Abstract
Using a genetic screen we discovered that YGR198w (named YPP1), which is an essential Saccharomyces cerevisiae gene of unknown function, suppresses the toxicity of an α-synuclein (α-syn) mutant (A30P) that is associated with early onset Parkinson's disease. Here, we show that YPP1 suppresses lethality of A30P, but not of wild-type α-syn or the A53T mutant. The Ypp1 protein, when overexpressed, drives each of the three α-syns into vesicles that bud off the plasma membrane, but only A30P-containing vesicles traffick to and merge with the vacuole, where A30P is proteolytically degraded. We show that Ypp1p binds to A30P but not the other two α-syns; that YPP1 interacts with genes involved in endocytosis/actin dynamics (SLA1, SLA2, and END3), protein sorting (class E vps), and vesicle-vacuole fusion (MON1 and CCZ1) to dispose of A30P; and that YPP1 also participates in pheromone-triggered receptor-mediated endocytosis. Our data reveal that YPP1 mediates the trafficking of A30P to the vacuole via the endocytic pathway.
Introduction
Parkinson's disease is characterized by the selective degeneration of dopamine-producing neurons that comprise the substantia nigra pars compacta and the presence of proteinaceous inclusion bodies (Lewy bodies) in the affected neurons (Dawson and Dawson, 2003). The principal component of Lewy bodies is α-synuclein (α-syn), which is an intrinsically unfolded protein of unknown function (Weinreb et al., 1996; Uversky et al., 2000). Wild-type (WT) α-syn and two mutants, A30P and A53T (Polymeropoulos et al., 1997; Kruger et al., 1998), associated with early onset PD have been linked to a plethora of defects, including proteasomal and mitochondrial dysfunction (Tanaka et al., 2001; Martin et al., 2006), the accumulation of reactive oxygen species (ROS) (Xu et al., 2002), blockage of ER to Golgi traffic (Cooper et al., 2006), and histone acetylation inhibition (Kontopoulos et al., 2006).
Using a genetic screen that exploits the super sensitivity of α-syn–expressing yeast cells to killing by H2O2 (Flower et al., 2005), we discovered that YGR198w, which is an essential gene of unknown function, suppresses the toxicity of the mutant α-syn A30P. Because humans have several possible YGR198w orthologues, understanding the function of this gene may shed light on how human neurons protect themselves from α-syn. Given that YGR198w codes for an α-synuclein protective protein, YGR198w was named YPP1.
Until quite recently, other than its deletion kills cells (Rodriguez-Pena et al., 1998), scant information existed about YPP1. A recent study using a technique to probe the spectrum of synthetic genetic interactions among essential genes revealed that YPP1 interacts genetically with ACT1, ARP2, SEC1, SEC15, and SEC18 (Davierwala et al., 2005). Other studies have indicated that YPP1 functions in MAPK pathways (Roberts et al., 2000; Hazbun et al., 2003; Mnaimneh et al., 2004). Ypp1p-GFP localizes in a punctate pattern around the plasma membrane (Huh et al., 2003), although there is no indication from its sequence that Ypp1p is a membrane protein. Although information exists about its synthetic genetic interactions and localization, Ypp1p's function has remained obscure. Here, we show that Ypp1p binds A30P (but not WT or A53T) and mediates a sequence of events in which A30P is encapsulated into vesicles at the plasma membrane, and the vesicles then transit to and merge with the vacuole, where the A30P protein is proteolytically degraded.
Results
A genetic screen identifies YPP1 as a suppressor of A30P toxicity
A high copy yeast genomic library was used to identify suppressors of the super sensitivity of A30P expressing cells to killing by hydrogen peroxide (Fig. 1 A). Herein, we describe the characterization of one suppressor, YPP1, which is an essential gene of unknown function. Table I gives the strains and plasmids used in this study.
