Abstract
Three-dimensional images of the undercoat structure on the cytoplasmic surface of the upper cell membrane of normal rat kidney fibroblast (NRK) cells and fetal rat skin keratinocytes were reconstructed by electron tomography, with 0.85-nm–thick consecutive sections made ∼100 nm from the cytoplasmic surface using rapidly frozen, deeply etched, platinum-replicated plasma membranes. The membrane skeleton (MSK) primarily consists of actin filaments and associated proteins. The MSK covers the entire cytoplasmic surface and is closely linked to clathrin-coated pits and caveolae. The actin filaments that are closely apposed to the cytoplasmic surface of the plasma membrane (within 10.2 nm) are likely to form the boundaries of the membrane compartments responsible for the temporary confinement of membrane molecules, thus partitioning the plasma membrane with regard to their lateral diffusion. The distribution of the MSK mesh size as determined by electron tomography and that of the compartment size as determined from high speed single-particle tracking of phospholipid diffusion agree well in both cell types, supporting the MSK fence and MSK-anchored protein picket models.
Introduction
The portion of the cytoskeleton that is closely associated with the cytoplasmic surface of the plasma membrane is often called the membrane skeleton (MSK; Heuser and Kirschner, 1980; Hirokawa and Heuser, 1981; Bennett, 1990; Luna and Hitt, 1992). The term MSK is useful partly because this part of the cytoskeleton is expected to differ from the bulk cytoskeleton in terms of its structure and protein composition, for its interactions with the plasma membrane in general and with specific molecules in the plasma membrane, and also because it plays important roles in a variety of membrane functions. It is involved in the localization of transmembrane proteins at specific sites in the cell membrane (Bennett and Chen, 2001; Pan et al., 2006) and in endocytosis and exocytosis (Gaidarov et al., 1999; Valentijn et al., 2000) in various cell types. It also provides the plasma membrane with the mechanical strength and resilience to withstand the stress and shear forces from the outside environment, which is well established in the thick cortical actin layers in immune cells (Hartwig and Yin, 1988) and in the spectrin–actin network in red blood cells (Mohandas and Evans, 1994; Discher et al., 1995). Therefore, the MSK works as a part of the plasma membrane as well as a part of the cytoskeleton. It is a truly interfacial structure between the bulk cytoskeleton and the 2D bilayer of the plasma membrane.
Recently, a new function of the MSK has become apparent. It was proposed that a part of the MSK is directly and closely associated with the cytoplasmic surface of the plasma membrane, and this part induces partitioning of the cell membrane with regard to the translational diffusion of membrane molecules based on high speed single-particle tracking data on membrane proteins and lipids (Jacobson et al., 1995; Kusumi et al., 2005). In the short-time regime, these membrane molecules are temporarily confined within the compartments delimited by the MSK mesh, and, in the long-time regime, they undergo macroscopic diffusion by hopping between these compartments (MSK fence model). In the fence model, as a result of the collision of the cytoplasmic domains of transmembrane proteins with the MSK, transmembrane proteins are temporarily confined in the MSK mesh (Sheetz, 1983; Tsuji and Ohnishi, 1986; Tsuji et al., 1988; Saxton, 1989, 1990; Sako and Kusumi, 1994, 1995; Jacobson et al., 1995; Kusumi and Sako, 1996; Saxton and Jacobson, 1997; Sako et al., 1998; Tomishige et al., 1998; Suzuki et al., 2005).
Lipid molecules also undergo hop diffusion, which might be explained by the anchored protein picket model (Fujiwara et al., 2002; Murase et al., 2004; Kusumi et al., 2005). In this model, various transmembrane proteins anchored to the actin-based MSK might effectively act as rows of pickets against the free diffusion of all of the molecules incorporated in the cell membrane as a result of steric hindrance and circumferential slowing (a hydrodynamic frictionlike effect, which propagates quite far from the immobile protein surface; without this effect, pickets will not be effective for blocking diffusion; Bussell et al., 1994, 1995) of the immobile picket proteins anchored to and lined up along the MSK. Lipid movement is affected only by pickets, whereas both pickets and fences would act on transmembrane proteins. These MSK picket-fence effects would be dramatically enhanced when the membrane receptor molecules form signaling complexes upon ligand binding as a result of receptor oligomerization and/or binding of the cytoplasmic signaling molecules to the receptor, leading to the trapping of signaling complexes in the MSK mesh, where the extracellular signal is received. This would enable spatial confinement and regulation of the downstream signaling events (Kusumi and Sako, 1996; Iino et al., 2001).
Despite the importance of the MSK functions and the long history of its study using EM (Byers and Porter, 1977; Heuser and Kirschner, 1980; Hirokawa and Heuser, 1981; Heuser and Anderson, 1989; Hartwig and DeSisto, 1991; Rothberg et al., 1992), our knowledge of its structure and the overall distribution over the plasma membrane has been very limited. For example, we do not know whether the MSK exists everywhere on the cytoplasmic surface of the cell membrane, how extensive the spatial variations of MSK mesh size is, and whether and how MSK interacts with other structures in the plasma membrane such as clathrin-coated pits (CCPs), caveolae, and cell adhesion structures. Even the structure of the MSK of the human red blood cell ghost, a traditional paradigm for MSK studies, is not satisfactorily understood (Sheetz and Sawyer, 1978; Tsukita et al., 1980; Branton et al., 1981; Shen et al., 1986; Ursitti et al., 1991; Takeuchi et al., 1998).
In this study, to further advance our understanding of the MSK structure and function, we observed the undercoat structure on the cytoplasmic surface of the plasma membrane of cultured mammalian cells using rapid-freeze, deep-etch, immunoreplication EM. We paid special attention to the following three points.
First, we tried to consistently prepare and observe large plasma membrane fragments (>10 μm in diameter) to facilitate inspections of very large plasma membrane areas. Almost all of the previous MSK studies, including those cited above, investigated the ultrastructural features of the structure of interest, but within a very limited view field. By observing these large membrane surfaces, the spatial variations of the MSK mesh size and of the number density of CCPs and caveolae can be reliably examined.
Second, the 3D reconstruction of the undercoat structure within 100 nm from the cytoplasmic surface of the plasma membrane was performed using electron tomography for the platinum-replicated samples: 97–141 images for a specimen tilted at different angles (every 1°) with respect to the incident electron beam in the range of ±48–70° were obtained and then converted to 100–121 sliced images of every 0.85–1.34 nm for the 3D reconstructed images (Perkins et al., 1997; Medalia et al., 2002; Lucic et al., 2005; McIntosh et al., 2005; Zeuschner et al., 2006).
Third, using the 3D reconstructed images of the MSK structure within 13.6 nm (16 slices of 0.85-nm thickness) from the cytoplasmic surface, the MSK mesh size distribution on the cytoplasmic surface of the plasma membrane was determined. This part of the MSK, which is closely associated with the cytoplasmic surface of the plasma membrane, might form the compartment boundaries for partitioning of the plasma membrane for the diffusion of membrane molecules, thus determining the compartment size. Therefore, it is interesting to compare the distribution of the MSK mesh size on the membrane determined this way and that of the compartment size sensed by membrane molecules. Because the compartment size distributions for membrane molecules are very different between normal rat kidney fibroblast (NRK; median = 230 nm) and fetal rat skin keratinocyte (FRSK; median = 41 nm) cell lines (Fujiwara et al., 2002; Murase et al., 2004), the distribution of the MSK mesh size on the membrane surface was examined using these two cell lines. Although the compartment size is very different between these cell lines, within each cell type, the histogram for the MSK mesh size on the membrane surface is very similar to that for the diffusion compartment size. This strongly supports the MSK fence and MSK-anchored transmembrane protein picket models.
