Abstract
Stromal interacting molecule 1 (STIM1), reported to be an endoplasmic reticulum (ER) Ca2+ sensor controlling store-operated Ca2+ entry, redistributes from a diffuse ER localization into puncta at the cell periphery after store depletion. STIM1 redistribution is proposed to be necessary for Ca2+ release–activated Ca2+ (CRAC) channel activation, but it is unclear whether redistribution is rapid enough to play a causal role. Furthermore, the location of STIM1 puncta is uncertain, with recent reports supporting retention in the ER as well as insertion into the plasma membrane (PM). Using total internal reflection fluorescence (TIRF) microscopy and patch-clamp recording from single Jurkat cells, we show that STIM1 puncta form several seconds before CRAC channels open, supporting a causal role in channel activation. Fluorescence quenching and electron microscopy analysis reveal that puncta correspond to STIM1 accumulation in discrete subregions of junctional ER located 10–25 nm from the PM, without detectable insertion of STIM1 into the PM. Roughly one third of these ER–PM contacts form in response to store depletion. These studies identify an ER structure underlying store-operated Ca2+ entry, whose extreme proximity to the PM may enable STIM1 to interact with CRAC channels or associated proteins.
Introduction
In many cells, agonists trigger biphasic Ca2+ signals in which release of Ca2+ from the ER is followed by a sustained influx of Ca2+ through store-operated channels (SOCs) in the plasma membrane. SOCs are activated by the depletion of Ca2+ from the ER, and their activity is essential for a diversity of functions in electrically nonexcitable cells, including secretion, motility, and gene expression. In T cells, antigen binding to the T cell receptor (TCR) elevates inositol 1,4,5-trisphosphate (IP3), depleting intracellular Ca2+ stores and thereby activating a highly Ca2+-selective SOC called the Ca2+ release–activated Ca2+ (CRAC) channel (Lewis, 2001). CRAC channels play a critical role in promoting gene expression during T cell activation, as shown by spontaneous mutations that abrogate Ca2+ influx and cause severe combined immunodeficiency in human patients (Partiseti et al., 1994; Feske et al., 2005, 2006).
Despite persistent research efforts over the past 15 yr, there is still a lack of consensus as to the molecular mechanism underlying store-operated Ca2+ entry (for reviews see Venkatachalam et al., 2002; Parekh and Putney, 2005). A major stumbling block in this search has been the lack of identified proteins that could be shown to be unequivocally required for the process. In a major advance, two groups recently used RNAi-based screens to identify stromal interacting molecule 1 (STIM1) as an essential, conserved regulator of store-operated Ca2+ entry (Liou et al., 2005; Roos et al., 2005). Current evidence suggests that STIM1 is the Ca2+ sensor for store depletion. STIM1 has an EF-hand domain predicted to face the ER lumen, and mutations of residues that are expected to reduce STIM1's affinity for Ca2+ constitutively activate store-operated Ca2+ entry in cells with full Ca2+ stores (Liou et al., 2005; Zhang et al., 2005). Interestingly, store depletion causes wild-type STIM1 to redistribute from a diffuse ER localization into puncta located at the cell periphery. This redistribution is thought to be an important step in SOC activation for two reasons. First, STIM1 molecules bearing EF-hand mutations are constitutively localized in these puncta, even when stores are full, consistent with their ability to constitutively activate Ca2+ entry (Liou et al., 2005; Zhang et al., 2005). Second, redistribution brings STIM1 toward the plasma membrane, suggesting a means by which STIM1 could transmit a store depletion signal to CRAC channels.
Several issues must be addressed to determine whether and how the redistribution of STIM1 is involved in opening CRAC channels. First, does STIM1 accumulate in puncta rapidly enough to be causally linked to CRAC channel activation? The kinetics of STIM1 redistribution has been reported in HeLa cells and Jurkat cells using total internal reflection fluorescence (TIRF) and confocal microscopy (Liou et al., 2005; Zhang et al., 2005). Although puncta formation is generally slow, like CRAC current (ICRAC) activation, quantitative comparisons have not been made between redistribution and ICRAC activation in single cells, which is essential for testing causality. Another critical area of uncertainty is the precise location of STIM1 puncta within the cell. Liou et al. (2005) concluded that puncta formed in the ER near the plasma membrane. They based their conclusions on the fact that YFP-labeled STIM1 was within the evanescent TIRF field of 100–200 nm but could not be detected by anti-GFP antibodies applied to the cell exterior. On the other hand, Zhang et al. (2005) concluded that store depletion causes STIM1 to enter the plasma membrane, based on immuno-EM and surface biotinylation studies.
The precise location of STIM1 after store depletion is central to understanding how store depletion is linked to SOC activation. Over the past decade, several classes of models have been proposed to describe how this communication might occur (Berridge, 2004; Parekh and Putney, 2005). These include conformational coupling between proteins in the ER and plasma membranes, possibly between STIM1 and the CRAC channel itself. In an extension of this model, referred to as “induced-coupling” or “secretion-like coupling,” the depleted ER would move to the cell periphery to establish these physical coupling sites (Venkatachalam et al., 2002). Store depletion may also promote vesicle fusion and insertion of CRAC channels or STIM1 into the plasma membrane. Finally, depletion could activate CRAC channels at a distance through the release of a diffusible messenger from the ER. Despite extensive studies by many groups, no single model has achieved widespread acceptance. Resolving the precise location of STIM1 in the cell and, in particular, its relationship to the ER and the plasma membrane is needed to more fully define STIM1's role in the store-operated Ca2+ entry process.
Unfortunately, neither light microscopy nor immuno-EM provide sufficient spatial resolution to determine whether STIM1 puncta form in the plasma membrane or in ER directly beneath it. We therefore approached this question in two ways. First, we examined the quenching of a GFP-labeled STIM1 using TIRF microscopy as a real-time assay for plasma membrane–localized STIM1. Second, we used HRP-labeled STIM1 to visualize STIM1 at the EM level. HRP-labeled proteins have been used to study subcellular localization with greater spatial resolution and better preservation of membrane structure than is possible with immuno-EM (Connolly et al., 1994; Dubois et al., 2001).
We report that STIM1 puncta formation slightly precedes CRAC channel opening in single cells, strongly supporting the idea that STIM1 redistribution is a critical early step in channel activation. Store depletion causes STIM1 to redistribute within the ER to form puncta in tubules near the plasma membrane without any detectable insertion of STIM1 into the plasma membrane on the time scale of CRAC channel activation. At the EM level, STIM1 puncta are found in both preexisting and newly docked junctional ER located 10–25 nm from the plasma membrane. The distance between STIM1 puncta and the plasma membrane is short enough to allow for the possibility of direct interactions between STIM1 and proteins at the cell surface. In Luik et al. (this issue, p. 815), we demonstrate that CRAC channels and Ca2+ entry occur exclusively at these junctional sites containing STIM1, thereby establishing a structural and functional basis for the elementary unit of store-operated Ca2+ entry.
Results
STIM1 puncta formation precedes CRAC channel activation in store-depleted cells
The redistribution of STIM1 from a diffuse ER localization into puncta at the cell periphery is widely assumed to be an important step in opening CRAC channels. Although STIM1 redistribution measured in cell populations has been shown to occur on average with roughly the same time course as CRAC channel activation, a direct comparison of STIM1 puncta formation and ICRAC activation has not been done (Liou et al., 2005; Zhang et al., 2005). If the formation of STIM1 puncta is necessary for CRAC channel activation, it must precede channel opening. Because the kinetics of CRAC channel activation and STIM1 redistribution vary among cells (and will vary independently if they are not coupled), the relative timing of these two processes can only be unequivocally determined in single-cell measurements. We therefore applied TIRF microscopy and whole-cell patch-clamp recording to monitor STIM1 redistribution and ICRAC activation simultaneously in individual Jurkat T cells transiently expressing a GFP-STIM1 chimera. GFP-STIM1 was made by inserting GFP into the lumenal domain of STIM1, between the N-terminal signal sequence and the EF-hand. GFP-STIM1 behaved similarly to wild-type STIM1 and to the fluorescent protein–labeled STIM1 constructs used by others (Liou et al., 2005; Zhang et al., 2005) in that GFP-STIM1 was localized to the ER in resting cells (Fig. 1 A and Figs. S1 A and S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200604014/DC1) but redistributed into bright puncta located at the cell periphery upon store depletion (Fig. 1 A, Fig. S1 A, Fig. S2 B, and Video 1).