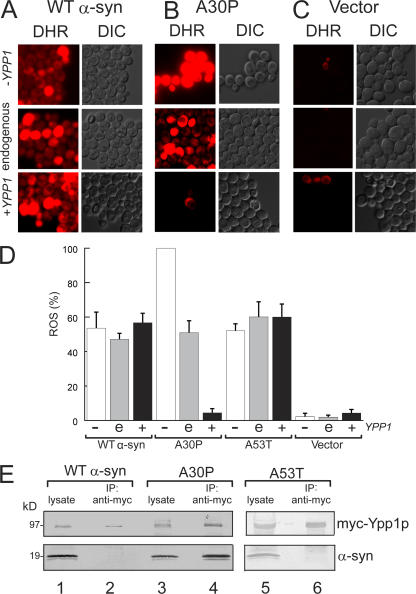
Figure 1.
Identification of YPP1 as a suppressor of A30P α-syn toxicity. Suppression of A30P toxicity by a high-copy yeast genomic library. FY23 cells transformed with pTF202 (A30P) and plated on SGal–Trp (left plate). FY23 cells transformed with pTF202 (A30P) and a 2-μm yeast genomic library and plated on SGal–Trp–Leu (right plate). 2-d incubation at 30°C. The disk in the center of each plate contained 10 μl of ∼8% hydrogen peroxide. (B) Western blot analysis of cells expressing α-syn with or without myc-Ypp1p overexpression. FY23 cells transformed with pTF201 (WT α-syn), pTF202 (A30P), or pTF203 (A53T) and pTF504 (myc-Ypp1) or pTF503 (empty vector) were pregrown in noninducing media to mod-log phase, shifted to inducing media, and incubated for 3 h at 30°C. Cell extracts were prepared and subjected to SDS-PAGE and immunoblotting. The proteins were visualized using monoclonal antibodies (anti-α-syn; anti-myc). Lane 1, WT α-syn; lane 2, myc-Ypp1p and WT α-syn; lane 3, A30P; lane 4, myc-Ypp1p and A30P. (C) Effect of Ypp1p overexpression on cell viability after 12 h of induction. The FUN1 dye stains metabolically active cells red and dead cells green. The top panel shows that cells expressing A30P (plus plasmid with no insert) are dead (green). The bottom panel shows that cells expressing A30P with Ypp1p overexpression are viable (red). The FY23 strain, harboring pTF302 (A30P) and pTF602 (YPP1) or pTF604 (empty vector), was used in these experiments. (D) Plot of percent viability (mean ± SD). The number of cells counted for each group was n = 400 to 1,400. *, P = 0.000013.
Table I.
Yeast strains and plasmids
| Strains | ||
|---|---|---|
| YPP1-GFP | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YPP1-GFP | MATa |
| FY23 | ura3-52 trp1Δ63 leu2Δ 1 | MATa |
| S288c | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gal2 | MATα |
| Tet-YPP1 | pYGR198W∷kanR-tet07-TATA URA3∷CMV-tTA his3-1 leu2-0 met15-0 | MATa |
| Tet-SEC1 | pSEC1∷kanR-tet07-TATA URA3∷CMV-tTA his3-1 leu2-0 met15-0 | MATa |
| Tet-SEC5 | pSEC5∷kanR-tet07-TATA URA3∷CMV-tTA his3-1 leu2-0 met15-0 | MATa |
| Tet-SEC12 | pSEC12∷kanR-tet07-TATA URA3∷CMV-tTA MATa his3-1 leu2-0 met15-0 | MATa |
| Plasmids | ||
| pRS313 | Low copy CEN HIS3 ARS Amp r | ATCC |
| pRS314 | Low copy CEN TRP1 ARS Ampr | ATCC |
| pRS316 | Low copy CEN URA3 ARS Ampr | ATCC |
| pRS326 | 2 μ URA3 Ampr | (Christianson et al., 1992) |
| pRS325 | 2 μ LEU2 Ampr | (Christianson et al., 1992) |
| pTF200 | GAL1 promoter in pRS314 | (Flower et al., 2005) |
| pTF201 | GAL1 promoter WT α-syn in pRS314 | (Flower et al., 2005) |
| pTF202 | GAL1 promoter A30P α-syn in pRS314 | (Flower et al., 2005) |
| pTF203 | GAL1 promoter A53T α-syn in pRS314 | (Flower et al., 2005) |
| pTF300 | GAL1 promoter in pRS316 | (Flower et al., 2005) |
| pTF301 | GAL1 promoter WT α-syn in pRS316 | (Flower et al., 2005) |
| pTF302 | GAL1 promoter A30P α-syn in pRS316 | (Flower et al., 2005) |
| pTF303 | GAL1 promoter A53T α-syn in pRS316 | (Flower et al., 2005) |
| pTF305 | GAL1 promoter GFP-WT α-syn in pRS316 | This study |
| pTF306 | GAL1 promoter GFP-A30P in pRS316 | This study |
| pTF307 | GAL1 promoter GFP-A53T in pRS316 | This study |
| pTF308 | GAL1 promoter GFP in pRS316 | This study |