Results
Bird's-eye view of the undercoat structure of the upper cell membrane
Glass coverslips precationized by a treatment with Alcian blue were placed on top of the cells cultured in 35-mm plastic dishes and were allowed to settle and attach to the upper cell membrane at 4°C for 15 min. The buffer containing 1% PFA and 0.25% glutaraldehyde was then added to the space between the coverslip and the plastic bottom of the culture dish. As the coverslip was floated apart, the cells were ruptured and the upper cell membrane was retained, still adhering to the overlaying Alcian blue–coated coverslip. The upper membrane was rapidly frozen by pressing its exposed cytoplasmic surface onto a pure copper block precooled by liquid helium. The frozen sample was deep etched, coated with platinum-carbon, and observed under an electron microscope. We have made extensive efforts to reproducibly prepare and observe large cell membrane fragments >10 μm in diameter.
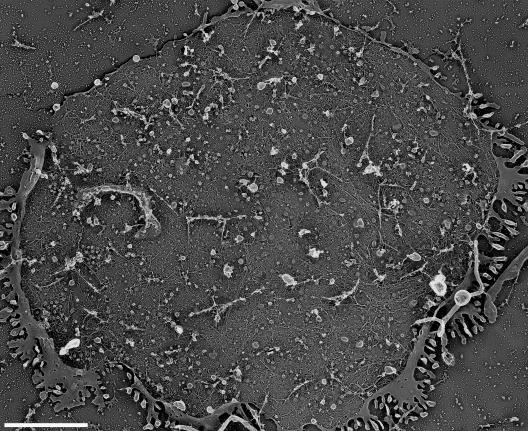
Fig. 1 is a typical electron micrograph providing a bird's-eye view of the cytoplasmic surface of a large area of the upper cell membrane of a cultured NRK cell. Many such EM images showing the cytoplasmic surfaces of large cell membrane fragments were obtained for NRK and FRSK cells, suggesting that the entire (upper) plasma membrane, except for the places where CCPs and caveolae exist, is coated with the filamentous netlike structure.
Figure 1.
A bird's-eye view of the large cytoplasmic surface of the upper cell membrane (the membrane facing the buffer rather than the coverslip) of an NRK cell observed by rapid-freeze, deep-etch, freeze-replica EM. Bar, 2.5 μm.
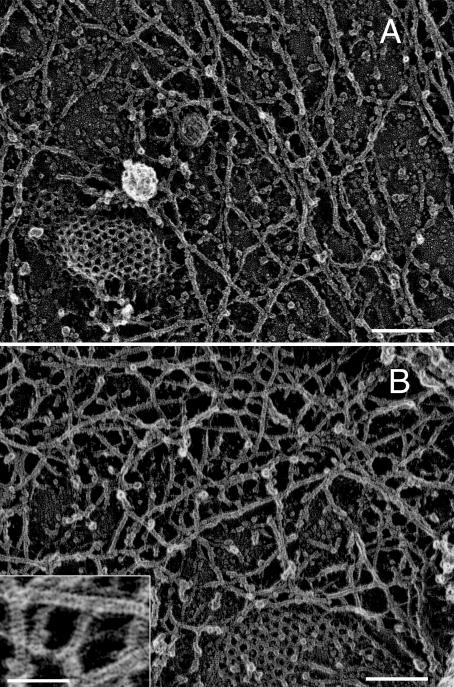
Fig. 2 (A and B), which was obtained for an NRK cell (Fig. 2 A) and an FRSK cell (Fig. 2 B), shows the magnified images of the cytoplasmic surface of the plasma membrane, exhibiting extensive filamentous netlike structures, which are the MSK. The presence of clathrin-coated structures shows that this is indeed the cytoplasmic surface. The striped banding patterns with a 5.5-nm periodicity on individual filaments are characteristic of actin filaments and, thus, indicate that these are actin filaments (Heuser and Kirschner, 1980; Heuser, 1983; Katayama, 1998; Schoenenberger et al., 1999). Because almost all of these filaments contain this striped pattern, it is concluded that the MSK is predominantly composed of actin filaments. This was also confirmed by immunogold staining (see Fig. 3 and related text).
Figure 2.
Magnified MSK images of an NRK and FRSK cell on the cytoplasmic surface of the upper membrane. (A) NRK cell; (B) FRSK cell. Clathrin- coated structures (A and B) and a caveola (A) show the cytoplasmic surface. The striped banding patterns with the 5.5-nm periodicity on individual filaments are characteristic of actin filaments. These images also reveal close links of the MSK actin filaments with the clathrin-coated structures and caveolae. Bars (A and B), 100 nm; (inset) 50 nm.
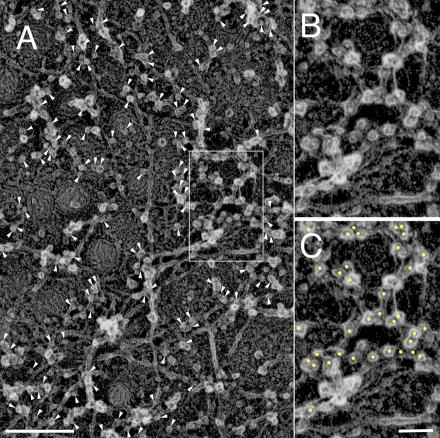
Figure 3.
Immunogold labeling also indicates that the major component of MSK is actin filaments (NRK cell). Actin filaments were indirectly immunolabeled with 5-nm colloidal gold particles coated with secondary antibodies. Each gold particle can be identified as a clear white spot (colloidal gold particle) surrounded by a fuzzy gray ring, which is caused by the platinum rotary shadowing around the secondary antibody coating of the gold particle. Representative probe images are indicated by arrowheads. (A) Most of the filamentous structures are labeled by colloidal gold probes. (B and C) The boxed area in A is shown at a higher magnification. In C, the gold particles are marked by yellow dots. The filaments with the 5.5-nm striped banding pattern, which is characteristic of the actin filament, are labeled with these gold probes. Bars (A), 200 nm; (B and C) 50 nm.
The electron micrograph shown in the inset in Fig. 2 B indicates the spatial resolution: because each band in the striped pattern with a 5.5-nm periodicity is visibly separated, the effective resolution is thought to be ∼2 nm (both the thickness of the platinum coating and the platinum granule size are ≤2 nm; Heuser and Kirschner, 1980; Heuser, 1983). The MSK structure observed here on the upper cell membrane is similar to that on the bottom cell membrane (the part of the cell membrane facing the coverslip) observed previously (Heuser and Anderson, 1989).
These results suggest that the cytoplasmic surface of a portion of the upper cell membrane >10 μm in diameter was visualized with a spatial resolution of ∼2 nm, which is much smaller than the width of a single actin filament or the repeat distance of the stripes. As shown in Figs. 1 and 2 (A and B), the MSK is likely to cover the entire cytoplasmic surface of the upper cell membrane except for the places where CCPs and caveolae are present in both NRK and FRSK cells. Such a notion of the complete coverage of the cytoplasmic surface of the plasma membrane by actin filaments might have existed for >30 yr in a part of the EM community (Byers and Porter, 1977; for review see Sheetz et al., 2006), but the data specifically indicating that the actin filaments of the MSK may cover the entire cell membrane has not been presented in the literature, as done here, nor shared in the cell biology community. The EM observations shown in this study are consistent with the MSK fence and anchored transmembrane protein picket models, in which the entire plasma membrane except for the specific membrane domains is partitioned into many small compartments with regard to lateral diffusion of the molecules incorporated in the plasma membrane.
The MSK predominantly consists of actin filaments: immunogold labeling of actin and actin-binding proteins
To further examine whether the MSK is predominantly composed of actin filaments (and partly because the 5.5-nm periodicity of the banding pattern is somewhat difficult to discern in some of the filaments), we examined it using an indirect immunolabeling method with 5-nm-diameter colloidal gold particles (see Materials and methods; Fig. 3). On the filaments with striped patterns, the enlarged images (Fig. 3, B and C) show the presence of many colloidal gold actin probes, which appear as distinct white spots surrounded by somewhat blurred white halos, reflecting the platinum shadow over the antibody molecules attached to the gold particle. The electron micrographs in Fig. 3 revealed that almost all of the colloidal gold probes are bound to the filaments located on the cytoplasmic surface (yellow dots). Therefore, it is concluded that actin is the main constituent molecule of the MSK.
