocation to the cytoplasm is not observed when the FRLα N terminus is coexpressed with a mutant WASP-GBD that cannot bind Cdc42 (Figs. 4 H and S1). These results suggest that membrane localization activity of the FRLα N terminus derives, in part, from direct binding to Cdc42 and also from an interaction with some other membrane-associated factor that lies downstream of Cdc42.
Although our data suggest a direct interaction between Cdc42 and the N terminus of FRLα, we have been unsuccessful in demonstrating a direct interaction in pull-down or fluorescence-based binding assays using bacterially expressed recombinant proteins (unpublished data). However, when cotransfected into mammalian 293T cells, constitutively active Cdc42 and the wild-type FRLα N terminus can be coimmunoprecipitated (Fig. 4 J, lanes a–c). The N terminus does not coimmunoprecipitate with constitutively active Rac1 or RhoA (Fig. 4 J, lanes g–i and j–l, respectively), suggesting specificity for Cdc42 over these other two Rho family members. One caveat here is that despite significant effort, the level of Rac expression was always lower than that of Cdc42 in these experiments. However, along with the biochemistry and aforementioned localization data, these results do support a much greater role for Cdc42 than Rac in regulating FRLα. The T126D mutant N terminus does not coimmunoprecipitate with Cdc42, suggesting that in vitro and in cells, the Cdc42 responsiveness of FRLα is mediated by the same set of interactions (Fig. 4 J, lanes d–f). Furthermore, full-length FRLα and mini- FRLα do not coimmunoprecipitate with Cdc42 even though they localize at the plasma membrane when coexpressed with constitutively active Cdc42 (Fig. 4 J, lanes m–o and p–r, respectively). These results suggest that Cdc42 has higher affinity for the FRLα N terminus as compared with an autoinhibited FRLα molecule, which is consistent with the idea that Cdc42 must compete with the autoinhibitory interactions present in full-length or mini-FRLα.
Our in vitro and in vivo data suggest that FRLα is a direct effector of Cdc42 and that the interactions between FRLα and Cdc42 may be similar to those between mDia1 and Rho. Cdc42 relieves the autoinhibition of FRLα actin assembly activity in vitro and relieves the autoinhibition of FRLα membrane localization in macrophages. The affinity of Cdc42 for FRLα appears to be weak, suggesting that Cdc42 may have to cooperate with other factors to regulate FRLα in vivo.
Autoinhibitory control of mDia1 intracellular localization
As in FRLα, a GFP fusion of the N terminus of mDia1(residues 1–570) is localized at the plasma membrane (Fig. 5, A and F). Both full-length mDia1-GFP and mini-mDia1–GFP, analogous to our mini-FRLα construct, are cytoplasmic and are excluded from the nucleus of RAW cells (Fig. 5, B, C, and F). Thus, autoinhibition controls the localization of mDia1 in a manner analogous to FRLα. Also, like FRLα, the membrane localization activity in the mDia1 N terminus has GTPase-dependent and -independent components. The V161D mutant N-terminal fragment of mDia1 (analogous to the aforementioned FRLα T126D mutant) is strongly defective in Rho binding (Otomo et al., 2005). This protein localizes to the plasma membrane in macrophages but to a lesser degree than the wild-type N terminus (Fig. 5, D and F). Interestingly, the degree of membrane enrichment of this protein decreases as expression level increases (Fig. 5, D and F), suggesting that the GTPase-independent membrane interaction is saturable. A nearly identical behavior is also observed for a fragment of the mDia1 N terminus lacking the G region entirely (Fig. 5, E and F; mDia1_ΔG_N-term), which also is severely impaired in Rho binding (Otomo et al., 2005; Rose et al., 2005). Analogous fragments of FRLα are not expressed in bacteria (in contrast to mDia1_ΔG_N-term, which expresses well; unpublished data), are expressed only weakly and inconsistently in macrophages, and were not analyzed in detail. Together, our data suggest that for both FRLα and mDia1, plasma membrane localization of the N terminus is mediated by GTPase-dependent and -independent interactions and that these are controlled by autoinhibitory contacts within the formins.
Figure 5.
Autoinhibition controls localization of the Rho-regulated DRF mDia1. (A–D) Confocal images of RAW cells coexpressing mRFP and mDia1 N terminus–GFP (A), full-length mDia1-GFP (B), mini-mDia1–GFP (C), or mDia1 V161D N terminus–GFP (D). Three different cells are shown to illustrate the different patterns observed for this construct. The GFP fusion is localized at the plasma membrane in the first cell (top) and in the cytosol of the third cell (bottom). The second cell (middle) has an intermediate distribution between membrane and cytosol. (E) mDia1 ΔG–N terminus–GFP. Three different cells are shown to illustrate the different localization patterns observed with this construct. (F) Membrane enrichment versus expression level for the indicated constructs. Each data point represents one transfected cell.