| pTF503 | GAL1 promoter cMyc (9 repeats) in pRS326 | This study |
| pTF504 | GAL1 promoter cMyc (9 repeats) YPP1 in pRS326 | This study |
| pTF601 | GAL1 promoter YPP1 in pRS326 | This study |
| pTF602 | GAL1 promoter YPP1 in pRS325 | This study |
| pTF603 | GAL1 promoter in pRS326 | This study |
| pTF604 | GAL1 promoter in pRS325 | This study |
| pTF700 | GAL1 promoter MON1 in pRS313 | This study |
| pTF701 | GAL1 promoter SLA1 in pRS313 | This study |
YPP1 codes for a protein with a theoretical molecular mass of 95.4 kD. Cells expressing α-syn and transformed with a plasmid harboring a myc-tagged YPP1 fusion indeed expressed myc-Ypp1p, as judged by Western blot analysis and staining with an anti-myc antibody (Fig. 1 B). Specifically, myc-Ypp1p was detected in cells that coexpressed WT α-syn or A30P (lanes 2 and 4); however, in each case, a ladder of bands ranging from 95 to >200 kD was observed. We attribute the band at 95 kD to myc-Ypp1p and suggest that the bands at higher molecular mass are posttranslationally modified forms of Ypp1p. The four major bands of myc-tagged Ypp1p were absent in lysates of cells that did not harbor the MYC-YPP1 plasmid (lanes 1 and 3).
The effect of Ypp1p overexpression on the viability of cells expressing the various α-syns was also evaluated. A viability assay (Fannjiang et al., 2004) was conducted using the dye FUN1 on cells induced for 12 h. Dead cells stained green; metabolically active, and hence viable cells, stained red; and a small percentage of cells (∼10%) failed to stain. The percentage of red cells indicated viability. For cells expressing A30P with Ypp1p overexpression, 90% of the cells were viable, whereas only 10% of cells were viable when A30P was expressed without Ypp1p overexpression (Fig. 1, C and D). In contrast, for cells expressing WT α-syn or A53T, with or without Ypp1p overexpression, only ∼10% of the cells were viable (Fig. 1 D). This viability assay demonstrated that YPP1 in high copy specifically enhances the viability of A30P expressing cells but not of cells expressing the other two α-syns (WT or A53T).
YPP1 suppresses ROS accumulation in A30P expressing cells
A signature feature of α-syn is that it induces oxidative stress in various types of cells (Xu et al., 2002; Flower et al., 2005; Smith et al., 2005). Given that YPP1 in high copy suppresses the super sensitivity of A30P expressing cells to killing by hydrogen peroxide, we expected that Ypp1p overexpression would abolish ROS production in A30P expressing yeast cells (Flower et al., 2005), but not in cells expressing WT or A53T α-syn. To test this hypothesis, cells expressing α-syn (WT, A30P, or A53T) with decreased, endogenous, or increased levels of Ypp1p were stained with the cell-permeant dye DHR 123. This dye enters cells, and when oxidized by free radicals yields a fluorescent product (Schulz et al., 1996). For these experiments a strain from the “Hughes collection” of titratable promoter alleles was used (Mnaimneh et al., 2004) in which a kanR-tetO7-TATA cassette is integrated into the promoter of YPP1. Repression is controlled by adding doxycycline, which has no appreciable effect on global gene expression at the concentrations (20 μg/ml) used for the promoter shut off.