Electron tomography of the undercoat structure on the cytoplasmic surface of the plasma membrane
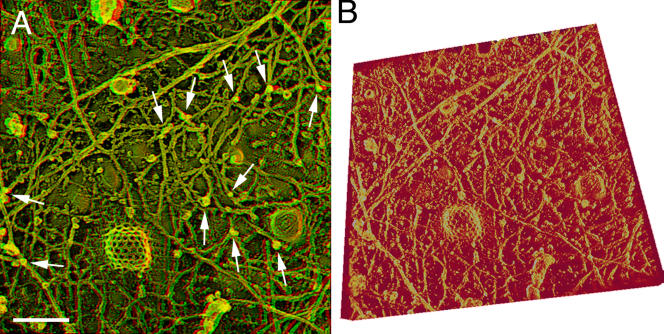
The 3D structure of the undercoat within 100–134 nm from the cytoplasmic surface of the plasma membrane, which includes CCPs, caveolae, and the actin-based MSK, was reconstructed using electron tomography for the platinum-replicated samples. Based on the 97–141 tilt images acquired in the range of ±48–70° every 1° step for a single EM view field, 100–121 sliced images of every 0.85–1.34 nm perpendicular to the z axis (parallel to the image obtained at 0° of the tilt angle) were calculated by a computer (long wavelength [≥∼500 nm] undulations of the cell membrane were corrected by the 3D reconstruction software IMOD). The 3D image was reconstructed based on these serial thin slices. Representative images obtained for an EM view field are shown in Video 1 (131 tilt images; an anaglyph produced from images taken at ±12° is shown in Fig. 4 A) and Video 2 (showing the 3D image by rotating the 3D reconstructed undercoat structure; a typical view is shown in Fig. 4 B; videos are available at http://www.jcb.org/cgi/content/full/jcb.200606007/DC1). Throughout the present research, this protocol was used to obtain 3D images.
Figure 4.
Stereo electron micrographs and 3D reconstructed images of the undercoat structure, CCPs, and caveolae in NRK cells. (A) An EM anaglyph of the undercoat structure generated at ±12° of the tilt angle among the 131 tilt images (acquired in the range of ±65° with 1° steps). Use view glasses for the 3D structure (left = red). See Video 1 for all 131 of the tilt images. Arrows indicate actin filaments protruding from the membrane cytoplasmic surface toward the cytoplasm. The arrows point to the places where the protruding actin filaments intersect with the MSK meshwork located close to the membrane. (B) The 3D undercoat structure reconstructed from the tilt images shown in Video 1. See Video 2, where the 3D structure is rotated (available at http://www.jcb.org/cgi/content/full/jcb.200606007/DC1). Bar, 100 nm.
In these images, because of their 3D representation, it is especially clear that the MSK, which is mostly composed of actin filaments, generally spreads along the membrane, covering almost the entire cytoplasmic surface of the upper membrane except for the places with caveolae and CCPs. In addition, CCPs and caveolae are very closely associated with the actin filaments in the MSK, as seen in these images and also in Figs. 2 (A and B) and 3. These results are consistent with Rothberg et al. (1992), Fujimoto et al. (2000), and Parton (2003), but in NRK cells studied here, many more actin filaments were found to be associated with each CCP or caveola. Furthermore, 92 and 93% of CCPs and caveolae (n = 200) were bound by the actin filaments. These results are consistent with the requirement of filamentous actin for CCP internalization (Qualmann et al., 2000; Merrifield et al., 2002).
Many short, thin filaments protrude toward the cytoplasm, mostly perpendicularly, from the membrane surface (they were short probably because they were broken at the time of the membrane rip off; Fig. 4 A, arrows). Note that these perpendicular filaments are almost always connected to the MSK network lying on the cytoplasmic surface (see the tips of the arrows; Fig. 4 A). Thus, the part of the MSK that is located on the cytoplasmic surface is connected three dimensionally to the cytoskeleton. Together, they will provide mechanical support for the membrane and the force for deforming the membrane.
3D reconstruction of the MSK structure
The part of the actin-based MSK that is in contact with the cytoplasmic surface of the cell membrane has been proposed to partition the cell membrane into 30–230-nm compartments by the fence and picket effect (Edidin et al., 1991; Kusumi and Sako, 1996; Kusumi et al., 2005). If these fence and picket models are correct, the distribution of the mesh size of the MSK on the cytoplasmic surface of the plasma membrane would be practically the same as that of the compartment size determined by diffusion measurements of membrane molecules. To carry out this examination, the 3D reconstruction of MSK by electron tomography provides a unique opportunity because the obtained images provide quantitative data on how far the individual filaments are located from the membrane surface.
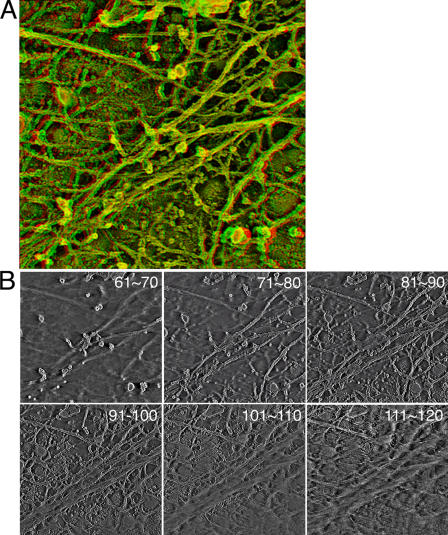
In Fig. 5 A, a typical MSK structure quantitatively analyzed in this study is shown in an anaglyph, and its 8.5-nm–thick sections (created by superimposing 10 0.85-nm sections) of the MSK of an NRK cell, starting from the cytoplasmic side toward the membrane, are shown (Fig. 5 B; a series of the original tilt images is shown in Video 3, and a series of sliced images of every 0.85 nm is shown in Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200606007/DC1). The actin-based MSK is visible on image sections 81–110. Individual actin filaments, forming a network as well as bundles, can be identified. Given the high density of the actin filament meshwork, which is much smaller than the optical resolution, conventional fluorescence microscopy will be unable to visualize the MSK meshwork and can visualize only the bundles of actin filaments.
Figure 5.
A series of sliced images of the actin MSK on the plasma membrane cytoplasmic surface of an NRK cell. (A) A typical actin MSK structure used for analysis of the mesh size on the cytoplasmic surface of the plasma membrane using computed tomography. An anaglyph of the undercoat structure generated at ±12° of the tilt angle among the 97 tilt images (acquired in the range of ±48° with 1° steps). See Video 3 for all 97 of the tilt images. (B) 10 consecutive sections, each 0.85-nm thick, are superimposed, and six of these superimposed images, which represent 60 image sections out of 121 image sections, are shown from the cytoplasmic side toward the plasma membrane side. The numbers here indicate the number of slices counted from the cytoplasmic side. The actin-based MSK near the cytoplasmic surface of the plasma membrane is visible on images 81–110. All 121 of the sliced images of every 0.85 nm are shown in Video 4 (available at http://www.jcb.org/cgi/content/full/jcb.200606007/DC1).
Interface structure of MSK on the cytoplasmic surface of the plasma membrane
The filaments of the MSK that are directly associated with the cytoplasmic surface of the plasma membrane and may be involved in partitioning the plasma membrane were systematically determined. Out of the stack of 121 image slices taken every 0.85 nm from the cytoplasmic surface (∼100-nm thick altogether), 16 consecutive image slices from the membrane surface (∼13.6-nm thick altogether) were used for this analysis (Fig. 6, A and B).
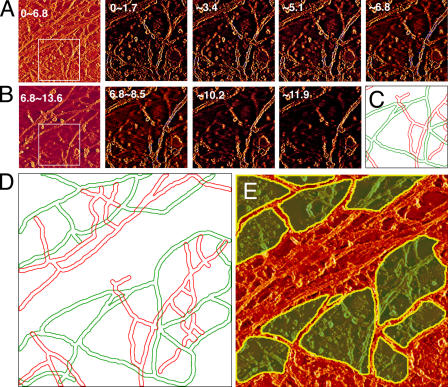
Figure 6.