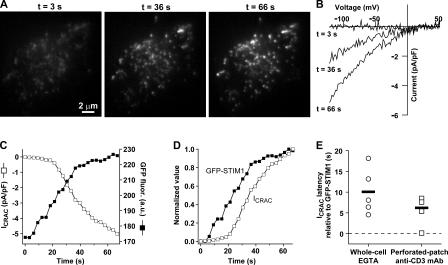
Figure 1.
STIM1 accumulates near the plasma membrane before CRAC channels open. (A) Jurkat cell expressing GFP-STIM1 imaged by TIRF microscopy during whole-cell recording with a pipette solution containing 1 mM EGTA. As stores become depleted, GFP-STIM1 redistributes into puncta near the plasma membrane as shown at the indicated times. (B) CRAC currents elicited by voltage ramps from −122 to +50 mV during the image acquisitions shown in A. Data were corrected for the leak current recorded at t = 0 s and expressed relative to cell size (capacitance). (C) Time courses of ICRAC development (measured at −122 mV) and GFP-STIM1 fluorescence (averaged across the entire cell footprint). (D) To compare the time courses of ICRAC activation and GFP-STIM1 fluorescence, the data from C were normalized to the same minimum and maximum values. (E) Accumulation of GFP-STIM1 near the plasma membrane precedes CRAC channel activation. The difference in times for ICRAC and GFP-STIM1 fluorescence to reach 20% of their maximum values is plotted for individual cells in response to passive depletion during whole-cell recording (left) or IP3-induced Ca2+ release during perforated-patch recording (right). IP3-induced Ca2+ release was triggered by cross-linking TCR with anti-CD3 mAb. The individual latencies (open circles; n = 5 cells) and mean latency (black bar) of 10 ± 2.5 s during passive depletion was comparable to the individual latencies (open squares; n = 4 cells) and mean latency (black bar) of 6.2 ± 1.8 s during IP3-induced Ca2+ release. The difference in the means was not statistically significant (t test: P = 0.28). The complete image and ICRAC sequences for A–D are shown in Video 2 (available at http://www.jcb.org/cgi/content/full/jcb.200604014/DC1). An individual cell response to TCR cross-linking is shown in Fig. S1.
1 mM EGTA was introduced into the GFP-STIM1–expressing cell through the recording pipette to deplete Ca2+ stores and activate ICRAC. The time course of ICRAC development was monitored during brief voltage steps to −122 mV delivered every 3 s. During each voltage step, a TIRF image was acquired to monitor GFP-STIM1 accumulation at sites within ∼200 nm of the plasma membrane. As reported previously in intact cells (Liou et al., 2005; Zhang et al., 2005), passive depletion of Ca2+ stores during whole-cell recording led to the appearance of GFP-STIM1 puncta near the plasma membrane (Fig. 1 A and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200604014/DC1). Inward current was activated in parallel and was identified as ICRAC on the basis of its characteristic slow kinetics, dependence on extracellular Ca2+, and inwardly rectifying current–voltage relation (Fig. 1 B). The mean density of ICRAC was increased by 78 ± 30% (transfected, n = 6; control, n = 14) in cells expressing low to moderate levels of GFP-STIM1 (or Cherry-STIM1; Luik et al., 2006), suggesting that the abundance of endogenous STIM1 may normally limit the extent of CRAC channel activation. GFP-STIM1 redistribution and ICRAC activation occurred over a period of ∼60 s after break-in to the whole-cell configuration (Fig. 1 C). A plot of each parameter on a normalized scale illustrates that GFP-STIM1 accumulated near the plasma membrane in this cell before CRAC channels began to open (Fig. 1 D). The relative timing of STIM1 accumulation and ICRAC induction was assessed from the times for each parameter to reach 20% of its maximal value. In five cells examined in this way, GFP-STIM1 fluorescence began to increase 10 ± 2.5 s before ICRAC increased, and in every cell, GFP-STIM1 redistribution preceded the initial increase in ICRAC (Fig. 1 E). Similar results were obtained using perforated-patch recording to avoid washout of intracellular constituents and a more physiological mode of stimulation in which an anti-CD3 mAb was used to cross-link the TCR and deplete stores via IP3-induced Ca2+ release (Fig. 1 E and Fig. S1). Under these conditions as well, STIM1 redistribution always preceded CRAC channel activation (mean latency of 6.2 ± 1.8 s; n = 4 cells). These results support the idea that STIM1 redistribution is required for CRAC channel activation.
STIM1 puncta that form after store depletion are intracellular
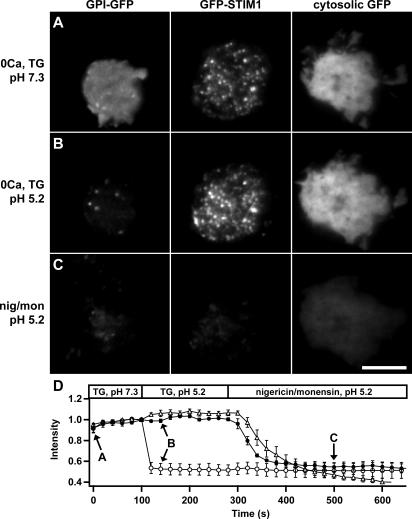
Determining the location of STIM1 after store depletion is critical to understanding how it activates CRAC channels. A recent study reported that store depletion causes STIM1 to translocate from the ER into the plasma membrane, where it might directly interact with or form CRAC channels (Zhang et al., 2005). To test whether STIM1 is inserted into the plasma membrane, we performed fluorescence-quenching experiments with GFP-STIM1. GFP fluorescence is pH sensitive, decreasing by >80% from pH 7.0 to 5.0 (Elsliger et al., 1999). If store depletion triggers GFP-STIM1 insertion into the plasma membrane, then the GFP will be exposed extracellularly and therefore be susceptible to quenching at low pH. We used TIRF imaging to compare the acid-induced quenching of GFP-STIM1 to that of cytosolic GFP and a glycosylphosphatidylinositol (GPI)-anchored GFP targeted to the outer leaflet of the plasma membrane. Fig. 2 and Video 3 (available at http://www.jcb.org/cgi/content/full/jcb.200604014/DC1) show TIRF images of Jurkat cells expressing GPI-GFP, GFP-STIM1, or cytosolic GFP. Ca2+ stores were depleted by incubating the cells in 0-Ca2+ Ringer's solution containing thapsigargin (TG), an ER Ca2+ pump inhibitor, for 10–15 min before the start of the experiment. Store depletion caused GFP-STIM1 to accumulate in bright puncta at the cell periphery, whereas the distributions of GPI-GFP and cytosolic GFP were unaffected (Fig. 2 A and Video 3). Acidifying the extracellular solution from pH 7.3 to 5.2 immediately quenched the fluorescence of the extracellular-facing GPI-GFP but had no effect on the fluorescence of GFP-STIM1 or cytosolic GFP (Fig. 2, B and D). GFP-STIM1 and cytosolic GFP fluorescence were quenched by low extracellular pH only after exposure to the ionophores nigericin and monensin, which equilibrate intracellular and extracellular pH (Fig. 2 C). Because significant quenching of GFP-STIM1 only occurred in the presence of ionophores (Fig. 2 D), we conclude that virtually all of the GFP-STIM1 is intracellular. Similar GFP-quenching experiments using wide-field epifluorescence microscopy confirmed the TIRF results (Fig. S2 and Video 4). To address the possibility that the large (239 amino acid) GFP tag may have interfered with trafficking to the plasma membrane, we performed similar experiments using STIM1 labeled with a much smaller (41 amino acid) N-terminal HA tag. After store depletion, HA immunofluorescence was detected in permeabilized cells but not in intact cells, demonstrating that HA-STIM1 is not inserted into the plasma membrane to any significant degree (Fig. S3). Therefore, we conclude that the STIM1 puncta that form after store depletion are intracellular and located near but not in the plasma membrane.