FRLα is required for Fc-γ receptor–mediated phagocytosis and is recruited to the phagocytic cup by Cdc42
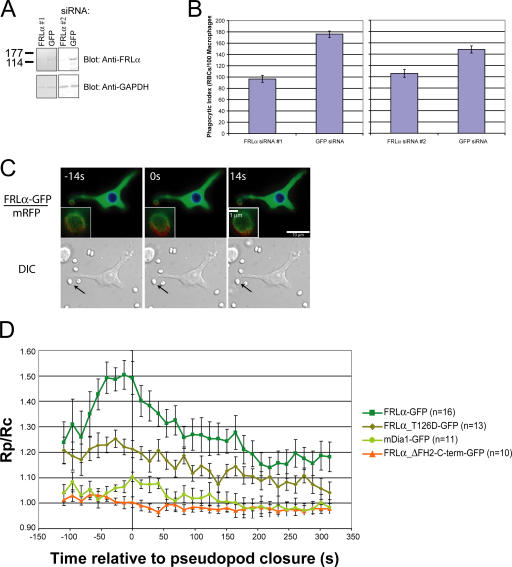
We next sought to determine whether FRLα is involved in Fc-γ receptor–mediated phagocytosis. This process is Cdc42 dependent and requires extensive actin rearrangement at the cell surface of macrophages (Caron and Hall, 1998). Using two different sets of siRNAs directed against FRLα, we achieved significant knockdown of endogenous FRLα in RAW macrophages (Fig. 6 A). These reduced Fc-γ receptor–mediated phagocytosis of IgG-opsonized RBCs by 45 ± 4 and 29 ± 2%, respectively, compared with cells expressing control siRNAs (Fig. 6 B). Overall cell appearance, adhesion, and filamentous actin content judged by phalloidin staining are all normal in the siRNA-treated cells (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200605006/DC1). Moreover, complement receptor-mediated phagocytosis, a process that proceeds through a Rho–mDia1 pathway (Colucci-Guyon et al., 2005), is also unaffected by siRNA treatment (Fig. S3 B). These results demonstrate that the knockdown of FRLα does not lead to generalized cytoskeletal defects in RAW cells and establish an important role for FRLα specifically in Fc-γ receptor–mediated phagocytosis.
Figure 6.
Cdc42 recruits FRLα to the phagocytic cup during Fc-γ receptor–mediated phagocytosis. (A) Western blot of lysates prepared from cells transfected with siRNAs directed against FRLα or GFP (control) and probed with anti-FRLα or anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH; loading control) antibodies. (B) Phagocytic index (number of RBCs/100 macrophages) of siRNA-transfected RAW macrophages. Experiments were performed in duplicate and repeated three times. Data are the means based on counting at least 300 macrophages per experiment. (C) FRLα-GFP–expressing cell undergoing Fc-γ receptor–mediated phagocytosis. The zero time point is a reference for when the phagosome closes around the RBC being engulfed. The top panels are pseudocolored to represent the GFP/mRFP ratio at each pixel in the cell. Low ratios are represented by blue or cool colors, whereas higher ratios are represented by increasingly red or warmer colors. Bottom panels are the corresponding DIC images for each time point. The ingested RBCs are indicated by arrows. The insets depict additional magnification of the phagocytic cup. (D) Time course of formin accumulation during Fc-γ receptor–mediated phagocytosis. n indicates the number of phagocytic events analyzed for each construct. For each time point, the GFP/mRFP ratio at the cell surface in contact with the RBC (Rp) was divided by the ratio in the cell cytoplasm (Rc). Error bars represent SEM.
We used time-lapse microscopy to analyze phagocytic events in live RAW macrophages incubated with IgG-opsonized RBCs at 37°C. Under our assay conditions, macrophages readily phagocytose the RBCs with a time course of ∼90 s from cell–cell contact to complete RBC internalization as judged by light microscopy. For RBCs that encounter macrophages from the side, the extension of macrophage pseudopods around the RBC is readily observable in fluorescently labeled cells. The pseudopods eventually meet at the top of the RBC and fuse to close the phagosome, which is then internalized into the main cell body.
In cells coexpressing FRLα-GFP and mRFP, we used ratiometric imaging to determine specific sites of FRLα accumulation. During phagocytosis, FRLα-GFP accumulates in the developing phagocytic cup and appears to concentrate at the tips of the extending pseudopods (Fig. 6 C and Videos 1–3, available at http://www.jcb.org/cgi/content/full/jcb.200605006/DC1). FRLα-GFP accumulation is maximal immediately before pseudopod fusion, which we set as a reference time point to compare multiple phagocytic events, and rapidly dissipates as the RBC is internalized (Fig. 6, C and D). This spatial and temporal pattern is reproducibly observed in different transfected cells; additional phagocytic events are shown in Fig. S4. A control GFP construct fused to the C-terminal 101 residues of FRLα does not accumulate at the phagocytic cup (Fig. 6 D). Consistent with previous findings that Rho is not involved in Fc-γ receptor–mediated phagocytosis (Caron and Hall, 1998), mDia1-GFP does not localize to the cup during this process (Fig. 6 D). The T126D mutant of FRLα shows a consistent modest increase in localization at the cup relative to mDia1 and the FRL C terminus, but its enrichment does not change appreciably over the course of internalization (Fig. 6 D). This enrichment is likely caused by the ability of the T126D mutant N terminus to interact with membranes (Fig. 4, F and I), which accumulate at the cup as a result of its topology. Importantly, the wild-type protein shows much different behavior, with a greater degree of accumulation and substantial changes over the course of the process, suggesting recruitment to the cup by an active mechanism.