The method for determining the MSK mesh on the cytoplasmic surface of the plasma membrane, which possibly delimits the compartments of the plasma membrane, using the 3D reconstructed images of the MSK (an NRK cell). (A and B) The images on the far left are the ∼0–6.8- or ∼6.8–13.6-nm sections, each of which is a stack of eight 0.85-nm sections of 670 × 670 nm. These are from a series of 121 image sections (0.85-nm thick) from the cytoplasmic surface after the tilt and long wavelength undulation of the cell surface were corrected. The boxed areas in A and B (330 × 330 nm) are expanded on the right of these image stacks, with a section thickness of 1.7 nm (two 0.85-nm sections are superimposed; 330 × 330 nm for each image). (C) The outline of each actin filament adjacent to the membrane surface (green, which could not be observed above 10.2 nm) and that of each actin filament that could be observed above 10.2 nm (red). The same view field and magnification as those for the thinner sections shown in A and B (330 × 330 nm). See Materials and methods for details. (D) The outline of actin filaments in a greater view field, which is the same as those in the thick sections (∼0–6.8 and ∼6.8–13.6 nm) in A and B (670 × 670 nm, expanded here). (E) The image of the ∼0–6.8-nm sections (670 × 670 nm) superimposed by the image of areas surrounded by the filaments outlined in green in D (green areas with yellow outlines). According to the fence and picket models, these areas are likely to be the compartments where membrane molecules are temporarily confined.
In Fig. 6 A (four images on the right) and Fig. 6 B (the second to fourth images), the boxed areas in the left-most images were expanded, and the sections of every 1.7 nm (superposition of two 0.85-nm–thick slices; 330 × 330 nm) are displayed between 0 and 11.9 nm. Using these sections, the filaments that are closely associated with the cytoplasmic surface of the cell membrane were determined. Because the thickness (width in the image) of the actin filament after platinum shadowing is between 9 and 11 nm (consistent with Heuser, 1983) and the thickness of the platinum replica is ≤2 nm (consistent with Heuser, 1983 and Moritz et al., 2000), the height of the actin filament that is associated with the membrane will be 7–9 nm (because the height is given by the actin thickness and one replica thickness, whereas the width in the image is determined by the actin thickness plus two replica thicknesses), with 8 nm being a reasonable estimate. In the series of electron tomography sections shown in Fig. 6 (A and B), the existence of three major classes of filaments with regard to the distance from the membrane surface can be discerned (for details of this analysis, see Materials and methods).
The first class of filaments is distinct in computer-reconstructed sections close to the cytoplasmic surface of the plasma membrane, even in the first ∼0–1.7-nm section (because the contrast is reversed in these micrographs, they look more lucent or white), but fade out of the reconstructions 8–10 nm away from the membrane surface (for details, see Materials and methods). These filaments are drawn in green in Fig. 6 C. We interpret that these filaments are in true contact with the plasma membrane (the gap between the filament and the inner membrane surface is <0.85 nm) because they can be seen clearly even in the first 0.85-nm section. These filaments are likely to be the significant ones for generating membrane corrals.
The second class of filaments also looks clear in sections very close to the membrane surface but does not fade out until ∼14 nm away from it. We interpret that these may be the actin filaments that had platinum coatings all around their surfaces because they stood off the surface somewhat, which slightly exaggerated their thickness and made them appear as though they were in contact with the plasma membrane when in fact they probably were not quite in direct contact. We did not consider these filaments to be close enough to generate membrane corrals.
The third class of filaments is not apparent in sections closest to the plasma membrane but becomes clear some distance away from it (>2–4 nm) and also does not fade out until ∼14 nm. We interpret these as being filaments that definitely do not contact the plasma membrane directly and, thus, should not contribute to forming corrals. The second and third classes of filaments are drawn in red in Fig. 6 C.
Therefore, we considered that only the first class of filaments (those drawn in green in Fig. 6, C and D) forms the MSK fences and pickets, and the area surrounded by these filaments is colored green in the 0–6.8-nm section shown in Fig. 6 E. Note that areas are excluded from this analysis in which bundles of actin filaments are present (e.g., the structure crossing diagonally from the bottom left to the top right in Fig. 5), actin filaments are too crowded to be individually discerned, an actin filament is terminated in the middle of a domain (domains that contain a loose end of an actin filament), or CCPs, caveolae, and the smooth surface membrane invaginations are present (the white regions in Fig. 7 C).
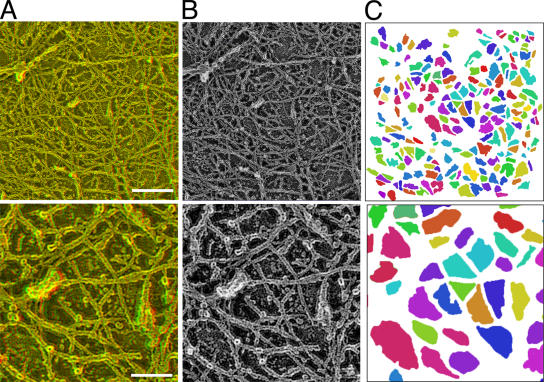
Figure 7.
The MSK meshwork directly on the cytoplasmic surface of the plasma membrane. The central parts of the figures in the top row (bar, 300 nm) are magnified by a factor of three and are shown in the bottom row (bar, 100 nm). (A) Typical stereo views of the plasma membrane specimen (anaglyph; left = red). (B) Normal electron micrographs of the plasma membrane samples. The same view fields as those in A. (C) The areas delimited by the actin filaments closely apposed to the cytoplasmic surface of the cell membrane are shown. Different colors are shown to help the eye.
Comparison of the MSK mesh size on the plasma membrane determined by electron tomography with the compartment size for the diffusion of membrane molecules
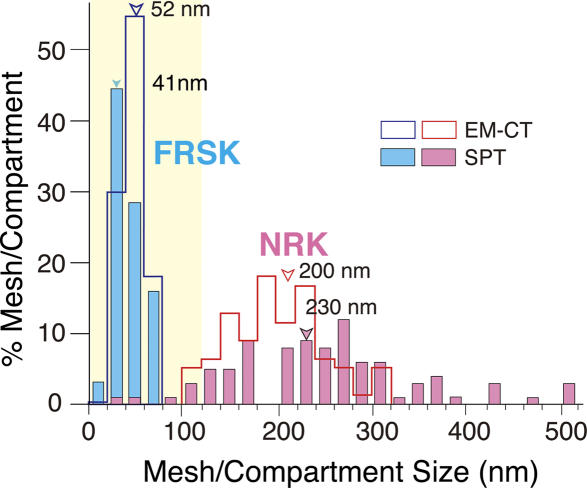
Similar determination of the MSK meshwork was performed for FRSK cells. Representative meshes of the MSK are shown in Fig. 7 (for an FRSK cell, colored to aid in visualization). We performed such analyses for 10 representative stacks of image sections (1,290 × 1,290-nm plane) each for NRK cells and FRSK cells (eight different cell membrane sheets for each cell type) and identified 76 and 1,300 areas bounded by the MSK meshwork, respectively (excluding the regions occupied by stress fibers and other membrane undercoat structures such as CCPs and caveolae; about the same total membrane areas were examined for each cell type, and, thus, the difference in the number of identified areas represents the difference in the area size between these two cell lines). The 2D area size for each domain was measured by Amira software. The distributions of the square root of the area size (the side length, assuming a square shape for the area) for NRK (Fig. 8, pink open bars) and FRSK (blue open bars) cells are shown in Fig. 8. The median values of the area and its square root are 3.9 × 104 nm2 and 200 nm, respectively, for NRK cells and 2.7 × 103 nm2 and 52 nm, respectively, for FRSK cells.
Figure 8.
Comparison of the distributions of the MSK mesh size on the cytoplasmic surface of the plasma membrane estimated by electron tomography with that of the compartment size determined from the phospholipid diffusion data for NRK and FRSK cells. Electron tomography, open bars; phospholipid diffusion data, closed bars (adapted from Fujiwara et al., 2002 and Murase et al., 2004). NRK, pink; FRSK, blue. Within the same cell type, the MSK mesh size and diffusion compartment size exhibited similar distributions (compare the open and closed bars with the same color). The actual sizes are quite different between NRK and FRSK cells. EM-CT, EM-based computer tomograph; SPT, single-particle tracking.