Figure 2.
GFP-STIM1 puncta in store-depleted cells are intracellular. Three Jurkat cells expressing GPI-GFP, GFP-STIM1, or cytosolic GFP were imaged at 20-s intervals by TIRF microscopy. The images in A–C were collected at the time points indicated by the arrows in D. (A) Cells were pretreated for 10–15 min with 0-Ca2+ Ringer's solution and 1 μM TG, pH 7.3, to deplete Ca2+ stores and redistribute GFP-STIM1 into puncta. (B) Perfusion with pH 5.2 solution immediately quenches the GPI-GFP fluorescence without changing GFP-STIM1 and cytosolic GFP fluorescence. (C) GFP-STIM1 and cytosolic GFP are quenched by the pH 5.2 solution only in the presence of nigericin and monensin, which permeabilize the cells and equilibrate the intracellular and extracellular pH. (D) The time course of mean fluorescence changes for cells expressing GPI-GFP (open circles; n = 7), GFP-STIM1 (closed circles; n = 5), or cytosolic GFP (open triangles; n = 3). Values are normalized to fluorescence at t = 100 s. The complete image sequence for the experiment in A–C is shown in Video 3 (available at http://www.jcb.org/cgi/content/full/jcb.200604014/DC1). Bar, 5 μm.
STIM1 puncta are located within the ER
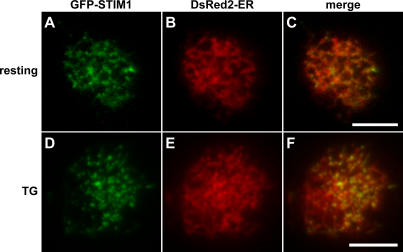
Given that the STIM1 puncta are intracellular, where do they form? To address this, we compared the localization of GFP-STIM1 to that of an ER marker (DsRed2-ER) in resting and store-depleted cells using TIRF microscopy. In Jurkat cells with full Ca2+ stores, GFP-STIM1 colocalized with the ER marker (Fig. 3, A–C), consistent with previous work (Liou et al., 2005; Zhang et al., 2005). After store depletion, GFP-STIM1 accumulated in puncta close to the plasma membrane (Fig. 3 D), and these puncta also overlapped with the ER marker (Fig. 3, E and F). These results suggest that rather than exiting the ER upon store depletion, GFP-STIM1 moves from being diffusely distributed within the ER to accumulate in puncta in areas of the ER near the plasma membrane.
Figure 3.
GFP-STIM1 colocalizes with an ER marker in control and store-depleted cells. Jurkat cells coexpressing GFP-STIM1 and the ER marker DsRed2-ER were imaged with TIRF microscopy before (A–C) and after (D–F) store depletion. In cells bathed in control Ringer's solution, GFP-STIM1 (A) colocalized with the ER marker (B) as shown in the merged image (C). In cells treated with 0-Ca2+ Ringer's containing 1 μM TG for 5 min to deplete Ca2+ stores, GFP-STIM1 redistributed into puncta near the plasma membrane (D). The GFP-STIM1 puncta also overlapped with DsRed2-ER (E) as shown in the merged image (F). Similar results were seen in 13 out of 13 cells. Bars, 5 μm.
STIM1 redistribution occurs without bulk movement of the ER
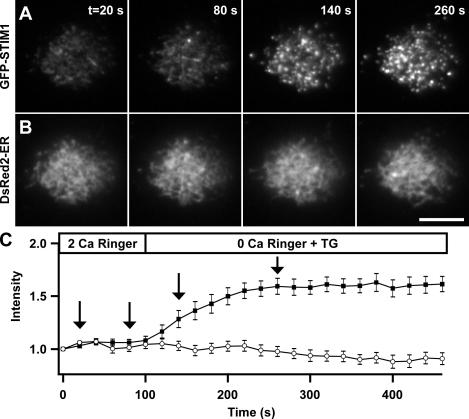
The formation of STIM1 puncta in areas of the ER close to the plasma membrane led us to consider the “induced coupling” model of store-operated Ca2+ entry previously proposed (Patterson et al., 1999; Venkatachalam et al., 2002). This model states that the ER moves to the plasma membrane to establish protein–protein interactions necessary for SOC activation. We used TIRF microscopy to image cells coexpressing GFP-STIM1 and DsRed2-ER to test this hypothesis. Because the intensity of the evanescent field decays exponentially with distance from the coverslip, any oriented movement of the ER toward the plasma membrane would be marked by increased DsRed fluorescence. After store depletion with TG, there was no detectable increase in DsRed2-ER fluorescence over the time period when GFP-STIM1 fluorescence increased dramatically (Fig. 4, A and B). The quantitation of results from 14 cells (Fig. 4 C) suggests that STIM1 accumulation near the plasma membrane does not involve bulk movements of the ER toward the plasma membrane. Thus, at the resolution of the light microscope, store depletion appears primarily to induce a redistribution of STIM1 within the ER.
Figure 4.
Store depletion drives STIM1 puncta formation without bulk ER movement toward the plasma membrane. TIRF images of a Jurkat cell coexpressing GFP-STIM1 and DsRed2-ER before and after store depletion. The images shown were collected at the times indicated by the arrows in C. Treatment with 1 μM TG in 0-Ca2+ Ringer's solution depleted Ca2+ stores and caused GFP-STIM1 to redistribute from a diffuse ER localization into bright puncta near the plasma membrane (A) without a significant increase in DsRed2-ER fluorescence (B). (C) After store depletion, GFP-STIM1 fluorescence (closed squares) increased without a corresponding increase in DsRed2-ER fluorescence (open circles) in the same cells (n = 14). Bar, 5 μm.
Ultrastructural analysis of STIM1 and ER distribution in resting and store-depleted cells
The TIRF imaging experiments suggest that store depletion causes GFP-STIM1 to redistribute from a diffuse localization in the ER to accumulate locally in the ER close to the plasma membrane, whereas the overall structure of the ER itself does not change. However, the resolution of light microscopy imposes certain limits on conclusions about localization. Even with the narrow optical sectioning afforded by TIRF microscopy, small rearrangements in ER structure may be difficult to detect against a large volume of fluorescent ER. Immuno-EM offers much better spatial resolution but often low contrast and poor preservation of membrane structure if cells are permeabilized to provide access to antibodies. Alternatively, the localization of HRP fusion proteins in the ER, Golgi, and transport vesicles has been achieved with very high contrast and without permeabilization, using peroxidase cytochemistry and conventional transmission EM (Connolly et al., 1994; Dubois et al., 2001). Because the HRP reaction product is restricted to the lumen of the HRP-containing compartment, ER-localized STIM1 should be clearly distinguishable from STIM1 in the plasma membrane. Therefore, we examined the subcellular distribution of STIM1 and the ER at the ultrastructural level using HRP-labeled STIM1 (HRP-STIM1) and an ER-targeted HRP (HRP-ER).
In a previous study, an ER-targeted HRP in transfected fibroblast and epithelial cells appeared in the nuclear envelope and in serpentine cytoplasmic tubules, consistent with localization in the ER (Connolly et al., 1994). In transfected Jurkat cells with full Ca2+ stores, both HRP-ER (Fig. 5, A and B) and HRP-STIM1 (Fig. 6, A–C) displayed a similar distribution, producing electron-dense HRP reaction product in the nuclear envelope as well as in tubules throughout the cytosol. These results confirm observations with light microscopy indicating that GFP-STIM1 is localized in the ER in cells with full stores (Fig. 3; Liou et al., 2005; Zhang et al., 2005) and confirm that the N terminus of STIM1 faces the ER lumen. Store depletion with TG did not grossly alter the ER morphology visualized with HRP-ER (Fig. 5, compare A and B with C and D), in agreement with our conclusions from TIRF measurements (Fig. 4).