Active Cdc42 has also been shown to accumulate transiently at the extending pseudopod tips of the phagocytic cup (Hoppe and Swanson, 2004). We monitored the localization of active Cdc42 during phagocytosis using the WASP-GBD fused to GFP, a reagent with high (19 nM) affinity for active Cdc42 (Kim et al., 2000b; Cannon et al., 2001; Labno et al., 2003; Seth et al., 2003). We note that at high levels of expression, this reagent blocked phagocytosis (unpublished data), but, at lower levels, it had no effect on phagocytic efficiency, and we could use it to visualize active Cdc42 during this process. As a control, we used a mutant GBD-GFP construct that is unable to bind Cdc42. We find that Cdc42, like FRLα, is recruited transiently to the phagocytic cup and that its accumulation peaks immediately before pseudopod fusion (Fig. 7, A and B; and Videos 4 and 5, available at http://www.jcb.org/cgi/content/full/jcb.200605006/DC1). The similarities in timing and localization of active Cdc42 and FRLα support the idea that FRLα function during phagocytosis is linked to Cdc42 signaling.
Figure 7.
Recruitment of active Cdc42 to the phagocytic cup during Fc-γ receptor–mediated phagocytosis. (A) GBD-GFP–expressing cell undergoing Fc-γ receptor–mediated phagocytosis. Cells were transfected with plasmids encoding mRFP and the indicated GBD-GFP construct. 2.5 μg of each plasmid (5 μg of total DNA) was used for each transfection. As in Fig. 6 C, the GFP/mRFP ratio is pseudocolored in the top panel to reflect the relative accumulation of GBD-GFP over mRFP. Localization of the GBD-GFP protein reflects the distribution of Cdc42-GTP during phagocytosis. Arrows in the bottom panels of DIC images indicate the ingested RBCs. Insets depict additional magnifications of the phagocytic cup. (B) Time course of active Cdc42 accumulation during Fc-γ receptor–mediated phagocytosis. Multiple phagocytic events were analyzed as in Fig. 6 B in cells expressing mRFP and GBD-GFP or the mutant GBD-GFP (as a negative control). Error bars represent SEM.
Discussion
Autoinhibition of DRF actin assembly activity
We have generated N-terminal fragments of FRLα based on recent crystal structures of the mDia1 N terminus (Otomo et al., 2005; Rose et al., 2005) to investigate the mechanisms of autoinhibition and activation in FRLα. Based on fragmented DID constructs, earlier work suggested that FRLα may bind Rac (Yayoshi-Yamamoto et al., 2000). However, using constructs containing intact structural domains that are biochemically well behaved, we now show that FRLα is a Cdc42-specific effector. Several lines of evidence support this assertion. Cdc42 is able to relieve the autoinhibition of FH2-mediated actin assembly by the wild-type N terminus but not by the T126D mutant N terminus. Mutation of the analogous residue in mDia1 prevents direct Rho binding and Rho-mediated activation (Otomo et al., 2005). Furthermore, the wild-type FRLα N terminus coimmunoprecipitates with active Cdc42 from cell lysates, but the point mutant does not. In addition, the timing and localization of Cdc42 and wild-type FRLα are largely coincident during phagocytosis, and the recruitment of FRLα to the phagocytic cup is blocked by the T126D mutation. Similar biochemical, coimmunoprecipitation, and localization assays fail to show a link between Rac1 activity and FRLα function. However, the interaction between Cdc42 and FRLα appears to be of low affinity, and the interaction between them may be modulated by cellular factors.
Autoinhibition of DRF localization: a membrane factor X?
The functional significance of the N terminus of DRFs has been thought to be limited to mediating the autoinhibition of the C terminus (Evangelista et al., 2003; Wallar and Alberts, 2003; Higgs, 2005). We now show for the DRFs FRLα and mDia1 that interactions between the N and C termini mediate the mutual autoinhibition of DRF localization and biochemical activity. The membrane localization activity of the DRF N termini derives, in part, from interactions with Rho GTPases. However, our results suggest that the N termini can also bind an additional membrane-associated factor. This factor binds the N termini competitively with the DAD and, thus, should contribute to DRF activation. Depending on the structural details of the interaction, the factor may act cooperatively with GTPases to drive DRF membrane localization and activation.
Coordinated cytoskeletal remodeling during Fc-γ receptor–mediated phagocytosis
Cdc42 plays an essential role in Fc-γ receptor–mediated phagocytosis (Caron and Hall, 1998). Formins have not previously been implicated in this process, although mDia1 has recently been shown to be involved in complement-mediated phagocytosis, a Rho-mediated process (Colucci-Guyon et al., 2005). Our identification of FRLα as a Cdc42 effector led us to test whether this macrophage-enriched DRF is involved in Fc-γ receptor–mediated phagocytosis. Two previous studies on Fc receptor–mediated phagocytosis reported that dominant-negative Cdc42 inhibits phagocytosis by ∼40–70% (Caron and Hall, 1998; Massol et al., 1998), which is similar to what we observe for our strongest FRLα knockdown (45%). The consistent degree of inhibition by dominant-negative Cdc42 and FRLα RNAi suggests that FRLα is an important effector of Cdc42-mediated signaling during Fc-γ receptor–mediated phagocytosis.