Fig. 2 (A–C) shows ROS accumulation in cells expressing WT α-syn, A30P, or empty vector controls. ROS accumulation in cells expressing WT α-syn (or A53T) was insensitive to variations in the level of Ypp1p (compare the three vertical panels labeled DHR in Fig. 2 A). In contrast, ROS accumulation in cells expressing A30P was exquisitively sensitive to the level of Ypp1p: 100% of the cells exhibited intense red fluorescence when the level of Ypp1p was decreased (+doxy), whereas only ∼5% of the cells exhibited red fluorescence when Ypp1p was overexpressed (compare the three vertical panels labeled DHR in Fig. 2 B). Identically treated control cells harboring the plasmid with no insert exhibited no appreciable ROS accumulation (Fig. 2 C). These experiments revealed that Ypp1p, when overexpressed, abolished ROS accumulation in cells expressing A30P—but not in cells expressing WT α-syn or A53T (Fig. 2 D).
Figure 2.
YPP1 suppresses ROS accumulation in A30P expressing cells. ROS accumulation in cells expressing A30P (pTF302) (A), WT α-syn (pTF301) (B), or vector control cells (pTF300) (C). The “Hughes” YPP1 strain that contains a titratable promoter was used in these experiments. This strain transformed with pTF602 (YPP1) or pTF604 (empty vector) was used for these experiments. For shutting off YPP1 (+doxy), cells were pregrown in noninducing media with 20 μM doxycycline for 7 h, and then shifted into inducing media with doxycycline. For experiments with Ypp1p overexpression, cells were pregrown in noninducing media to mid-log, and then shifted into inducing media. The images shown in all panels were obtained at 3 h in inducing media. The dye DHR123 was added after 2 h in inducing media to give 5 μg/ml, and the cells were then washed before image acquisition. DHR, rhodamine 123. DIC, differential interference contrast. (D) Plot of the percentage of cells that exhibited red fluorescence (mean ± SD). The number of cells counted for each group was n = 640 to 1,200. The symbols "−", “e”, and “+” indicate decreased (with doxycycline), endogenous, and increased levels of Ypp1p, respectively. (E) Coimmunoprecipitation shows selective binding of myc-Ypp1p to A30P. Cells expressing the various α-syns and overexpressing myc-tagged Ypp1p were lysed, the extract was incubated with an anti-myc antibody-protein A resin, and the resin was boiled and reduced and subjected to SDS-PAGE and immunoblotting. The top panels show the ∼95-kD band due to myc-Ypp1p; the bottom panels show α-syn. Lane 1, WT α-syn lysate; lane 2, myc-Ypp1p fails to pull down WT α-syn; lane 3, A30P lysate; lane 4, myc-Ypp1p pulls down A30P; lane 5, A53T lysate; lane 6, myc-Ypp1p fails to pull down A53T. The FY23 strain was transformed with pTF201 (WT α-syn), pTF202 (A30P), or pTF203 (A53T) and with pTF504 (myc-YPP1) or pTF503 (empty vector).
Ypp1p binds to A30P—but not to WT or A53T
Coimmunoprecipitation experiments were performed to determine whether Ypp1p physically associated with A30P. Coimmunoprecipitations of cells overexpressing Ypp1p with WT α-syn, A30P, or A53T coexpressed are shown in Fig. 2 E (lanes 1–6). For each sample, one lane contained the lysate and the other lane contained the myc-Ypp1p pull down. The myc-Ypp1p (∼95-kD band) was visualized with an anti-myc antibody and the α-syns (∼19 kD) with an anti-α-syn antibody. A comparison of the two blots showed that Ypp1p pulled down A30P (lane 4), but not WT α-syn or A53T (lanes 2 and 6). The experiments revealed that, when overexpressed, Ypp1p associates with A30P.
We also found that in high copy YPP1 permitted normal growth of cells expressing A30P but not of cells expressing WT or A53T (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200610071/DC1), and that in high copy YPP1 failed to protect yeast cells from hydrogen peroxide-induced ROS (Fig. S2). YPP1 is thus unlikely to code for an enzyme that inactivates hydrogen peroxide.