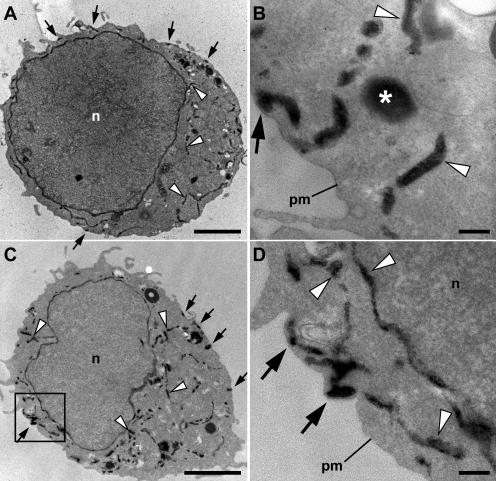
Figure 5.
Ultrastructural localization of ER in Jurkat cells before and after store depletion. Jurkat cells were transiently transfected with HRP-ER plasmid and processed for HRP cytochemistry and EM. (A and B) In resting cells with full Ca2+ stores, HRP-ER was located in the nuclear envelope and numerous cytoplasmic tubules (arrowheads), as well as in tubules closely associated with the plasma membrane (arrows). Large, round electron-dense structures (asterisks) were present in HRP-expressing cells as well as in untransfected cells, indicating that they are probably DAB-reactive peroxisomes or lysosomes. (C and D) In cells treated with 1 μM TG for 15 min to deplete stores, the overall distribution of HRP-ER was similar to that of resting cells, with dense reaction product present in both cytosolic and plasma membrane–associated tubules. The area outlined by the box in C is shown at higher magnification in D. n, nucleus; pm, plasma membrane. Bars: (A and C) 2 μm; (B and D) 200 nm.
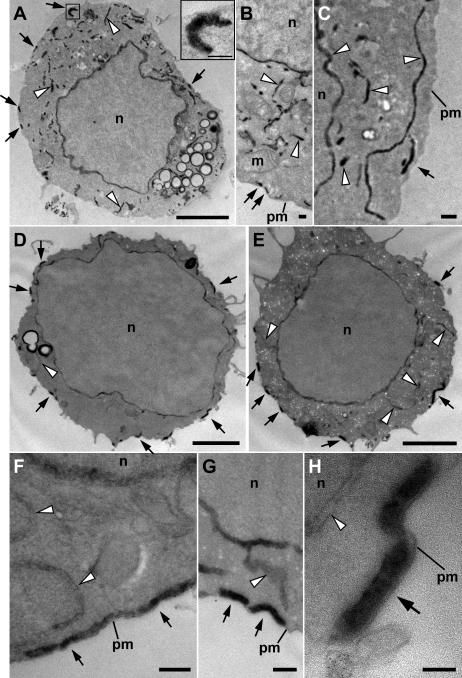
Figure 6.
Ultrastructural localization of HRP-STIM1 in control and store-depleted Jurkat cells. (A–C) In resting cells with full Ca2+ stores, HRP-STIM1 was localized in tubules throughout the cytoplasm, having a similar distribution as tubules marked by HRP-ER (Fig. 5, A and B). (D–H) In cells treated with 1 μM TG for 15 min to deplete stores, HRP-STIM1 was concentrated in tubules closely associated with the plasma membrane (arrows), whereas weak, diffuse staining was observed in cytoplasmic ER tubules (arrowheads). m, mitochondrion; n, nucleus; pm, plasma membrane. Bars: (A, D, and E) 2 μm; (A [inset], B, C, F, and G) 200 nm; (H) 100 nm.
In contrast, store depletion dramatically changed the distribution of HRP-STIM1. After treatment with TG, HRP-STIM1 shifted from being located in tubules throughout the cytoplasm (Fig. 6, A–C) to being concentrated in tubule segments that were closely associated with the plasma membrane (Fig. 6, D–H, arrows). HRP-STIM1 was clearly located in tubules rather than vesicles, and these tubules were similar in width and location to plasma membrane–associated ER tubules visualized with HRP-ER (Fig. 5, arrows). Consistent with a redistribution toward plasma membrane–associated ER, the residual HRP reaction product in cytosolic ER tubules after store depletion (Fig. 6, D–H, arrowheads) was greatly reduced compared with resting cells (Fig. 6, A–C, arrowheads). Importantly, in both resting and store-depleted cells, we failed to detect any extracellular HRP reaction product indicative of HRP-STIM1 in the plasma membrane, providing further evidence that STIM1 is not inserted into the plasma membrane in response to store depletion (Fig. 2, Fig. S2, and Fig. S3). Together, these results show that the peripheral STIM1 puncta seen by light microscopy in store-depleted cells correspond to the accumulation of STIM1 in plasma membrane–associated ER at the ultrastructural level.
Interestingly, we consistently observed in resting cells a small fraction of both HRP-ER and -STIM1 tubules in close apposition to the plasma membrane (Fig. 5, A and B; and Fig. 6, A–C, arrows). In terms of width and proximity to the plasma membrane, these ER–plasma membrane contact sites resemble the “junctional ER” described in many cell types (Gardiner and Grey, 1983; Takeshima et al., 2000; Baumann and Walz, 2001; Levine, 2004). The presence of junctional ER in resting Jurkat cells raises the question of whether STIM1 puncta form at these preexisting sites of ER–plasma membrane contact.
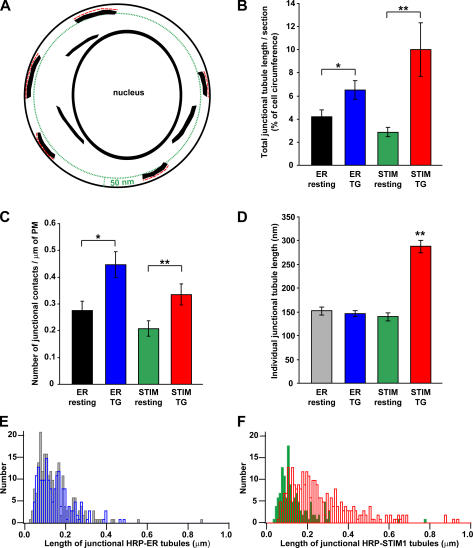
To begin to address this question, we asked whether store depletion affects the number or length of junctional ER structures, which we identified as HRP-ER tubules located within 50 nm of the plasma membrane (Fig. 7 A). Store depletion increased the total length of junctional ER by 55% (Fig. 7 B, blue vs. black), and this effect resulted from a 62% increase in the number of junctional ER contacts (Fig. 7 C) without a significant change in the lengths of individual contacts (Fig. 7, D and E). In cells overexpressing HRP-STIM1, depletion also increased the total length of junctional ER (Fig. 7 B, red vs. green); however, in these cells, the effect was attributed to a combined increase in both the number (Fig. 7 C) and the length of individual contacts (Fig. 7, D and F). Together, these results indicate that store depletion promotes the formation of new contact sites between the ER and the plasma membrane and causes STIM1 to accumulate at these sites as well as in preexisting regions of junctional ER.
Figure 7.
Store depletion enhances junctional contacts between the ER and the plasma membrane. Quantitative analysis of EM data from experiments like those in Figs. 5 and 6. (A) For each section where a nucleus was clearly visible, the lengths (red dashed lines) of all HRP-containing tubules located within 50 nm of the plasma membrane (green dotted line) were measured. (B) The total length of junctional HRP-tubules per section (normalized to the plasma membrane circumference) was averaged in HRP-ER–expressing cells with full (black; n = 17 cell sections) or depleted stores (blue; n = 15) and in HRP-STIM1–expressing cells with full (green; n = 16) or depleted stores (red; n = 21). The total length of junctional contacts increased after store depletion (t test: *, P = 0.02; **, P = 0.007). HRP-STIM1 overexpression did not significantly change the amount of junctional ER in resting cells (black vs. green). (C) Store depletion also increased the number of junctional contact sites (as measured by the number per micrometer of plasma membrane; t test: *, P = 0.006; **, P = 0.02). (D) Store depletion significantly increased the mean junctional tubule length in HRP-STIM1–expressing cells (ANOVA: **, P = 10−29; gray, n = 183 tubules; blue, n = 228; green, n = 133; red, n = 288; a total of 832 tubule lengths from 69 micrographs of cell sections were measured). (E) The distributions of the lengths of junctional HRP-ER tubules in resting cells (gray) and store-depleted cells (blue) were similar. (F) The lengths of junctional HRP-STIM1 tubules increased after store depletion (green, control; red, store depleted). All error bars show SEM. PM, plasma membrane.