It is useful to distinguish three phases of Fc-γ receptor–mediated phagocytosis: pseudopod extension, pseudopod fusion, and particle internalization (Castellano et al., 2001; May and Machesky, 2001; Swanson and Hoppe, 2004). Time-lapse imaging shows that fluorescently labeled actin accumulates at the base and extending pseudopods of the phagocytic cup (Araki et al., 2003; Scott et al., 2005). At closure, actin forms a sphere around the phagosome (Scott et al., 2005). After closure, actin dissipates from the phagosome starting at the base, resulting in a short-lived actin cap structure at the top of the phagosome (Scott et al., 2005). The actin architectures at each phase of phagocytosis and the signaling pathways that control their formation have yet to be determined.
Recent studies have started to define the spatio-temporal patterns and functional roles of different actin regulatory proteins during Fc-γ receptor–mediated phagocytosis, focusing mostly on Cdc42 and Rac (Caron and Hall, 1998; Massol et al., 1998; Castellano et al., 1999; Castellano and Chavrier, 2000; Hoppe and Swanson, 2004). The model that has emerged from these studies suggests that active Cdc42 localizes primarily at the tips of the extending pseudopods, mediating their extension and dissipating rapidly after their fusion. In contrast, active Rac1 is localized at the base and throughout the pseudopods during extension and then concentrates at the base to control fusion and internalization. Thus, Cdc42 and Rac likely play different roles during Fc-γ receptor–mediated phagocytosis.
In contrast to the Rho GTPases, the actions of downstream signaling molecules during phagocytosis have been studied less extensively. WASP family proteins are important for Arp2/3 complex activation during cell migration (Pollard and Borisy, 2003), and static images reveal that the WASP and Arp2/3 complex are recruited to the phagocytic cup (May et al., 2000; Coppolino et al., 2001). However, the detailed spatio-temporal patterns of these molecules during phagocytosis have not been reported.
Fc-γ receptor–mediated phagocytosis represents one of the first isolated cytoskeletal systems in higher eukaryotes in which both Arp2/3 (May et al., 2000) and formins (this study) have essential functions. Why might two different actin nucleation machines be required for Fc-γ phagocytosis? Although both the Arp2/3 complex and FRLα can nucleate actin filaments, there are important differences in their resulting actin networks. Arp2/3 networks are branched and sensitive to capping proteins, and, therefore, the behavior of the Arp2/3 network depends on the balance of capping and nucleation. FRLα-generated filaments are unbranched and resistant to capping proteins. Based on its linkage to Cdc42, FRLα function may be important primarily for pseudopod extension, whereas the Arp2/3 complex may be required for the other phases of phagocytosis. In addition to nucleating new filaments, the FRLα FH2 domain can also sever actin filaments (Harris et al., 2004). This activity could lead to an increase in ATP-capped filament ends, which appear to be preferential sites of Arp2/3-mediated branching (Ichetovkin et al., 2002). Indeed, FRLα may not serve a nucleation function during phagocytosis at all but rather may facilitate pseudopod extension by functioning as an anticapping or severing protein to modify the Arp2/3 actin network. Currently, the specific biochemical and biological roles of FRLα and Arp2/3 complex during Fc-γ receptor–mediated phagocytosis remain elusive. Understanding the detailed molecular mechanisms of phagocytosis will shed light on this essential function of the immune system and will help determine the relative contributions of different actin regulatory elements to cytoskeletal dynamics in higher eukaryotes.
Materials and methods
Constructs
Full-length FRLα (residues 1–1,094), the FRLα N terminus (residues 1–450), the FRLα C terminus (residues 612–1,094), and the FRLα ΔFH2–C terminus (residues 994–1,094) were cloned into pET vectors containing N-terminal tobacco etch virus protease-cleavable GST or MBP affinity tags. FRLα cDNA was provided by T. Watanabe (Kyushu University, Fukuoka, Japan). Full-length mDia1 (resides 1–1,255), the mDia1 N terminus (residues 1–570), and the mDia1 ΔG–N terminus (residues 131–570) were cloned by PCR. All FRLα and mDia1 constructs were cloned into a modified pCMV-Script vector containing C-terminal EGFP or mRFP. Template cDNA for mRFP was provided by R.Y. Tsien (University of California, San Diego, San Diego, CA). Mini-FRLα (residues 1–536 and 994–1,094 tethered by a GGSGGS linker) and mini-mDia1 (residues 1–457 and 1,168–1,255 tethered by a GGSGGS linker) were generated using overlap extension PCR. Myc-tagged GTPase constructs were provided by M.H. Cobb (University of Texas Southwestern, Dallas, TX). The WASP-GBD comprises residues 230–288 of the human protein. The WASP-GBD mutant was generated by scrambling the amino acid sequence of the internal Cdc42/Rac interactive binding motif (IGAPSGFKHVSHVGWD mutated to KWDVPIHHGGFGASVS), which is necessary for high affinity interactions with Cdc42 (Rudolph et al., 1998).