Ypp1p alters GFP-α-syn localization
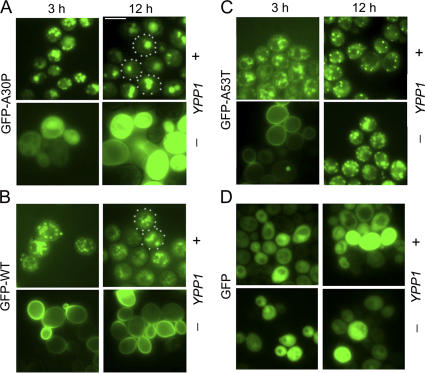
To gain insight into the mechanism of suppression, fluorescence microscopy studies were conducted to determine the effect of Ypp1p overexpression on the localization of the various GFP-tagged α-syns. We found that Ypp1p overexpression altered the localization of each of the three GFP-tagged α-syns (WT, A30P, or A53T), but in subtly distinct ways compared with control cells without Ypp1p overexpression (Fig. 3, A–C). For example, cells expressing GFP-A30P with Ypp1p overexpression exhibited 3 to 6 inclusions per cell at 3 h of induction, whereas at 12 h the inclusions had coalesced into 1 to 2 larger inclusions (Fig. 3 A). Without Ypp1p overexpression, GFP-A30P expressing cells exhibited diffuse green fluorescence at 3 and 12 h. In contrast, Ypp1p, when overexpressed, even drove GFP-WT α-syn and GFP-A53T from the plasma membrane into inclusions, but in each case the inclusions failed to efficiently merge with one another (Fig. 3, B and C). Notice that control cells expressing GFP exhibited diffuse green fluorescence that was unaffected by increased levels of Ypp1p (Fig. 3 D). The experiments revealed that Ypp1p when overexpressed drives the various α-syns into inclusions. That Ypp1p concentrated GFP-A30P into 1 to 2 inclusions per cell raised the possibility that Ypp1p also drives A30P into a compartment such as the vacuole.
Figure 3.
Ypp1p alters GFP-α-syn localization. The effect of Ypp1p overexpression on the localization of (A) GFP-A30P, (B) GFP-WT α-syn, (C) GFP-A53T, and (D) GFP at 3 and 12 h of induction. In each group of four panels, the top and bottom rows are of cells expressing GFP-α-syn with and without Ypp1p overexpressed, respectively. S288c cells were transformed with pTF305 (WT α-syn), pTF306 (A30P), pTF307 (A53T), or pTF308 (GFP) and pTF602 (YPP1) or pTF604 (empty vector). Bar, 5 μm.
Ypp1p drives A30P to the vacuole
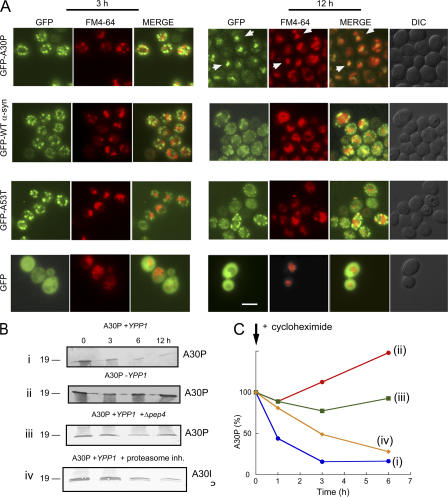
To test the hypothesis that Ypp1p drives A30P to the lysosome/vacuole, two-color fluorescence microscopy experiments were conducted using cells expressing the various GFP-tagged α-syns and the lipophilic dye FM4-64, which stains vacuoles (Vida and Emr, 1995). At various times (3 or 12 h) after induction, cells expressing GFP-α-syn were incubated with the FM4-64 dye and microscopy images were acquired (Fig. 4 A). If GFP-α-syn enters the vacuole then the green fluorescence from GFP and the red fluorescence from FM4-64 should overlap. No overlap between any of the GFP-α-syn inclusions with the red structures occurred after 3 h of induction, whereas almost every one of the GFP-A30P inclusions overlapped with the red structures after 12 h of induction. No such overlap occurred for GFP-WT α-syn or GFP-A53T structures with vacuoles. However, because vacuoles were not prominent in the DIC images (12 h), these experiments do not prove that Ypp1p drove A30P into the vacuole. For this reason, the following biochemical analysis was conducted.
Figure 4.