The minimum distance between HRP-containing tubules and the plasma membrane was quantified in cells expressing HRP-ER and -STIM1 before and after store depletion. For every 200-nm length of tubule located within 50 nm of the plasma membrane, we measured the minimum distance from the proximal side of the tubule to the plasma membrane. Because there was no significant difference in these values among the different cells and conditions, the measurements were pooled, giving a mean minimum distance between junctional ER tubules and the plasma membrane of 17 ± 10 nm (mean ± SD; n = 293 measurements from 61 micrographs).
Discussion
Using a combination of TIRF microscopy and EM, we have determined the kinetics and location of STIM1 redistribution in response to Ca2+ store depletion in Jurkat T cells. STIM1 puncta formation always precedes the induction of ICRAC, consistent with a causal role for STIM1 redistribution in activating CRAC channels. We have established that STIM1 redistributes by moving within the ER to accumulate in discrete junctional ER tubules located within 10–25 nm of the plasma membrane. Store depletion increases the number of junctional ER contacts by 62% (such that one third of junctions in depleted cells are from newly formed and two thirds from preexisting contacts) but does not lead to any detectable insertion of STIM1 into the plasma membrane. These measurements describe a critical step in the control of store-operated Ca2+ entry and at the ultrastructural level define a structure that mediates the activation of CRAC channels in the plasma membrane (Luik et al., 2006).
The kinetics of STIM1 redistribution support a causal role in CRAC channel activation
For STIM1 redistribution to be directly involved in CRAC channel activation, it must precede channel opening. Previous studies have shown that the mean time course of STIM1 puncta formation in cell populations is slow and roughly parallels the activation of SOCs measured in different cells (Liou et al., 2005; Zhang et al., 2005). However, the precise timing of STIM1 accumulation and ICRAC could easily be obscured by the cell-to-cell variation within populations; for this reason, it is important to measure the two simultaneously in the same cell. In this study, combined patch-clamp and TIRF microscopy demonstrated that GFP-STIM1 puncta form with a time course slightly faster than that of ICRAC induction in the same cell (Fig. 1). These results support the idea that STIM1 redistribution is an early step in the CRAC activation process. Given that the increase in the TIRF signal reflects both the accumulation of STIM1 in junctional ER and to a lesser extent the formation of new junctional ER sites (Fig. 7), both the mobility of STIM1 within the ER membrane and the dynamics of ER movement to the plasma membrane may limit the speed with which CRAC channels respond to changes in ER [Ca2+], which takes tens of seconds even in response to rapid IP3-induced Ca2+ release (Hoth and Penner, 1993; Prakriya and Lewis, 2001). The 6–10-s latency between STIM1 accumulation and ICRAC activation suggests that there may be additional channel activation steps subsequent to redistribution or that a critical level of STIM1 or Ora1 (Luik et al., 2006) must be reached before activation can occur.
STIM1 redistributes into puncta within the ER without entering the plasma membrane
Initial studies came to two different conclusions regarding the redistribution of STIM1 after store depletion. Liou et al. (2005) reported that STIM1 accumulates in ER near the plasma membrane. Their conclusion was based on the fact that YFP-labeled STIM1 puncta were visible with TIRF microscopy, thus locating them within 100–200 nm of the plasma membrane. In addition, they failed to detect staining of the cell surface with antibodies that would be expected to react with YFP-STIM1 at the cell surface, although the data were not published. A second study, by Zhang et al. (2005), concluded that store depletion causes significant translocation of STIM1 from the ER into the plasma membrane, where it was proposed to stimulate Ca2+ entry by activating CRAC channels or by assembling with additional STIM1 monomers or other proteins to form functional CRAC channels at the cell surface.
In the present study, TIRF microscopy revealed colocalization of STIM1 puncta and ER labels, and at the ultrastructural level HRP-STIM1 was present in tubules having similar distribution, dimensions, and abundance to ER labeled with a targeted HRP. The EM results reveal directly for the first time the orientation of STIM1 in the ER, with the N-terminal half of the protein (containing HRP) clearly exposed to the ER lumen. In cells expressing GFP-, HA-, or HRP-STIM1, insertion of any of these proteins into the plasma membrane would have exposed the label (GFP, HA, or HRP) to the extracellular side. Thus, in cells treated with TG, the lack of any detectable low-pH quench of GFP (Fig. 2), the lack of positive anti-HA staining in unpermeabilized cells (Fig. S3), and the lack of HRP reaction product in the extracellular space detectable by EM (Fig. 6) all argue against a significant insertion of STIM1 into the plasma membrane in response to store depletion. For these reasons, we believe our results provide strong evidence that after store depletion, STIM1 remains in the ER and does not translocate into the plasma membrane.
Several possibilities may reconcile our conclusions with the results of Zhang et al. (2005), who based their argument for insertion on three lines of evidence. The first was immunofluorescence, in which the overlap of STIM1 with ER and cytosolic markers appeared to decrease after store depletion. A reduction of apparent colocalization would actually be expected after redistribution of STIM1 from central to peripheral ER, where the cell is thinner and the cytosol and ER less abundant than in the center of the cell. Second, immuno-EM with quantum dot–labeled secondary antibodies showed STIM1 reactivity over the plasma membrane. However, the spatial resolution offered by indirect immunolabeling is at most 25–30 nm (Jorgensen and Jones, 1987; Kimura et al., 1999), insufficient to rule out protein locations in ER located within 10–25 nm of the plasma membrane. Third, the ability of avidin to pull down STIM1 after biotinylation of surface proteins seemed to indicate that store depletion increased the amount of STIM1 in the plasma membrane. However, biotinylation of STIM1 itself was not verified; it is possible that STIM1 in the ER was binding to biotinylated plasma membrane proteins, and these interactions might be expected to increase as STIM1 accumulates in junctional ER. In addition, the biotinylation was performed after 30 min of treatment with sarco(endo)plasmic reticulum Ca2+ ATPase inhibitors, and prolonged depletion of stores is known to impair retention of ER resident proteins (Booth and Koch, 1989; Suzuki et al., 1991; Egan et al., 2002). There is evidence for STIM1 expression in the plasma membrane based on immunolabeling and biotinylation of K562 cells, although the same caveats about biotinylation apply to this study (Manji et al., 2000). It is important to note, however, that although some STIM1 may be expressed at the cell surface, our results show no detectable insertion of STIM1 into the plasma membrane on the time scale of CRAC channel opening, making it unlikely that insertion is a required step in the activation of store-operated Ca2+ entry.
STIM1 accumulates in preexisting and newly formed junctional ER
STIM1 puncta in store-depleted cells were identified at the EM level as regions of HRP-STIM1 accumulation in ER tubules lying in close proximity to the plasma membrane. Junctional ER has been observed before in many cell types, including Jurkat cells, Xenopus laevis oocytes, smooth muscle, neurons, and yeast, though a role in store-operated Ca2+ entry has never been established (Henkart et al., 1976; Gardiner and Grey, 1983; Takeshima et al., 2000; Pichler et al., 2001; Dadsetan et al., 2005). We found that store depletion causes STIM1 to accumulate in preexisting junctions (comprising approximately two thirds of the total junctions in depleted cells) as well as in newly formed junctions (comprising approximately one third; Fig. 7 C). Store depletion–induced formation of new junctional ER is reminiscent of the “induced coupling” or “secretion-like coupling” model for store-operated Ca2+ entry, in which depletion is thought to trigger movement of peripheral ER tubules to the plasma membrane, to allow ER proteins to activate plasma membrane SOCs through physical coupling (Patterson et al., 1999; Venkatachalam et al., 2002). This hypothesis was originally proposed to explain the ability of jasplakinolide and the phosphatase inhibitor calyculin A to block store-operated Ca2+ entry in smooth muscle cells (Patterson et al., 1999). Both compounds promote polymerization of cortical actin, which was proposed to create a physical barrier preventing the movement of ER to docking sites at the plasma membrane. Although jasplakinolide and calyculin A produce similar inhibitory effects in a variety of cells, including platelets, corneal epithelial cells, and pancreatic acinar cells (Rosado et al., 2005), they do not affect ICRAC in rat basophlic leukemia cells (Bakowski et al., 2001). Our observation of depletion-dependent formation of ER–plasma membrane contacts in Jurkat cells provides the first direct evidence in favor of the induced-coupling hypothesis. However, only about one third of junctional ER contacts in store-depleted Jurkat cells arise from this “induced coupling”; the majority of junctional sites are already present in resting cells, implying that most of the STIM1 puncta localize to preexisting junctional ER. Differences among cells in the relative amounts of preexisting and newly formed junctional ER may help explain why cortical actin polymerization inhibits SOC activation in some but not all cell types.