The following siRNA oligonucleotides were used for RNAi experiments: FRL2614 sense (CAGACGUUGUUGCACUACUTT), FRL2614 antisense (AGUAGUGCAACAACGUCUGTT), GFP sense (GCAGAAGAACGGCAUCAAGTT), and GFP antisense (CUUGAUGCCGUUCUUCUGCTT). The second set of FRL siRNAs, which was purchased from Dharmacon, was a pool of four duplexes that did not overlap with the sequence targeted by the first siRNA duplex.
Bacterial expression and protein purification
All proteins were expressed in the Escherichia coli strain BL21(DE3). FRLα N-terminal constructs were expressed as GST fusion proteins, whereas C-terminal constructs were expressed as MBP fusions. N-terminal constructs were purified using glutathione–Sepharose beads (GE Healthcare), anion exchange chromatography, and gel filtration. C-terminal constructs were purified by cation exchange chromatography and gel filtration. Before some assays, the affinity tags were cleaved using tobacco etch virus protease, and the tag-free proteins were purified either with anion or cation exchange chromatography. Cdc42 (residues 1–179) was prepared and loaded with GMPPNP as previously described (Abdul-Manan et al., 1999; Kim et al., 2000a). Actin was purified from rabbit skeletal muscle and labeled with pyrene as described previously (Pollard and Cooper, 1984).
Biochemical assays
Pull-down experiments.
600 pmol GST-FRLα N terminus was loaded onto glutathione–Sepharose beads (GE Healthcare) and incubated with 600 pmol FRLα C terminus in 200 μl of binding buffer (20 mM Tris, pH 8.0, 100 mM NaCl, and 1 mM DTT). Beads were washed three times with 200 μl of the binding buffer containing 0.1% Triton X-100, washed twice with 200 μl of the binding buffer without detergent, and analyzed by SDS-PAGE and Coomassie blue staining. The same protocol was also performed with 600 pmol MBP–C terminus on amylose resin (New England Biolabs, Inc.) incubated with 600 pmol of GST–N terminus.
Actin assembly assays.
Experiments were performed as described previously (Higgs et al., 1999). Reactions contained 4 μM actin (5% pyrene labeled), 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM EGTA, and 1 mM MgCl2. Pyrene fluorescence (λex = 365 nm and λem = 407 nm) was recorded over time to monitor actin assembly.
Cell culture and transfection
RAW 264.7 cells were purchased from the American Type Culture Collection and maintained in DME with 10% FBS and 1 mM sodium pyruvate (Invitrogen). 106 cells were transfected with 5 μg DNA using LipofectAMINE 2000 (Invitrogen). For siRNA transfection, 2 × 105 cells were transfected with siRNAs using 1.5 μl siQuest reagent (Mirus Bio Corporation) and 250 nM siRNAs. Under these conditions, nearly 100% of RAW cells were transfected by siRNAs as judged by experiments using LabelIT Fluorescent RNA delivery controls (Mirus Bio Corporation). To determine the efficiency of FRLα knockdown, cell lysates were prepared 48 h after transfection. Western blots were performed using anti-FRLα antibodies provided by H.N. Higgs (Dartmouth College, Hanover, NH).
HEK293T cells were maintained in 10% calf serum (American Type Culture Collection) and 1 mM penicillin/streptomycin (Invitrogen). For coimmunoprecipitation, 106 cells plated in 10-cm dishes were transfected with 10 μg DNA using the calcium phosphate precipitation method.
Coimmunoprecipitation
HEK293T cells were cotransfected with expression vectors for each formin-GFP and myc-GTPase construct. 36 h after transfection, cell lysates were prepared in lysis buffer (20 mM Tris, pH 7.4, 100 mM NaCl, 1 mM DTT, 1 mM MgCl2, 0.2% NP-40, 1 mM PMSF, 1 mM benzamidine, 1 μg/ml leupeptin, 1 μg/ml antipain, 0.5 μg/ml pepstatin, 20 mM NaF, and 0.5 mM NaVO3). The cleared lysate was incubated with anti-GFP antibody (Invitrogen) for 1 h at 4°C. Immune complexes were precipitated with UltraLink immobilized protein A/G beads (Pierce Chemical Co.) and analyzed by Western blotting.
Confocal microscopy
24 h after transfection, cells were replated onto 12-mm glass coverslips (Fisher Scientific), allowed to adhere for 6–8 h, and were fixed. Imaging was performed on a laser scanning microscope (LSM510 META; Carl Zeiss MicroImaging, Inc.) using a 63× oil immersion objective (Carl Zeiss MicroImaging, Inc.). Images were analyzed using Slidebook software (Intelligent Imaging Innovations). Membrane and cytosolic intensities were determined by calculating the mean fluorescence in masks placed either at the membrane or in the cytoplasm. The expression level was expressed as the sum of the fluorescence intensity across a given confocal plane divided by the cross-sectional area.
Phagocytosis assays for live cell imaging
Image acquisition.