The size distributions of the compartments for the diffusion of membrane molecules were obtained for an unsaturated phospholipid, l-α-dioleoylphosphatidylethanolamine, by Fujiwara et al. (2002) and Murase et al. (2004) for NRK and FRSK cells, respectively. The distributions of the side lengths for NRK (Fig. 8, pink closed bars) and FRSK (blue closed bars) cells are shown in the histograms in Fig. 8. The median values of the compartment area and the side length are 4.3 × 104 nm2 and 230 nm, respectively, for NRK cells and 2.1 × 103 nm2 and 41 nm, respectively, for FRSK cells (Murase et al., 2004).
These results indicate that in the same cell line (for both the NRK and FRSK cases), the MSK mesh size determined by electron tomography and the diffusion compartment size determined by the high speed single-particle tracking of a phospholipid are similar to each other. However, between these two cell lines, both the MSK mesh and the diffusion compartment sizes differ greatly. The similarities between the MSK mesh sizes and the diffusion compartment sizes in cell lines that exhibit quite different distributions strongly support the MSK fence and picket models.
Discussion
We performed quantitative analyses of the undercoat structure of the cytoplasmic surface of the plasma membrane using electron tomography for samples prepared by a rapid-freeze, deep-etch, platinum replication technique. One of the most important limitations of this technique is that the cell has to be placed in a hypotonic medium at 4°C for 5–15 min to remove the upper cell membrane. However, with this method, large membrane fragments that were covered by the dense MSK meshwork could be obtained, which was important for the purpose of the present research.
We obtained the results by specifically addressing the following three questions. (1) Does the dense meshwork of the MSK exist everywhere on the cytoplasmic surface of the cell membrane, and, if so, how is it linked to the bulk cytoskeleton? (2) If so, what is its relationship with other structures of the plasma membrane, such as CCPs and caveolae? (3) How is the distribution of the MSK mesh size right on the cytoplasmic surface of the plasma membrane?
The final point is important because this part of the MSK might form the corrals of the plasma membrane for the diffusion of membrane molecules. Therefore, it is interesting to compare the distribution of the mesh size of the MSK directly attached to the cytoplasmic surface of the plasma membrane, as determined by an EM method, with that of the compartment size for the diffusion of membrane molecules. NRK (median size = 230 nm) and FRSK (41 nm) cell lines were selected for such a comparison because their compartment sizes are very different (Fujiwara et al., 2002; Murase et al., 2004). This will be an interesting test for the MSK fence and MSK-anchored transmembrane protein picket models and became possible by obtaining the 3D reconstructed images of the MSK structure on the cytoplasmic surface of the plasma membrane.
Does the dense meshwork of the MSK exist everywhere on the cytoplasmic surface of the cell membrane, and, if so, how is it linked to the bulk cytoskeleton?
The cytoplasmic surface of the plasma membrane has been observed by EM for >30 yr (Byers and Porter, 1977). Stunning high resolution EM images of the structures near the cytoplasmic surface have been published previously (Heuser and Kirschner, 1980; Hirokawa and Heuser, 1981; Heuser and Anderson, 1989; Hartwig and DeSisto, 1991; Rothberg et al., 1992), suggesting that the plasma membranes of animal cells are shaped by cytoskeletal interactions. However, despite these interesting and important studies, we felt that more extensive studies addressing the questions posed in the title of this subsection are necessary in particular for cells in culture.
We found that almost the entire cytoplasmic surface of the upper cell membrane is covered with the actin-based MSK except for the places where CCPs, caveolae, and noncoated membrane invaginations exist. In addition, this extensive pseudo-2D–type MSK network is linked to many actin filaments that extend from the membrane cytoplasmic surface toward the cytoplasm (Fig. 4 A) and is probably connected to the bulk cytoskeleton. This is consistent with the quantitative analysis of bleb formation, in which the density of the MSK filaments must be higher than one every 0.5–1 μm to avoid blebbing (Sheetz et al., 2006).
How closely is the MSK associated with CCPs and caveolae?
Almost all of the CCPs and caveolae are extensively linked to the MSK meshwork. As found in Figs. 2–4 , many actin filaments that come out of the MSK meshwork are connected to these structures at their polygonal and striated patterns, respectively, and, in the cell types examined here, this occurs much more extensively than found previously (Rothberg et al., 1992; Fujimoto et al., 2000; Parton, 2003). Merrifield et al. (2002) found that actin rapidly becomes more concentrated at the CCPs right before they are internalized, but from the structures found in the present research, we could not tell what kind of actin–CCP interactions may be involved in such concentrations.
Is the distribution of the MSK mesh size right on the cytoplasmic surface of the plasma membrane consistent with that for the compartment size determined by a phospholipid diffusion probe?
3D reconstruction of the MSK using electron tomography allowed the determination of the MSK meshwork directly situated on the cytoplasmic surface of the plasma membrane. This meshwork is likely to partition the plasma membrane into many small compartments with regard to the lateral diffusion of membrane molecules. The size distribution of the areas bounded by the MSK meshwork agreed well with that determined for an unsaturated phospholipid undergoing hop diffusion in high speed single-particle tracking experiments in both NRK and FRSK cells, which have quite different mesh sizes (Fig. 8). These results support the MSK fence (corralling) model and the anchored transmembrane protein picket model.
How the MSK is attached to the cytoplasmic surface of the plasma membrane is unknown. Specific proteins that link the membrane and actin filaments at their barbed ends, such as gelsolin and villin (Pollard and Cooper, 1986; Hartwig et al., 1989), and at their sides, such as ponticulin (Wuestehube and Luna, 1987) and ezrin/radixin/moesin family proteins (Tsukita et al., 1997), exist. However, the weak nonspecific binding of actin filaments to membrane lipids and proteins may greatly contribute to the interactions of the actin filaments with the membrane. Although transmembrane proteins are temporarily trapped within a compartment, they hop to adjacent compartments in longer terms, suggesting that the link between the membrane and actin filaments may break at a rate comparable with the hop rate, which is once every several to several hundred milliseconds (depending on the molecule and cell type).
In addition to actin and actin-associated proteins, some other proteins may contribute to forming the MSK and membrane corrals. For example, septin, which is ∼7-nm thick in the negatively stained images (Kinoshita et al., 2002), and agorin (Apgar and Mescher, 1986) may play some roles. Further characterization of the proteins involved in the MSK and its interaction with the membrane components and further analysis of the dynamics of membrane–MSK interactions will be important for understanding the roles played by the MSK–plasma membrane interactions in signal transduction, domain formation in the plasma membrane, and reorganization of the cytoskeleton.
Materials and methods
Antibodies and other reagents
Rabbit anti-actin IgG was obtained from Biomedical Technologies, and colloidal gold probes (5-nm diameter) coated with secondary antibodies (produced in goat) were purchased from GE Healthcare.