It is not known how junctional ER contacts are formed and maintained. However, the lengthening of junctional tubules by store depletion in cells overexpressing STIM1 (Fig. 7 D) suggests that STIM1 may help stabilize contacts. After store depletion, Ca2+ release by the EF-hand is likely to cause a conformational change in STIM1, as it does in other EF-hand–containing proteins (Herzberg et al., 1986; Zhang et al., 1995). This conformational change could lead to “trapping” and accumulation of Ca2+-free STIM1 in junctional ER by increasing STIM1's affinity for a protein in the plasma membrane. Similarly, Ca2+-free STIM1 could promote formation of new junctional contact sites by stabilizing moving extensions of the ER when they come into contact with plasma membrane target sites. Because store depletion increases the number of contact sites similarly, regardless of whether STIM1 is expressed at endogenous or much higher levels (Fig. 7 C), it seems unlikely that STIM1 alone is involved in forming new contact sites. Thus, the abundance of other proteins such as STIM1-interacting partners in the plasma membrane may limit the number of junctional contact sites. The existence of junctions in resting cells also suggests a role for proteins that may be independent of Ca2+ store content; one such candidate is junctate. Overexpression of junctate induces and/or stabilizes ER–plasma membrane contact sites in HEK293 cells, and knock down of junctate by RNAi reduces the amount of junctional ER (Treves et al., 2004). Further studies will be needed to test whether junctate plays a role and to identify other proteins that may be involved in forming new sites of junctional ER and in trapping STIM1 in puncta.
The close proximity of STIM1 puncta to the plasma membrane: implications for CRAC channel activation
We measured the minimum distance between HRP-containing tubules and the plasma membrane in Jurkat cells to be 10–25 nm. The redistribution of STIM1 into junctional ER located such a short distance from the plasma membrane suggests that STIM1 may communicate store depletion to CRAC channels via a local signal. Two types of signaling mechanisms are possible: STIM1 accumulation could trigger the release of a diffusible messenger, or STIM1 may interact directly with plasma membrane proteins. It is noteworthy that the cytoplasmic domain of STIM1 contains several putative protein interaction domains, including two α-helical coiled-coil domains and a proline-rich domain (Manji et al., 2000; Williams et al., 2001). Proteins in opposing membrane compartments can form stable complexes through coiled-coil interactions; a classic example involves the SNARE proteins, which mediate vesicle docking and fusion through formation of a four-stranded coiled-coil complex measuring ∼12 nm in length (Sutton et al., 1998). Thus, the 10–25-nm separation we have measured between ER tubules and the plasma membrane may be small enough to permit interactions between proteins in the two opposing membranes as well as rapid diffusion of a soluble messenger. Regardless of the nature of the signal, our results define a structure, i.e., STIM1 concentrated within junctional ER, that provides a possible substrate for local communication between the ER Ca2+ store and CRAC channels in the plasma membrane. Studies detailed in Luik et al. (2006) address the functional aspects of this structure and define in detail its spatial relationship to open CRAC channels.
Materials and methods
Plasmids, cells, and solutions
The GPI-GFP construct was a gift from D. Legler (University of Konstanz, Konstanz, Germany), and cytosolic GFP (pEGFP-C2) and pDsRed2-ER were obtained from BD Biosciences. GFP- and HRP-STIM1 plasmids were made by inserting EGFP or HRP at amino acid 39, just after the signal sequence of human STIM1 in pCMV6-XL5 (Origene). EGFP was amplified by PCR from pEGFP-C2, and HRP was amplified from HRP-pCR (a gift from M. Meyer and S. Smith, Stanford University, Stanford, CA), with primers that appended NarI restriction sites to the ends. Amplified, NarI-digested EGFP and HRP inserts were each ligated into a unique NarI site, engineered into STIM1 by site-directed mutagenesis (Quikchange XL; Stratagene) to produce GFP- and HRP-STIM1. HRP-ER was made by amplifying HRP by PCR with primers that appended AgeI restriction sites to the ends. Amplified, AgeI-digested HRP was ligated into a unique AgeI site located just after the signal sequence in pDsRed2-ER. HA-STIM1 was constructed by inserting a 41-amino-acid sequence encoding three HA epitopes in a series (HA-glycine-HA-glycine-HA, flanked by short linkers on both ends) at amino acid 35 of hSTIM1 in pCMV6-XL5. HA-STIM1 was then cut and ligated into the BamHI site in pIRES2-EGFP (BD Biosciences) so that HA-STIM1–positive cells could be identified by GFP coexpression.
Jurkat E6-1 cells (American Type Culture Collection) were grown as previously described (Prakriya and Lewis, 2001). Cells were transiently transfected with a gene pulser (Bio-Rad Laboratories) 48–72 h before experiments. For each transfection, 107 cells were centrifuged, resuspended in 300 μl of Jurkat medium, and mixed with 10 μg of DNA in a 0.4-cm gap cuvette. Cells were incubated with DNA for 10 min at room temperature, electroporated at 0.24 kV and 960 μF, incubated again for 10 min, and resuspended in 10 ml of fresh medium.
All salts and chemicals were obtained from Sigma-Aldrich unless otherwise stated. Control Ringer's solution contained 155 mM NaCl, 4.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM d-glucose, and 5 mM Hepes, pH 7.3 with NaOH. 0-Ca2+ Ringer's solution contained 155 mM NaCl, 4.5 mM KCl, 3 mM MgCl2, 10 mM d-glucose, 5 mM Hepes, and 1 mM EGTA, pH 7.3. For pH 5.2 0-Ca2+ Ringer's solution, CaCl2 was replaced with 1 mM EGTA, and Hepes was replaced with MES. Nigericin/monensin solution contained 70 mM NaCl and 70 mM KCl (to equilibrate extracellular and intracellular H+), 1.5 mM K2HPO4, 2 mM CaCl2, 1 mM MgSO4, 10 mM glucose, 10 mM Hepes, 10 mM MES, 0.01 mM nigericin, and 0.01 mM monensin, pH 5.2. TG was obtained from LC Laboratories; for TCR cross-linking experiments, 10 μg/ml OKT3, a murine mAb against human CD3 (eBioscience), was added to the extracellular solution.
Patch-clamp electrophysiology
Patch-clamp experiments were conducted in the standard whole-cell recording and perforated-patch configurations as previously described (Zweifach and Lewis, 1995; Bautista et al., 2002). The internal solution for whole-cell recording contained 140 mM Cs aspartate, 3 mM MgCl2, 1 mM EGTA, and 10 mM Hepes, pH 7.2 with CsOH. For perforated-patch experiments, the internal solution contained 115 mM Cs aspartate, 1 mM CaCl2, 5 mM MgCl2, 10 mM NaCl, 10 mM Hepes, pH 7.2 with CsOH, and 100 μg/ml amphotericin B. For all experiments, currents were filtered at 1 kHz and sampled at 2 kHz without series resistance compensation. Unless indicated, all data were leak subtracted using currents collected in nominally Ca2+-free bath solution plus 10 μM LaCl3.
Simultaneous TIRF imaging (see the following section) and patch-clamp measurements were performed using in-house software developed on the IgorPro platform interfaced with an input/output board (ADAC 1300; Molecular Devices) that simultaneously controlled the charge-coupled device (CCD) camera, laser shutter, and amplifier (Axopatch 200B; Molecular Devices). Images were acquired simultaneously with a 100-ms step to −122 mV followed by a 100-ms ramp from −122 to +50 mV.