24 h after transfection, cells were replated onto 35-mm glass-bottom dishes and allowed to adhere for 6–8 h. For imaging, cells were incubated at 37°C on a heated stage connected to a humidifier module and covered with an optically clear foil cover (Carl Zeiss MicroImaging, Inc.). To start phagocytosis, 1 ml of 50-fold diluted opsonized RBCs prepared as described previously (Greenberg et al., 1990) was added dropwise to the macrophages. For each imaging run, 61 time points were collected. For each time point, a four-plane z stack of GFP and mRFP fluorescent images spaced 1.5 μm apart was acquired along with one differential interference contrast (DIC) image (50-ms exposure). A total of nine images per time point was collected at maximum speed, which, on our system, resulted in one time point per 13.7 s.
Imaging was performed using a 63× oil immersion objective (Carl Zeiss MicroImaging, Inc.) on an inverted microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) equipped with a 75-W Xenon lamp and CCD camera (Sensicam; PCO Computer Optics). Fluorescence images were acquired using FITC and Cy3 filter sets (Chroma Technology Corp.), 200-ms exposure times, 2 × 2 binning, and a 10% neural density filter.
Image analysis.
For each phagocytic event, the optimal plane (where the phagoyctic cup was in best focus) was determined at each time point. These planes were stitched together to create a 2D time-lapse series. For each event, the time point at which pseudopod fusion occurred, as seen in the fluorescent images, was set as the zero time reference. Only events in which the macrophage and RBC could be observed 8 time points before fusion and 23 time points after were further analyzed. Thus, for each event we have analyzed 32 time points, which is sufficient for complete analysis of the dynamics that we observe.
To analyze fusion protein accumulation during phagocytosis, we averaged the GFP/mRFP ratio at the site of phagocytosis (Rp) and normalized it to the GFP/mRFP ratio in the cytoplasm (Rc) in a manner similar to previously described methods (Hoppe and Swanson, 2004). Rp was calculated by creating a mask around the region of contact between the macrophage and the RBC. An Rc mask, which was placed in the cytoplasm, was also created. For each pixel in each mask at each time point, we calculated the ratio of GFP intensity to mRFP intensity. We then averaged the ratios for each mask to generate the GFP/mRFP ratio at the site of phagocytosis or in the cytoplasm.
Phagocytosis assays for siRNA-transfected cells
24 h after transfection, RAW cells were replated onto 12-mm glass coverslips. 48 h after transfection, cells were washed and returned to the 37°C incubator in 0.2 ml of buffer with divalents (20 mM Hepes, pH 7.4, 125 mM NaCl, 5 mM KCl, 5 mM glucose, 10 mM NaHCO3, 1 mM KH2PO4, 1 mM MgCl2, and 1 mM CaCl2). After 1 h, 20 μl of 10-fold diluted opsonized RBCs were added. For complement phagocytosis, cells were treated with 100 nM PMA 30 min before the addition of RBCs. After 1 h, cells were washed, incubated in distilled water to lyse extracellular RBCs, and fixed. Cells were permeabilized with PSG buffer (PBS, 0.01% saponin, 0.25% gelatin, and 0.02% NaN3). Intracellular RBCs were stained with 1:200 diluted FITC-conjugated anti–rabbit antibodies (Jackson ImmunoResearch Laboratories). Imaging was performed on an inverted microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) using a 63× oil immersion objective (Carl Zeiss MicroImaging, Inc.). The number of internalized RBCs was counted using the FITC fluorescence signal, and the number of macrophages was counted using cellular autofluorescence. For complement phagocytosis, internalized RBCs were counted using DIC. Each experiment was performed in duplicate, and a minimum of 300 macrophages was counted for every condition.
Online supplemental material
Table S1 provides the mean GFPm/GFPc ratios for all constructs and conditions tested. Fig. S1 shows that membrane localization of the FRLα N terminus is inhibited by DAD binding in trans. Fig. S2 shows the localization of endogenous FRLα and expression of myc-tagged Rho GTPase mutants in RAW cells. Fig. S3 shows the characterization of cells treated with FRLα siRNA, and Fig. S4 shows additional images of FRLα recruitment to the phagocytic cup. Videos 1 and 2 are DIC and fluorescence time lapses, respectively, of FRLα-GFP–expressing macrophages undergoing Fc-γ receptor–mediated phagocytosis. Video 3 shows a magnified phagocytic cup from Video 2. Videos 4 and 5 are DIC and fluorescence time lapses, respectively, of GBD-GFP–expressing macrophages undergoing Fc-γ receptor–mediated phagocytosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200605006/DC1.
Supplementary Material
Acknowledgments
We are indebted to Dr. Takanori Otomo for discussion and advice on actin and formin biochemistry; Drs. Simon Daefler, Kate Luby-Phelps, Rama Ranganathan, and Christoph Wulfing for discussions regarding image acquisition and processing; Jason R. Mock for advice on the phagocytosis assays; Rashu B. Seth for discussions about RNAi and coimmunoprecipitation; Song Huang for preparation of the initial mDia1 constructs and for assisting with the preliminary characterization of mDia1 localization; Neeta Mistry for technical support; Dr. Gaya K. Amarasinghe, Dr. Sanjay C. Panchal, Daisy W. Leung, and Hui-Chun Cheng for sharing reagents for the actin assembly assay; and Dr. Takeshi Watanabe, Dr. Henry N. Higgs, Dr. Roger Y. Tsien, and Dr. Melanie H. Cobb for providing reagents described in the Materials and methods section. Confocal microscopy was performed in the University of Texas Southwestern Live Cell Imaging Core Facility.