Ypp1p drives A30P to the vacuole. (A) Two-color fluorescence microscopy experiments of cells expressing GFP-α-syn with Ypp1p overexpression. S288c cells transformed with pTF305 (WT α-syn), pTF306 (A30P), pTF307 (A53T), or pTF308 (empty vector) and pTF602 (YPP1) or pTF604 (empty vector) were pregrown in noninducing media to mid-log phase, transferred to inducing media, and induced for 3 or 12 h. Before the analysis cells were stained with the dye FM4-64. Aliquots were removed, washed, resuspended in YPD, and incubated with 40 μM FM4-64 for 10 min at 30°C. Cells were then washed twice, resuspended in YPD, incubated an additional 30 min at 30°C, and then visualized by fluorescence microscopy. The white arrows (top right-hand panel, GFP-A30P) indicate areas of overlap between the GFP inclusions with the red structures. Bar, 5 μm. (B) Ypp1p promotes degradation of A30P. Western blot analysis was conducted to monitor the level of A30P in cells. Blots (i and ii): Ypp1p overexpression. 10 μM cycloheximide was used to halt protein synthesis in cells with (i) or without (ii) Ypp1p overexpression. S288c cells transformed with pTF202 (A30P) and pTF602 (YPP1) or pTF604 (empty vector) were pregrown in noninducing media to mid-log phase, transferred to inducing media, inhibited with cycloheximide, and induced for 12 h. Indicated times are after addition of cycloheximide. Blot (iii): Deletion of PEP4. A Δpep4 deletion strain transformed with pTF202 (A30P) and pTF602 (YPP1) was pregrown in noninducing media to mid-log phase, transferred to inducing media, inhibited with 10 μM cycloheximide, and induced for 12 h. Indicated times are after addition of cycloheximide. Blot (iv): Proteasome inhibition. S288c cells transformed with pTF202 (A30P) and pTF602 (YPP1) were pregrown in noninducing media to mid-log phase, transferred to inducing media, inhibited with 10 μM cycloheximide and 50 μM of MG132, and induced for 12 h. Indicated times are after addition of inhibitors. For each experiment, cell extracts were prepared, subjected to SDS-PAGE, and immunoblotted using a monoclonal antibody specific for α-syn. In each blot, 20 μg of protein was loaded per well. Experiments were conducted two or three times. (C) Degradation of A30P. Band intensities in the blots were determined using a scanner with image quantization software and plotted against time. (i) Blue, A30P + Ypp1p overexpression; (ii) red, A30P + endogenous level of Ypp1p; (iii) green, A30P + Ypp1p overexpression in Δpep4 strain; (iv) yellow, A30P + Ypp1 overexpression + 50 μM MG132.
Ypp1p-mediated transport of A30P to the vacuole was tested using Western blot analysis to monitor the level of the A30P protein. Cells transformed with the various plasmids were pregrown in noninducing media to mid-log phase, shifted to inducing media, inhibited with 10 μM cycloheximide to halt protein synthesis, and incubated for 12 h. Aliquots were removed at the indicated times and extracts were prepared, subjected to SDS-PAGE followed by Western blot analysis. First, we found that A30P was rapidly degraded (t1/2 ∼1 h) in cells overexpressing Ypp1p (Fig. 4 B i, and C), whereas in cells expressing A30P, but with no Ypp1p overexpression, no such degradation of A30P occurred (Fig. 4 B ii, and C). Second, if A30P is degraded in the vacuole, then A30P should fail to be degraded in a strain that lacks PEP4, which is a gene that codes for a protease that activates a variety of vacuolar proteases (Jones et al., 1982). To test this hypothesis, the level of A30P was monitored in a Δpep4 deletion strain, and the A30P protein showed no appreciable degradation in this strain (Fig. 4 B iii, and C). Third, to rule out the possibility that Ypp1p mediates the proteasomal degradation of A30P, experiments were also conducted using the proteasome inhibitor MG132 (50 μM). With protein synthesis inhibited and with proteasome function inhibited A30P was still degraded (Fig. 4 B iv, and C), albeit not with the exact kinetics as observed when only cycloheximide was used. The ability of the pep4 deletion to halt A30P degradation, combined with the lack of appreciable inhibition of A30P degradation by a proteasome inhibitor, indicated that Ypp1p mediates the transport of A30P to the vacuole, where it is proteolytically degraded.