Epifluorescence and TIRF microscopy
Transiently transfected Jurkat cells were plated onto poly-l-lysine–coated coverslip chambers and imaged with epifluorescence or through-the-objective TIRF microscopy using a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) with a Fluar 100×, 1.45 NA oil-immersion objective (Carl Zeiss MicroImaging, Inc.). Laser light (488 or 514.5 nm) from a 100-mW argon ion laser (Enterprise 622; Coherent) was coupled to the microscope via optical fiber (Point Source) and focused at the back focal plane of the objective using a TILL-TIRF condenser (Till Photonics) that employs a translatable 45° prism to control the incident angle of the laser beam. For epifluorescence microscopy, cells were excited with 488-nm (GFP) or 540-nm (rhodamine) light using a monochromator (Polychrome II; Till Photonics). GFP and DsRed/rhodamine images were acquired by switching between GFP (z488/10 excitation, Q505LP dichroic, and 535/30 emission) and DsRed/rhodamine (HQ535/45 excitation, Q570LP dichroic, and D660/50 emission) filter cubes (Chroma Technology Corp.). For simultaneous patch-clamp and TIRF imaging experiments, a 1.0 neutral density filter was added in front of the GFP excitation filter and the 535/30 emission filter was replaced with a 510LP. Images were acquired with a cooled CCD camera (ORCA-ER; Hamamatsu). Filter switching and image acquisition were controlled by scripts written in MetaMorph (Molecular Devices). All fluorescence images were acquired with 2 × 2 pixel binning except for those in Fig. 3 (1 × 1). Video 2 was acquired at 3-s frame intervals, and other videos were acquired at 20-s frame intervals. All experiments were performed at room temperature (22–25°C).
EM
After transient transfection with HRP-STIM1 or -ER plasmids, Jurkat cells were plated onto poly-l-lysine–coated coverslips and fixed with 2% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M Na cacodylate buffer (Electron Microscopy Sciences) in a microwave oven (EM3450; Pelco) equipped with a cold spot set at 15°C. Fixed cells were amplified with a TSA-biotin system (PerkinElmer) and ABC kit (Vector Laboratories) for 30 min each before being prereacted with 1 mg/ml DAB (Sigma-Aldrich) in Tris-buffered saline for 10 min and then reacted with DAB with 0.01% H2O2 for 30 min. After postfixation with 1% OsO4 and en bloc stain with 1% uranyl acetate, cells were further processed as previously described (Ahmari et al., 2000) before embedding in Embed 812 (Electron Microscopy Sciences). Glass coverslips were removed, cells were located by light microscopy and punched out, and 50–90-nm sections were cut and mounted on formvar-coated grids and viewed with an 80-kV transmission electron microscope (model 1230; JEOL) equipped with a slow-scan cooled CCD camera (model 967; Gatan).
Immunofluorescence
HA-STIM1 and untransfected Jurkat cells were plated onto poly-l-lysine–coated coverslips for 45 min at 37°C, treated with either control Ringer's or 0-Ca Ringer's plus 1 μM TG for 15 min at room temperature, fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in 10 mM PBS (Sigma-Aldrich) for 30 min at room temperature, and washed with PBS plus 20 mM glycine. Selected coverslips were permeabilized with 0.1% Triton X-100 for 5 min before blocking for 1.5 h in PBS plus 2% BSA, 20 mM glycine, and 75 mM NH4Cl. Cells were incubated with each antibody diluted in blocking buffer (1 μg/ml 12ca5 mouse anti-HA [a gift from A. Venteicher, Stanford University, Stanford, CA] and 30 μg/ml rhodamine red-X goat anti–mouse IgG [Jackson ImmunoResearch Laboratories]) for 1 h each at room temperature and washed thoroughly with blocking buffer in between incubations. Coverslips were mounted onto glass slides with Vectashield (Vector Laboratories).
Image analysis
Image analysis was performed using NIH ImageJ software; figures were prepared using Photoshop and Illustrator (Adobe). All raw fluorescence images were dark-noise subtracted before analysis. Images in Fig. 1 and Fig. S1 A were thresholded to three to four times the remaining background. To quantitate fluorescence intensities of GFP- and DsRed-labeled proteins, images were thresholded, and only data from pixels above the threshold were quantified. The threshold was set at the minimum pixel intensity + 10% of the range of pixel intensities for that image (threshold = min + 10%[max – min]). Because the footprints of Jurkat cells occasionally increased during an experiment, we limited intensity measurements to an outline drawn around the cell footprint in the first frame of a time-lapse experiment. For each GFP-quenching video, the mean fluorescence intensity of a cell in each frame of the video was normalized to the mean fluorescence intensity of that cell in the frame just before the switch from pH 7.3 to 5.2 (Fig. 2, t = 100 s; Fig. S2, t = 540 s). In Fig. 4, the mean fluorescence intensity of a cell in each frame of a video was normalized to the mean fluorescence intensity of that cell in the first frame of the movie. Any adjustments made to brightness and contrast levels were made across the entire fluorescence or EM image or the entire video.
To measure lengths of HRP-tubules in close contact with the plasma membrane (≤50 nm; Fig. 7 A), we selected only micrographs where the nucleus and entire cell circumference were visible (taken at 8,000–25,000× magnification), to restrict our analysis to sections cut through the middle rather than the edges of cells. To estimate the mean minimum distance between HRP-tubules and the cell membrane, we measured the distances between the proximal edge of HRP-tubules and the cell membrane using micrographs taken at 25,000–150,000×. We measured the minimum distance from the plasma membrane for every 200 nm of HRP-containing tubule located within 50 nm of the plasma membrane. Means are expressed ±SEM unless otherwise stated; statistical significance was measured by the t test or analysis of variance (ANOVA).
Online supplemental material
Fig. S1 shows that STIM1 accumulation precedes ICRAC activation during IP3-induced Ca2+ release. Fig. S2 shows fluorescence-quenching measurements using wide-field epifluorescence microscopy to assay for plasma membrane–localized GFP-STIM1. Fig. S3 shows immunofluorescence staining of HA-tagged STIM1 in Jurkat cells before and after store depletion. Video 1 shows GFP-STIM1 redistribution in an intact cell after store depletion with TG. Video 2 shows STIM1 accumulation near the plasma membrane during ICRAC activation. Video 3 shows assaying for plasma membrane–localized GFP-STIM1 puncta by fluorescence quenching and TIRF microscopy. Video 4 shows assaying for plasma membrane–localized GFP-STIM1 puncta by fluorescence quenching and wide-field epifluorescence microscopy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200604014/DC1.
Supplementary Material
Acknowledgments
We thank Stephen Smith and his laboratory and John Perrino and the Beckman Center EM core facility for help with EM experiments; Daniel Legler, Martin Meyer, Stephen Smith, and Paul Hoover for plasmids; Andrew Venteicher for the anti-HA antibody; Pam Pappone and Brian Peter for comments on the manuscript; and members of the Lewis Laboratory for stimulating discussions.
This work was supported by National Institutes of Health National Research Service Award grant 5T32AI07290-21 (to M.M. Wu), Stanford Graduate Fellowship and National Institutes of Health training grant GM007276 (to R.M. Luik), National Institutes of Health grant GM45374 (to R.S. Lewis), and a gift from the Mathers Charitable Foundation (to R.S. Lewis).
Abbreviations used in this paper: CCD, charge-coupled device; CRAC, Ca2+ release–activated Ca2+; GPI, glycosylphosphatidylinositol; ICRAC, CRAC current; IP3, inositol 1,4,5-trisphosphate; SOC, store-operated channel; STIM1, stromal interacting molecule 1; TCR, T cell receptor; TG, thapsigargin; TIRF, total internal reflection fluorescence.