A. Seth is supported by the University of Texas Southwestern Medical Scientist Training Program. Research was supported by grants from the National Institutes of Health (GM56322) and Welch Foundation (I–1544).
Abbreviations used in this paper: Arp2/3, actin-related protein 2/3; CC, coiled coil; DAD, diaphanous autoregulatory domain; DD, dimerization domain; DIC, differential interference contrast; DID, diaphanous inhibitory domain; DRF, diaphanous-related formin; FH, formin homology; FRL, formin-related gene in leukocytes; GBD, GTPase-binding domain; GMPPNP, β,γ-imidoguanosine 5′-triphosphate; MBP, maltose-binding protein; mDia1, mouse diaphanous 1; WASP, Wiskott-Aldrich syndrome protein.
References
- Abdul-Manan, N., B. Aghazadeh, G.A. Liu, A. Majumdar, O. Ouerfelli, K.A. Siminovitch, and M.K. Rosen. 1999. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 399:379–383. [DOI] [PubMed] [Google Scholar]
- Alberts, A.S. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276:2824–2830. [DOI] [PubMed] [Google Scholar]
- Araki, N., T. Hatae, A. Furukawa, and J.A. Swanson. 2003. Phosphoinositide-3-kinase-independent contractile activities associated with Fcgamma-receptor-mediated phagocytosis and macropinocytosis in macrophages. J. Cell Sci. 116:247–257. [DOI] [PubMed] [Google Scholar]
- Cannon, J.L., C.M. Labno, G. Bosco, A. Seth, M.H. McGavin, K.A. Siminovitch, M.K. Rosen, and J.K. Burkhardt. 2001. Wasp recruitment to the T cell:APC contact site occurs independently of Cdc42 activation. Immunity. 15:249–259. [DOI] [PubMed] [Google Scholar]
- Caron, E., and A. Hall. 1998. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 282:1717–1721. [DOI] [PubMed] [Google Scholar]
- Castellano, F., and P. Chavrier. 2000. Inducible membrane recruitment of small GTP-binding proteins by rapamycin-based system in living cells. Methods Enzymol. 325:285–295. [DOI] [PubMed] [Google Scholar]
- Castellano, F., P. Montcourrier, J.C. Guillemot, E. Gouin, L. Machesky, P. Cossart, and P. Chavrier. 1999. Inducible recruitment of Cdc42 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr. Biol. 9:351–360. [DOI] [PubMed] [Google Scholar]
- Castellano, F., P. Chavrier, and E. Caron. 2001. Actin dynamics during phagocytosis. Semin. Immunol. 13:347–355. [DOI] [PubMed] [Google Scholar]
- Colucci-Guyon, E., F. Niedergang, B.J. Wallar, J. Peng, A.S. Alberts, and P. Chavrier. 2005. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr. Biol. 15:2007–2012. [DOI] [PubMed] [Google Scholar]
- Coppolino, M.G., M. Krause, P. Hagendorff, D.A. Monner, W. Trimble, S. Grinstein, J. Wehland, and A.S. Sechi. 2001. Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcgamma receptor signalling during phagocytosis. J. Cell Sci. 114:4307–4318. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., D. Pruyne, D.C. Amberg, C. Boone, and A. Bretscher. 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4:260–269. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., S. Zigmond, and C. Boone. 2003. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116:2603–2611. [DOI] [PubMed] [Google Scholar]
- Greenberg, S., K. Burridge, and S.C. Silverstein. 1990. Colocalization of F-actin and talin during Fc receptor-mediated phagocytosis in mouse macrophages. J. Exp. Med. 172:1853–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.S., F. Li, and H.N. Higgs. 2004. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279:20076–20087. [DOI] [PubMed] [Google Scholar]
- Higgs, H.N. 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30:342–353. [DOI] [PubMed] [Google Scholar]
- Higgs, H.N., and T.D. Pollard. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70:649–676. [DOI] [PubMed] [Google Scholar]
- Higgs, H.N., and K.J. Peterson. 2005. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell. 16:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, H.N., L. Blanchoin, and T.D. Pollard. 1999. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 38:15212–15222. [DOI] [PubMed] [Google Scholar]
- Hoppe, A.D., and J.A. Swanson. 2004. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell. 15:3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichetovkin, I., W. Grant, and J. Condeelis. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84. [DOI] [PubMed] [Google Scholar]
- Ishizaki, T., Y. Morishima, M. Okamoto, T. Furuyashiki, T. Kato, and S. Narumiya. 2001. Coordination of microtubules and the actin cyto- skeleton by the Rho effector mDia1. Nat. Cell Biol. 3:8–14. [DOI] [PubMed] [Google Scholar]
- Kato, T., N. Watanabe, Y. Morishima, A. Fujita, T. Ishizaki, and S. Narumiya. 2001. Localization of a mammalian homolog of diaphanous, mDia1, to the mitotic spindle in HeLa cells. J. Cell Sci. 114:775–784. [DOI] [PubMed] [Google Scholar]
- Kim, A.S., L.T. Kakalis, N. Abdul-Manan, G.A. Liu, and M.K. Rosen. 2000. a. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 404:151–158. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Z. Li, and D.B. Sacks. 2000. b. E-cadherin-mediated cell-cell attachment activates Cdc42. J. Biol. Chem. 275:36999–37005. [DOI] [PubMed] [Google Scholar]