Rapid-freeze, deep-etch, immunoreplica EM of the cytoplasmic cell surface
NRK and FRSK cells were maintained in HAM-F12 or DME mediums, respectively, supplemented with 10% FBS (Sigma-Aldrich) under a 5% CO2 atmosphere at 37°C. The cells used for the experiments were grown in 35-mm plastic dishes to ∼70% confluency, usually 2 d after inoculation. The cells were washed three times with ice-cold Pipes buffer (10 mM Pipes, 100 mM KCl, 5 mM MgCl2, and 3 mM EGTA, pH 6.8, which mimics the environment in the cytoplasm somewhat but is slightly hypotonic) and were exposed for 15–30 s to an ice-cold 70% Pipes buffer (the Pipes buffer diluted by a factor of 1.43 with water, making this solution more hypotonic; Rutter et al., 1988). After the buffer on the coverslip was drained, the remaining excess water was removed by filter paper. To expose the cytoplasmic surface of the upper cell membrane, the upper cell membrane was removed from the rest of the cell after it was adhered to a coverslip placed on top of the cell layer (Rutter et al., 1988; Sanan and Anderson, 1991). 5 × 5-mm coverslips (Matsunami) coated with positively charged Alcian blue 8GX (Wako; Alcian blue–coated coverslips were prepared by first immersing them in 1% Alcian blue in distilled water at room temperature for 10 min, washing them with distilled water, and drying them in the air) were placed on top of the cells (upper surface facing the medium rather than the coverslip) and incubated at 4°C for 5–15 min. During this period, good contact between the cell surface and the coverslip was developed. Then, the coverslips were gently floated off from the cell using the surface tension of the buffer by slowly adding ice-cold Pipes buffer containing 1% PFA/0.25% glutaraldehyde into the space between the culture dish and the coverslip. When the coverslip floated off, the cells were cleaved, and the upper cell membrane came off with the coverslip. Then, the cells were further fixed by incubation in fresh ice-cold 1% PFA/0.25% glutaraldehyde in Pipes buffer for 10 min. After fixation, the coverslips were washed three times, for 10 min each time, with PBS (8.10 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl, and 2.68 mM KCl, pH 7.4).
To identify the actin filaments on the cytoplasmic surface of the cell membrane, immunogold labeling was performed after fixation. The fixed upper cell membranes on the coverslips with their cytoplasmic surfaces exposed toward the buffer were incubated at 4°C for 2 h in PBS containing 10 μg/ml rabbit anti-actin IgG and 1% BSA (Sigma-Aldrich), and, after three washes for 10 min each in 25 mM Tris-buffer, pH 8.0, the cell membranes were incubated in Tris-buffer containing 1% BSA and 5-nm–diameter colloidal gold conjugated with anti–rabbit IgG goat antibodies (GE Healthcare) at 4°C for 2 h. After three washes in PBS, these labeled membranes were further fixed in 2% glutaraldehyde in PBS on ice for 5 min. Finally, the coverslips were washed in distilled water for 1 min before rapid freezing.
Each coverslip was placed on the plunger tip of the rapid-freezing device (Eiko; Usukura and Yamada, 1987) with the cytoplasmic surface of the membrane facing down. The specimen was slammed down (free fall) onto a polished pure copper block, which was precooled by direct immersion in liquid helium. The frozen coverslip was placed in liquid nitrogen and was transferred into the freeze-etching shadowing chamber (FR7000-S; Hitachi). The excess ice covering the cytoplasmic surface of the membrane was shaved off with a prechilled glass knife using a microtome placed in the chamber at −140°C or below. The cytoplasmic surface was then etched for ∼10 min after the specimen temperature was raised to −90°C. The etched specimen surfaces were then rotary shadowed with platinum at an angle of 22.5° from the surface and with carbon from the top. The molecules as well as the gold probes localized on the cytoplasmic surface of the cell membrane were immobilized to the deposited platinum (Fujimoto, 1995; Fujimoto et al., 1996).
Collodion was applied immediately after the platinum-carbon replicas were removed from the cold chamber to fortify them. The platinum-carbon replica was removed from the glass coverslip in 1% hydrofluoric acid in distilled water. After the replicas were successfully removed from the glass surface and mounted on the grid, the collodion coat was dissolved away in n-pentyl acetate. In this protocol, the sodium hypochlorite solution, which is generally used to remove the replicas from the coverslip and also to clear the membrane and the undercoat structure of the replicas, was replaced with 1% hydrofluoric acid to keep the cell membrane, the undercoat structure, and the immunogold probes that had been attached to these structures on the platinum replicas (1% hydrofluoric acid is likely to only dissolve the glass, leaving the cell membrane molecules bound to the platinum replica; Fujimoto, 1995; Fujimoto et al., 1996). An additional advantage of using 1% hydrofluoric acid is that the platinum replicas break less often, probably because it does not remove the membrane components and, thus, leaves the replicas rather intact. In addition, to keep as many colloidal gold particles and membrane molecules attached to the platinum replicas as possible, we included Photo-Flo 200 (Kodak), a detergent used to prevent water-drop stains on photographic film in all of the solutions used here (advice given by J. Heuser). After the replicas were washed with distilled water, they were mounted on 100–200 mesh copper grids (Ted Pella) coated with polyvinyl formvar (Nisshin EM) and observed at magnifications of ∼10,000–70,000 with a transmission electron microscope (1200EX; JEOL).
The following methodological precautions and improvements were made to reproducibly produce large cell membranes and replicas without excessive fragmentation. (1) An Alcian blue coat rather than poly-l-lysine coat was used (Rutter et al., 1988; Sanan and Anderson, 1991). (2) Before overlaying the coverslips, excess water was removed from the specimen, leaving just enough buffer to cover the cell. (3) To cleave off the upper membrane attached to the overlaid coverslip, the coverslip was floated off very gently by adding cleavage medium (using the surface tension of the buffer to float the coverslip). If this was not performed gently enough, the membrane was fragmented. (4) The frozen sample was shaved with a glass knife, with the angle between the knife and the coverglass adjusted to a shallow angle (<6°) so that most of the excess water and the cytoplasm were removed and the cytoplasmic surface of the cell membrane could be exposed after light etching. Because replicas with too many variations in height tend to break when they are removed from the coverslip and placed on the water surface, removal of the excess cytoplasm helps to avoid replica breakage. (5) Collodion was applied immediately after the replicas were removed from the cold chamber (before the replicas were removed from the coverslip on the water surface) to fortify the replica (a technique learned from T. Baba and S. Ohno). This step also helped to prevent replica breakage when the replicas were removed from the coverslip. After the large replicas were removed from the glass surface, the collodion coat was dissolved away in n-pentyl acetate. (6) A solution of 1% hydrofluoric acid was used to slightly dissolve the glass surface to facilitate the removal of replicas from the coverslip. (7) A detergent, Photo-Flo 200 (Kodak), was included in all of the solutions that contacted platinum replicas.
Electron microscope tomography
For 3D reconstruction, the replica was imaged at tilt angles of every 1.0° in the range between ±48 and 70° (total of 97–141 images) for a single field by a transmission electron microscope (Tecnai Sphera F20; FEI) equipped with a CCD camera (1,024 × 1,024 pixels). The pixel size at the specimen was 0.85 nm. The image acquisition was fully automated as previously described (Medalia et al., 2002). The 100–121 image sections of every 0.85–1.34 nm were obtained by a calculation based on the set of 97–141 tilt images using an IMOD software package running on Linux (University of Colorado; Kremer et al., 1996) . Corrections for the tilt of the specimen and the long wavelength undulations of the membrane were also achieved with IMOD software. 3D rendering (displaying 3D images in different ways) was performed using the Amira DEV software package (Mercury Computer Systems) operating on a Linux system.
Analysis of the 3D reconstructed images of the MSK
In the series of electron tomography sections shown in Fig. 6 (A and B), the existence of three major classes of filaments with regard to the distance from the membrane surface was found in the following way. The first class is the filaments that are highly electron dense in the first ∼0–1.7-nm section (because the contrast is reversed in these micrographs, they look more lucent or white) and are continuously seen in the image sections up to the ∼6.8–8.5-nm section, which then dim rapidly in the ∼8.5–10.2- and ∼10.2–11.9-nm sections. To quantitatively evaluate such signal intensity changes within individual filaments, we selected points that are clearly seen in the image of the first ∼0–1.7-nm section every 100–250 nm on each filament, measured the signal intensity on each spot (five pixels), and looked for the section where the signal intensity on the spot decreases by >25% from that for the adjacent section closer to the membrane (the signal intensity tends to drop very rapidly around the threshold sections). If the 25% decrease in the signal intensity occurred between the sections of ∼6.8–8.5 and ∼8.5–10.2 nm or between the sections of ∼8.5–10.2 and ∼10.2–11.9 nm, these filaments were categorized into the first class (i.e., those closely associated with the cytoplasmic surface of the cell membrane). These filaments are drawn in green in Fig. 6 C (different regions within a single filament might become dim in either of these two sections).