References
- Ahmari, S.E., J. Buchanan, and S.J. Smith. 2000. Assembly of presynaptic active zones from cytoplasmic transport packets. Nat. Neurosci. 3:445–451. [DOI] [PubMed] [Google Scholar]
- Bakowski, D., M.D. Glitsch, and A.B. Parekh. 2001. An examination of the secretion-like coupling model for the activation of the Ca2+ release-activated Ca2+ current ICRAC in RBL-1 cells. J. Physiol. 532:55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, O., and B. Walz. 2001. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205:149–214. [DOI] [PubMed] [Google Scholar]
- Bautista, D.M., M. Hoth, and R.S. Lewis. 2002. Enhancement of calcium signalling dynamics and stability by delayed modulation of the plasma-membrane calcium-ATPase in human T cells. J. Physiol. 541:877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge, M. 2004. Conformational coupling: a physiological calcium entry mechanism. Sci. STKE. 2004:pe33. [DOI] [PubMed] [Google Scholar]
- Booth, C., and G.L. Koch. 1989. Perturbation of cellular calcium induces secretion of luminal ER proteins. Cell. 59:729–737. [DOI] [PubMed] [Google Scholar]
- Connolly, C.N., C.E. Futter, A. Gibson, C.R. Hopkins, and D.F. Cutler. 1994. Transport into and out of the Golgi complex studied by transfecting cells with cDNAs encoding horseradish peroxidase. J. Cell Biol. 127:641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadsetan, S., V. Shishkin, and A.F. Fomina. 2005. Intracellular Ca2+ release triggers translocation of membrane marker FM1-43 from the extracellular leaflet of plasma membrane into endoplasmic reticulum in T lymphocytes. J. Biol. Chem. 280:16377–16382. [DOI] [PubMed] [Google Scholar]
- Dubois, L., M. Lecourtois, C. Alexandre, E. Hirst, and J.P. Vincent. 2001. Regulated endocytic routing modulates Wingless signaling in Drosophila embryos. Cell. 105:613–624. [DOI] [PubMed] [Google Scholar]
- Egan, M.E., J. Glockner-Pagel, C. Ambrose, P.A. Cahill, L. Pappoe, N. Balamuth, E. Cho, S. Canny, C.A. Wagner, J. Geibel, and M.J. Caplan. 2002. Calcium-pump inhibitors induce functional surface expression of ΔF508-CFTR protein in cystic fibrosis epithelial cells. Nat. Med. 8:485–492. [DOI] [PubMed] [Google Scholar]
- Elsliger, M.A., R.M. Wachter, G.T. Hanson, K. Kallio, and S.J. Remington. 1999. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry. 38:5296–5301. [DOI] [PubMed] [Google Scholar]
- Feske, S., M. Prakriya, A. Rao, and R.S. Lewis. 2005. A severe defect in CRAC Ca2+ channel activation and altered K+ channel gating in T cells from immunodeficient patients. J. Exp. Med. 202:651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske, S., Y. Gwack, M. Prakriya, S. Srikanth, S.H. Puppel, B. Tanasa, P.G. Hogan, R.S. Lewis, M. Daly, and A. Rao. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 441:179–185. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M., and R.D. Grey. 1983. Membrane junctions in Xenopus eggs: their distribution suggests a role in calcium regulation. J. Cell Biol. 96:1159–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkart, M., D.M. Landis, and T.S. Reese. 1976. Similarity of junctions between plasma membranes and endoplasmic reticulum in muscle and neurons. J. Cell Biol. 70:338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg, O., J. Moult, and M.N. James. 1986. A model for the Ca2+-induced conformational transition of troponin C. A trigger for muscle contraction. J. Biol. Chem. 261:2638–2644. [PubMed] [Google Scholar]
- Hoth, M., and R. Penner. 1993. Calcium release-activated calcium current in rat mast cells. J. Physiol. 465:359–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, A.O., and L.R. Jones. 1987. Immunoelectron microscopical localization of phospholamban in adult canine ventricular muscle. J. Cell Biol. 104:1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, S., W. Laosinchai, T. Itoh, X. Cui, C.R. Linder, and R.M. Brown Jr. 1999. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell. 11:2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, T. 2004. Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol. 14:483–490. [DOI] [PubMed] [Google Scholar]
- Lewis, R.S. 2001. Calcium signaling mechanisms in T lymphocytes. Annu. Rev. Immunol. 19:497–521. [DOI] [PubMed] [Google Scholar]
- Liou, J., M.L. Kim, W. Do Heo, J.T. Jones, J.W. Myers, J.E. Ferrell Jr., and T. Meyer. 2005. STIM Is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15:1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik, R.M., M.M. Wu, J. Buchanan, and R.S. Lewis. 2006. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J. Cell Biol. 174:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji, S.S., N.J. Parker, R.T. Williams, L. van Stekelenburg, R.B. Pearson, M. Dziadek, and P.J. Smith. 2000. STIM1: a novel phosphoprotein located at the cell surface. Biochim. Biophys. Acta. 1481:147–155. [DOI] [PubMed] [Google Scholar]
- Parekh, A.B., and J.W. Putney Jr. 2005. Store-operated calcium channels. Physiol. Rev. 85:757–810. [DOI] [PubMed] [Google Scholar]
- Partiseti, M., F. Le Deist, C. Hivroz, A. Fischer, H. Korn, and D. Choquet. 1994. The calcium current activated by T cell receptor and store depletion in human lymphocytes is absent in a primary immunodeficiency. J. Biol. Chem. 269:32327–32335. [PubMed] [Google Scholar]
- Patterson, R.L., D.B. van Rossum, and D.L. Gill. 1999. Store-operated Ca2+ entry: evidence for a secretion-like coupling model. Cell. 98:487–499. [DOI] [PubMed] [Google Scholar]
- Pichler, H., B. Gaigg, C. Hrastnik, G. Achleitner, S.D. Kohlwein, G. Zellnig, A. Perktold, and G. Daum. 2001. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268:2351–2361. [DOI] [PubMed] [Google Scholar]
- Prakriya, M., and R.S. Lewis. 2001. Potentiation and inhibition of Ca2+ release-activated Ca2+ channels by 2-aminoethyldiphenyl borate (2-APB) occurs independently of IP3 receptors. J. Physiol. 536:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos, J., P.J. DiGregorio, A.V. Yeromin, K. Ohlsen, M. Lioudyno, S. Zhang, O. Safrina, J.A. Kozak, S.L. Wagner, M.D. Cahalan, et al. 2005. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado, J.A., P.C. Redondo, S.O. Sage, J.A. Pariente, and G.M. Salido. 2005. Store-operated Ca2+ entry: vesicle fusion or reversible trafficking and de novo conformational coupling? J. Cell. Physiol. 205:262–269. [DOI] [PubMed] [Google Scholar]
- Sutton, R.B., D. Fasshauer, R. Jahn, and A.T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature. 395:347–353. [DOI] [PubMed] [Google Scholar]
- Suzuki, C.K., J.S. Bonifacino, A.Y. Lin, M.M. Davis, and R.D. Klausner. 1991. Regulating the retention of T-cell receptor α chain variants within the endoplasmic reticulum: Ca2+-dependent association with BiP. J. Cell Biol. 114:189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima, H., S. Komazaki, M. Nishi, M. Iino, and K. Kangawa. 2000. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell. 6:11–22. [DOI] [PubMed] [Google Scholar]
- Treves, S., C. Franzini-Armstrong, L. Moccagatta, C. Arnoult, C. Grasso, A. Schrum, S. Ducreux, M.X. Zhu, K. Mikoshiba, T. Girard, et al. 2004. Junctate is a key element in calcium entry induced by activation of InsP3 receptors and/or calcium store depletion. J. Cell Biol. 166:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam, K., D.B. van Rossum, R.L. Patterson, H.T. Ma, and D.L. Gill. 2002. The cellular and molecular basis of store-operated calcium entry. Nat. Cell Biol. 4:E263–E272. [DOI] [PubMed] [Google Scholar]
- Williams, R.T., S.S. Manji, N.J. Parker, M.S. Hancock, L. Van Stekelenburg, J.P. Eid, P.V. Senior, J.S. Kazenwadel, T. Shandala, R. Saint, et al. 2001. Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem. J. 357:673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., T. Tanaka, and M. Ikura. 1995. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 2:758–767. [DOI] [PubMed] [Google Scholar]
- Zhang, S.L., Y. Yu, J. Roos, J.A. Kozak, T.J. Deerinck, M.H. Ellisman, K.A. Stauderman, and M.D. Cahalan. 2005. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 437:902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifach, A., and R.S. Lewis. 1995. Rapid inactivation of depletion-activated calcium current (ICRAC) due to local calcium feedback. J. Gen. Physiol. 105:209–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.