- Koka, S., C.L. Neudauer, X. Li, R.E. Lewis, J.B. McCarthy, and J.J. Westendorf. 2003. The formin-homology-domain-containing protein FHOD1 enhances cell migration. J. Cell Sci. 116:1745–1755. [DOI] [PubMed] [Google Scholar]
- Kovar, D.R., J.R. Kuhn, A.L. Tichy, and T.D. Pollard. 2003. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labno, C.M., C.M. Lewis, D. You, D.W. Leung, A. Takesono, N. Kamberos, A. Seth, L.D. Finkelstein, M.K. Rosen, P.L. Schwartzberg, and J.K. Burkhardt. 2003. Itk functions to control actin polymerization at the immune synapse through localized activation of Cdc42 and WASP. Curr. Biol. 13:1619–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers, M., R. Rose, A. Scrima, and A. Wittinghofer. 2005. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 24:4176–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., and H.N. Higgs. 2003. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13:1335–1340. [DOI] [PubMed] [Google Scholar]
- Li, F., and H.N. Higgs. 2005. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 280:6986–6992. [DOI] [PubMed] [Google Scholar]
- Massol, P., P. Montcourrier, J.C. Guillemot, and P. Chavrier. 1998. Fc receptor-mediated phagocytosis requires CDC42 and Rac1. EMBO J. 17:6219–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, R.C., and L.M. Machesky. 2001. Phagocytosis and the actin cytoskeleton. J. Cell Sci. 114:1061–1077. [DOI] [PubMed] [Google Scholar]
- May, R.C., E. Caron, A. Hall, and L.M. Machesky. 2000. Involvement of the Arp2/3 complex in phagocytosis mediated by FcgammaR or CR3. Nat. Cell Biol. 2:246–248. [DOI] [PubMed] [Google Scholar]
- Otomo, T., C. Otomo, D.R. Tomchick, M. Machius, and M.K. Rosen. 2005. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell. 18:273–281. [DOI] [PubMed] [Google Scholar]
- Ozaki-Kuroda, K., Y. Yamamoto, H. Nohara, M. Kinoshita, T. Fujiwara, K. Irie, and Y. Takai. 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, J., O. Nielsen, R. Egel, and I.M. Hagan. 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141:1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., and J.A. Cooper. 1984. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 23:6631–6641. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Pring, M., M. Evangelista, C. Boone, C. Yang, and S.H. Zigmond. 2003. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 42:486–496. [DOI] [PubMed] [Google Scholar]
- Romero, S., C. Le Clainche, D. Didry, C. Egile, D. Pantaloni, and M.F. Carlier. 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 119:419–429. [DOI] [PubMed] [Google Scholar]
- Rose, R., M. Weyand, M. Lammers, T. Ishizaki, M.R. Ahmadian, and A. Wittinghofer. 2005. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 435:513–518. [DOI] [PubMed] [Google Scholar]
- Rudolph, M.G., P. Bayer, A. Abo, J. Kuhlmann, I.R. Vetter, and A. Wittinghofer. 1998. The Cdc42/Rac interactive binding region motif of the Wiskott Aldrich syndrome protein (WASP) is necessary but not sufficient for tight binding to Cdc42 and structure formation. J. Biol. Chem. 273:18067–18076. [DOI] [PubMed] [Google Scholar]
- Sagot, I., S.K. Klee, and D. Pellman. 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42–50. [DOI] [PubMed] [Google Scholar]
- Scott, C.C., W. Dobson, R.J. Botelho, N. Coady-Osberg, P. Chavrier, D.A. Knecht, C. Heath, P. Stahl, and S. Grinstein. 2005. Phosphatidylinositol-4,5-bisphosphate hydrolysis directs actin remodeling during phagocytosis. J. Cell Biol. 169:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth, A., T. Otomo, H.L. Yin, and M.K. Rosen. 2003. Rational design of genetically encoded fluorescence resonance energy transfer-based sensors of cellular Cdc42 signaling. Biochemistry. 42:3997–4008. [DOI] [PubMed] [Google Scholar]
- Sharpless, K.E., and S.D. Harris. 2002. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell. 13:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, J.A., and A.D. Hoppe. 2004. The coordination of signaling during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 76:1093–1103. [DOI] [PubMed] [Google Scholar]
- Wallar, B.J., and A.S. Alberts. 2003. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13:435–446. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., and C. Higashida. 2004. Formins: processive cappers of growing actin filaments. Exp. Cell Res. 301:16–22. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., T. Kato, A. Fujita, T. Ishizaki, and S. Narumiya. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136–143. [DOI] [PubMed] [Google Scholar]
- Xu, Y., J.B. Moseley, I. Sagot, F. Poy, D. Pellman, B.L. Goode, and M.J. Eck. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723. [DOI] [PubMed] [Google Scholar]
- Yayoshi-Yamamoto, S., I. Taniuchi, and T. Watanabe. 2000. FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol. Cell. Biol. 20:6872–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, S.H. 2004. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 16:99–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.