The second class is similar to the first class but can be seen clearly even in the ∼10.2–11.9-nm section (and also in the ∼11.9–13.6-nm section, which usually looked similar to the ∼10.2–11.9-nm section). This may be the result of the actin filaments that are located several nanometers away from the cytoplasmic surface. Because the platinum rotary shadowing was performed at a low angle (22° from the surface), the platinum may have been deposited in the space between the filament and the membrane. Another possibility is that two filaments are stacked together for a long distance, but we do not think that this happens very often. Therefore, we categorized these filaments into those that stay near the membrane but do not closely associate with the membrane surface. These filaments were not considered to contribute to delimiting the membrane compartments for molecular diffusion in the plasma membrane.
The third class is the filaments that exhibit dim signals in the first ∼0–1.7-nm section and show higher electron densities in farther sections, at least up to the section of ∼10.2–11.9 nm, before fading out in the ∼11.9–13.6- and ∼13.6–15.3-nm sections (the latter two sections are not depicted). These filaments were again assumed not to contribute to forming membrane corrals. The second and third classes of filaments are drawn in red in Fig. 6 C.
There were regions that were not amenable to such analysis. They were the areas where bundles of actin filaments were present (e.g., the structure crossing diagonally from the bottom left to the top right in Fig. 5), actin filaments were too crowded to be individually discerned, an actin filament was terminated in the middle of a domain (domains that contain a loose end of an actin filament), or CCPs, caveolae, and the smooth surface membrane invaginations were present. They were excluded from this analysis (Fig. 7 C, white regions).
Online supplemental material
Video 1 shows a series of 131 tilt images of the undercoat structure on the cytoplasmic surface. Video 2 presents a 3D reconstructed image of the undercoat structure on the cytoplasmic surface of the plasma membrane, which is shown by rotating the reconstructed image. Video 3 shows a series of 97 tilt images of the MSK in an NRK cell, and Video 4 shows a series of 121 sliced images of every 0.85 nm of the MSK of an NRK cell calculated from the data shown in Video 3. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200606007/DC1.
Supplementary Material
Acknowledgments
We thank Y. Hirata (FEI) for her help in starting us with electron tomography, Drs. T. Baba and S. Ohno (Yamanashi University Medical School) for their help in preparing large platinum replicas, Dr. Mitsutoshi Setou (Mitsubishi Kagaku Institute of Life Sciences) for encouragement, and Dr. J. Heuser (Washington University) for helpful discussions and encouragement.
This research was supported, in part, by a Health Labor Sciences Research grant (nano-001 to N. Morone), a National Institute of Biomedical Innovation grant (05-32 to S. Yuasa), and Grants-in-Aid for Scientific Research and on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (to J. Usukura and A. Kusumi).
Abbreviations used in this paper: CCP, clathrin-coated pit; FRSK, fetal rat skin keratinocyte; MSK, membrane skeleton; NRK, normal rat kidney fibroblast.
References
- Apgar, J.R., and M.F. Mescher. 1986. Agorins: major structural proteins of the plasma membrane skeleton of P815 tumor cells. J. Cell Biol. 103:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, V. 1990. Spectrin-based membrane skeleton: a multipotential adaptor between plasma membrane and cytoplasm. Physiol. Rev. 70:1029–1065. [DOI] [PubMed] [Google Scholar]
- Bennett, V., and L. Chen. 2001. Ankyrins and cellular targeting of divers membrane proteins to physiological sites. Curr. Opin. Cell Biol. 13:61–67. [DOI] [PubMed] [Google Scholar]
- Branton, D., C.M. Cohen, and J. Tyler. 1981. Interaction of cytoskeletal proteins on the human erythrocyte membrane. Cell. 24:24–32. [DOI] [PubMed] [Google Scholar]
- Bussell, S.J., D.A. Hammer, and D.L. Koch. 1994. The effect of hydrodynamic interactions on the tracer and gradient diffusion of integral membrane-proteins in lipid bilayers. J. Fluid Mech. 258:167–190. [Google Scholar]
- Bussell, S.J., D.L. Koch, and D.A. Hammer. 1995. Effect of hydrodynamic interactions on the diffusion of integral membrane proteins: diffusion in plasma membranes. Biophys. J. 68:1836–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, H.R., and K.R. Porter. 1977. Transformations in the structure of the cytoplasmic ground substance in erythrophores during pigment aggregation and dispersion. I. A study using whole-cell preparations in stereo high voltage electron microscopy. J. Cell Biol. 75:541–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher, D.E., R. Winardi, P.O. Schischmanoff, M. Parra, J.G. Conboy, and N. Mohandas. 1995. Mechanochemistry of protein 4.1's spectrin- actin-binding domain: ternary complex interactions, membrane binding, network integration, structural strengthening. J. Cell Biol. 130:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edidin, M., S.C. Kuo, and M.P. Sheetz. 1991. Lateral movements of membrane glycoproteins restricted by dynamic cytoplasmic barriers. Science. 254:1379–1382. [DOI] [PubMed] [Google Scholar]
- Fujimoto, K. 1995. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 108:3443–3449. [DOI] [PubMed] [Google Scholar]
- Fujimoto, K., M. Umeda, and T. Fujimoto. 1996. Transmembrane phospholipid distribution revealed by freeze-fracture replica labeling. J. Cell Sci. 109:2453–2460. [DOI] [PubMed] [Google Scholar]
- Fujimoto, L.M., R. Roth, J.E. Heuser, and S.L. Schmid. 2000. Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis in mammalian cells. Traffic. 1:161–171. [DOI] [PubMed] [Google Scholar]
- Fujiwara, T., K. Ritchie, H. Murakoshi, K. Jacobson, and A. Kusumi. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157:1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov, I., F. Santini, R.A. Warren, and J.H. Keen. 1999. Spatial control of coated-pit dynamics in living cells. Nat. Cell Biol. 1:1–7. [DOI] [PubMed] [Google Scholar]
- Hartwig, J.H., and H.L. Yin. 1988. The organization and regulation of the macrophage actin skeleton. Cell Motil. Cytoskeleton. 10:117–125. [DOI] [PubMed] [Google Scholar]
- Hartwig, J.H., and M. DeSisto. 1991. The cytoskeleton of the resting human blood platelet: structure of the membrane skeleton and its attachment to actin filaments. J. Cell Biol. 112:407–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig, J.H., K.A. Chambers, and T.P. Stossel. 1989. Association of gelsolin with actin filaments and cell membranes of macrophages and platelets. J. Cell Biol. 108:467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J.E. 1983. Procedure for freeze-drying molecules adsorbed to mica flakes. J. Mol. Biol. 169:155–195. [DOI] [PubMed] [Google Scholar]
- Heuser, J.E., and M.W. Kirschner. 1980. Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J. Cell Biol. 86:212–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J.E., and R.G. Anderson. 1989. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J. Cell Biol. 108:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N., and J.E. Heuser. 1981. Quick-freeze, deep-etch visualization of the cytoskeleton beneath surface differentiations of intestinal epithelial cells. J. Cell Biol. 91:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino, R., I. Koyama, and A. Kusumi. 2001. Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophys. J. 80:2667–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, K., E.D. Sheets, and R. Simson. 1995. Revisiting the fluid mosaic model of membranes. Science. 268:1441–1442. [DOI] [PubMed] [Google Scholar]
- Katayama, E. 1998. Quick-freeze deep-etch electron microscopy of the actin-heavy meromyosin complex during the in vitro motility assay. J. Mol. Biol. 278:349–367. [DOI] [PubMed] [Google Scholar]
- Kinoshita, M., C.M. Field, M.L. Coughlin, A.F. Straight, and T.J. Mitchison. 2002. Self- and actin-templated assembly of Mammalian septins. Dev. Cell. 3:791–802. [DOI] [PubMed] [Google Scholar]