Ultra structural analysis
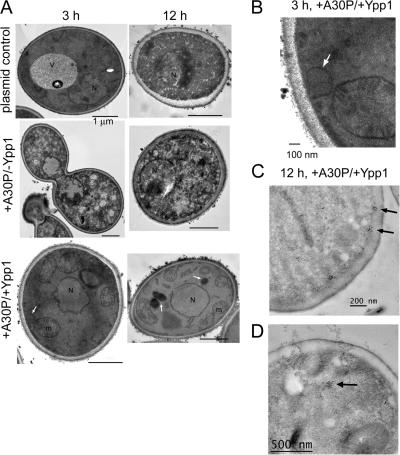
Transmission electron microscopy was conducted to gain insight into the nature of the α-syn inclusions in +A30P/+Ypp1 cells. A30P expression induced dramatic morphological changes in cells at only 3 h of induction compared with control cells (Fig. 5 A). Changes included granulation of the cytosol and chromatin condensation. In contrast, control cells containing two empty plasmids (−A30P/−Ypp1) had a well-delineated vacuole and cytoplasm. Cells expressing A30P with Ypp1p overexpressed exhibited smaller vacuoles than cells with the two empty plasmids and had numerous mitochondria per slice. Vesicles budding off of the plasma membrane were also observed in these cells. These vesicles were absent in cells expressing only A30P. Such a vesicle can be seen in the bottom left image in Fig. 5 A (denoted by the white arrow). At higher magnification the vesicle appears to be emerging from the plasma membrane (Fig. 5 B).
Figure 5.
Transmission electron microscopy images of cells expressing Ypp1p and A30P. (A) S288c cells transformed with pTF302 (A30P) or pTF300 (empty vector) and pTF602 (YPP1) or pTF604 (empty vector) were pregrown in noninducing media to mid-log phase, transferred to inducing media and induced for 3 or 12 h. Top: −A30P/−Ypp1 (cells contained two plasmids with no inserts). Middle: +A30P/−Ypp1 (cells contained two plasmids, one with no insert). Bottom: +A30P/+Ypp1 (cells contained two plasmids). Bars, 1 μm. (B) Magnification of +A30P/+Ypp1 cell at 3 h shows vesicle budding from the plasma membrane. (C and D) Immunogold labeling of cells expressing A30P and overexpressing Ypp1p at 12 h. A secondary antibody conjugated with 50-nm gold particles was used to detect A30P. Several clusters of gold particles occurred in the vicinity of the plasma membrane (C) and also in the interior of cells (D).
After 12 h of induction, cells expressing only A30P were characterized by a granulated cytosol and even more extensive chromatin condensation compared with 3 h induction. Comparing this TEM image to the images of the −A30P/−Ypp1 control cells and +A30P/+Ypp1 cells revealed the following differences: (1) control cells with two empty plasmids exhibited a granulated cytosol and chromatin condensation (Fig. 5 A). These changes did not occur when the strain was incubated in glucose or sucrose for 12 h. The changes occurred because of the abrupt shift from sucrose to galactose. It turns out that the wt S288c strain, but not the Resgen deletion collection, which is derived from the S288c strain, has a mutation in GAL2. After ∼15 generations WT S288c cells begin to lose viability. Using the FUN1 assay we found that 90% and 60% of the WT S288c cells were viable after 3 h and 12 h in galactose (unpublished data), respectively. It should be pointed out that YPP1 in high copy suppressed A30P-induced ROS at 12 h in strains FY23, S288c, and various S288c gene deletion strains. (2) The +A30P/+Ypp1 cells were characterized by a normal nucleus, numerous mitochondria per optical slice, and three large vacuoles (denoted by white arrows; Fig. 5 A). More than 90% of the EM sections examined contained such vacuoles.
Immunogold labeling was also performed to visualize the subcellular location of A30P. Cells expressing A30P and overexpressing Ypp1p were characterized by numerous clusters of gold particles in association with the plasma membrane (Fig. 5 C). In contrast, cells expressing A30P only, or harboring the two control plasmids, did not have gold particles in association with the plasma membrane. Therefore, we attributed the membrane-associated clusters to A30P molecules that were being packaged by Ypp1p into endocytic vesicles. Note that the membrane-associated clusters are smaller than the budding vesicle (compare Fig. 5, B and C); this could be because of the difficulty of preserving membrane structure for immunogold labeling and also because only a small portion of a vesicle contains A30P. Clusters of gold particles were also evident inside the cells (Fig. 5 D). The TEM results showed that YPP1 in high copy protects cells from A30P.