- Kremer, J.R., D.N. Mastronarde, and J.R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. [DOI] [PubMed] [Google Scholar]
- Kusumi, A., and Y. Sako. 1996. Cell surface organization by the membrane skeleton. Curr. Opin. Cell Biol. 8:566–574. [DOI] [PubMed] [Google Scholar]
- Kusumi, A., C. Nakada, K. Ritchie, K. Murase, K. Suzuki, H. Murakoshi, R.S. Kasai, J. Kondo, and T. Fujiwara. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. [DOI] [PubMed] [Google Scholar]
- Lucic, V., F. Forster, and W. Baumeister. 2005. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74:833–865. [DOI] [PubMed] [Google Scholar]
- Luna, E.J., and A.L. Hitt. 1992. Cytoskeleton–plasma membrane interactions. Science. 258:955–964. [DOI] [PubMed] [Google Scholar]
- McIntosh, R., D. Nicastro, and D. Mastronarde. 2005. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 15:43–51. [DOI] [PubMed] [Google Scholar]
- Medalia, O., I. Weber, A.S. Frangakis, D. Nicastro, G. Gerisch, and W. Baumeister. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 298:1209–1213. [DOI] [PubMed] [Google Scholar]
- Merrifield, C.J., M.E. Feldman, L. Wan, and W. Almers. 2002. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4:691–698. [DOI] [PubMed] [Google Scholar]
- Mohandas, N., and E. Evans. 1994. Mechanical properties of the red cell membrane in relation to molecular structure and genetic defects. Annu. Rev. Biophys. Biomol. Struct. 23:787–818. [DOI] [PubMed] [Google Scholar]
- Moritz, M., M.B. Braunfeld, V. Guenebaut, J. Heuser, and D.A. Agard. 2000. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2:365–370. [DOI] [PubMed] [Google Scholar]
- Murase, K., T. Fujiwara, Y. Umemura, K. Suzuki, R. Iino, H. Yamashita, M. Saito, H. Murakoshi, K. Ritchie, and A. Kusumi. 2004. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 86:4075–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Z., T. Kao, Z. Horvath, J. Lemos, J.Y. Sul, S.D. Cranstoun, V. Bennett, S.S. Scherer, and E.C. Cooper. 2006. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J. Neurosci. 26:599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton, R.G. 2003. Caveolae–from ultrastructure to molecular mechanisms. Nat. Rev. Mol. Cell Biol. 4:162–167. [DOI] [PubMed] [Google Scholar]
- Perkins, G.A., C.W. Renken, J.Y. Song, T.G. Frey, S.J. Young, S. Lamont, M.E. Martone, S. Lindsey, and M.H. Ellisman. 1997. Electron tomography of large, multicomponent biological structures. J. Struct. Biol. 120:219–227. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and J.A. Cooper. 1986. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55:987–1035. [DOI] [PubMed] [Google Scholar]
- Qualmann, B., M.M. Kessels, and R.B. Kelly. 2000. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 150:F111–F116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg, K.G., J.E. Heuser, W.C. Donzell, Y.S. Ying, J.R. Glenney, and R.G. Anderson. 1992. Caveolin, a protein component of caveolae membrane coats. Cell. 68:673–682. [DOI] [PubMed] [Google Scholar]
- Rutter, G., W. Bohn, H. Hohenberg, and K. Mannweiler. 1988. Demonstration of antigens at both sides of plasma membranes in one coincident electron microscopic image: a double-immunogold replica study of virus-infected cells. J. Histochem. Cytochem. 36:1015–1021. [DOI] [PubMed] [Google Scholar]
- Sako, Y., and A. Kusumi. 1994. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J. Cell Biol. 125:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako, Y., and A. Kusumi. 1995. Barriers for lateral diffusion of transferrin receptor in the plasma membrane as characterized by receptor dragging by laser tweezers: fence versus tether. J. Cell Biol. 129:1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako, Y., A. Nagafuchi, S. Tsukita, M. Takeichi, and A. Kusumi. 1998. Cytoplasmic regulation of the movement of E-cadherin on the free cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J. Cell Biol. 140:1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanan, D.A., and R.G. Anderson. 1991. Simultaneous visualization of LDL receptor distribution and clathrin lattices on membranes torn from the upper surface of cultured cells. J. Histochem. Cytochem. 39:1017–1024. [DOI] [PubMed] [Google Scholar]
- Saxton, M.J. 1989. The spectrin network as a barrier to lateral diffusion in erythrocytes. A percolation analysis. Biophys. J. 55:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton, M.J. 1990. The membrane skeleton of erythrocytes. A percolation model. Biophys. J. 57:1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton, M.J., and K. Jacobson. 1997. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26:373–399. [DOI] [PubMed] [Google Scholar]
- Schoenenberger, C.A., M.O. Steinmetz, D. Stoffler, A. Mandinova, and U. Aebi. 1999. Structure, assembly, and dynamics of actin filaments in situ and in vitro. Microsc. Res. Tech. 47:38–50. [DOI] [PubMed] [Google Scholar]
- Sheetz, M.P. 1983. Membrane skeletal dynamics: role in modulation of red cell deformability, mobility of transmembrane proteins, and shape. Semin. Hematol. 20:175–188. [PubMed] [Google Scholar]
- Sheetz, M.P., and D. Sawyer. 1978. Triton shells of intact erythrocytes. J. Supramol. Struct. 8:399–412. [DOI] [PubMed] [Google Scholar]
- Sheetz, M.P., J.E. Sable, and H.G. Dobereiner. 2006. Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 35:417–434. [DOI] [PubMed] [Google Scholar]
- Shen, B.W., R. Josephs, and T.L. Steck. 1986. Ultrastructure of the intact skeleton of the human erythrocyte membrane. J. Cell Biol. 102:997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., K. Ritchie, E. Kajikawa, T. Fujiwara, and A. Kusumi. 2005. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88:3659–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, M., H. Miyamoto, Y. Sako, H. Komizu, and A. Kusumi. 1998. Structure of the erythrocyte membrane skeleton as observed by atomic force microscopy. Biophys. J. 74:2171–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige, M., Y. Sako, and A. Kusumi. 1998. Regulation mechanism of the lateral diffusion of band 3 in erythrocyte membranes by the membrane skeleton. J. Cell Biol. 142:989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, A., and S. Ohnishi. 1986. Restriction of the lateral motion of band 3 in the erythrocyte membrane by the cytoskeletal network: dependence on spectrin association state. Biochemistry. 25:6133–6139. [DOI] [PubMed] [Google Scholar]
- Tsuji, A., K. Kawasaki, S. Ohnishi, H. Merkle, and A. Kusumi. 1988. Regulation of band 3 mobilities in erythrocyte ghost membranes by protein association and cytoskeletal meshwork. Biochemistry. 27:7447–7452. [DOI] [PubMed] [Google Scholar]
- Tsukita, S., S. Tsukita, and H. Ishikawa. 1980. Cytoskeletal network underlying the human erythrocyte membrane. Thin-section electron microscopy. J. Cell Biol. 85:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita, S., S. Yonemura, and S. Tsukita. 1997. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem. Sci. 22:53–58. [DOI] [PubMed] [Google Scholar]
- Ursitti, J.A., D.W. Pumplin, J.B. Wade, and R.J. Bloch. 1991. Ultrastructure of the human erythrocyte cytoskeleton and its attachment to the membrane. Cell Motil. Cytoskeleton. 19:227–243. [DOI] [PubMed] [Google Scholar]
- Usukura, J., and E. Yamada. 1987. Ultrastructure of the synaptic ribbons in photoreceptor cells of Rana catesbeiana revealed by freeze-etching and freeze-substitution. Cell Tissue Res. 247:483–488. [DOI] [PubMed] [Google Scholar]
- Valentijn, J.A., K. Valentijn, L.M. Pastore, and J.D. Jamieson. 2000. Actin coating of secretory granules during regulated exocytosis correlates with the release of rab3D. Proc. Natl. Acad. Sci. USA. 97:1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuestehube, L.J., and E.J. Luna. 1987. F-actin binds to the cytoplasmic surface of ponticulin, a 17-kD integral glycoprotein from Dictyostelium discoideum plasma membranes. J. Cell Biol. 105:1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuschner, D., W.J. Geerts, E. van Donselaar, B.M. Humbel, J.W. Slot, A.J. Koster, and J. Klumperman. 2006. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat. Cell Biol. 8:377–383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.