Genes that interact with YPP1
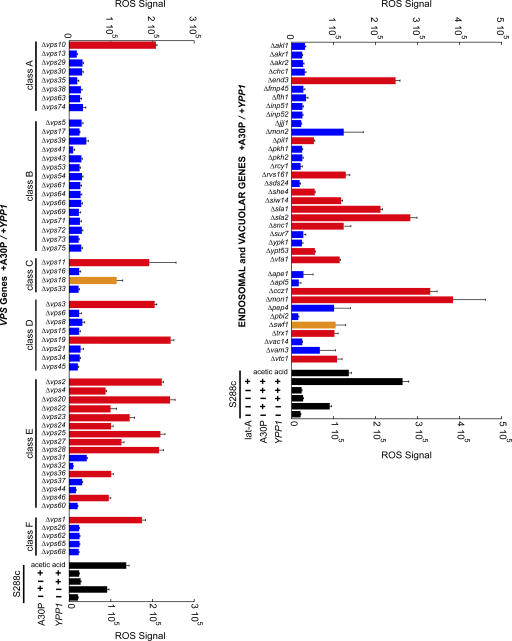
To begin to define the pathway by which YPP1 suppresses A30P toxicity, a targeted screen of 116 nonessential genes involved in autophagy, cytosol-to-vacuole transport (Cvt), endocytosis, and the vacuole protein sorting pathway (vps) was conducted (Fig. 6 and Table S1 for entire list, available at http://www.jcb.org/cgi/content/full/jcb.200610071/DC1). We reasoned that any gene that cooperates with YPP1 to protect against A30P, when deleted, should increase ROS accumulation. The ROS assay using the DHR 123 dye was conducted in 96-well plates on cells induced for 3 h using a plate reader with fluorescence detection. The various deletion strains were transformed with the pTF302 (A30P) and pTF602 (YPP1) plasmids.
Figure 6.
Screening for genes that interact withYPP1. The indicated deletion strains transformed with pTF302 (A30P) and pTF602 (YPP1) were pregrown in noninducing media to mid-log phase, transferred to inducing media, and induced for 3 h. Matched controls consisted of identically treated S288c cells transformed with the same two plasmids. The dye DHR 123 was added at 2 h after induction. After several washing steps, cells were aliquoted into 96-well plates and the DHR 123 fluorescent product was detected using a plate reader. Cells incubated in dilute acetic acid exhibit ROS (Ludovico et al., 2002), and this served as a positive control. Latrunculin A was used at 25 μM. Experiments were performed in two to five independent experiments, each in triplicate. Bars indicate mean ROS signal ± SD. Comparing each deletion strain to matched controls (S288c +A30P/+Ypp1) yielded P-values: red, P < 0.001; orange, P < 0.05; and blue, P > 0.05. Exact P values are given in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200610071/DC1).
The vacuolar protein sorting pathway (vps) is a network of genes involved in the transport of newly synthesized proteins from the late Golgi to the vacuole (Bowers and Stevens, 2005). 56 VPS deletion strains were analyzed, and of these 56 strains 16 gave statistically significant increases in ROS (Fig. 6). Curiously, 11 of the 16 hits clustered in the class E VPS genes. Many of the class E VPS genes code for ESCRT (Hurley and Emr, 2006) proteins that function to sort membrane-bound proteins into lumenal multivesicular body vesicles (MVB). Our interpretation of these findings is that the deletion of certain class E genes resulted in the failure of A30P to be sorted into lumenal MVB vesicles, and this prevented A30P from being delivered to and degraded within the vacuole. Failure to degrade A30P resulted in the accumulation of ROS.
Data obtained from selected deletion strains associated with actin organization, autophagy, Cvt, endocytosis, vacuolar biogenesis, and vesicle–vacuole fusion, are presented in Fig. 6 and Table S1. First, five deletion strains gave very large (>200,000 units; P < 0.001) increases in ROS compared with the WT control (25,818 ± 7005 units) (Fig. 6). These five strains were: Δsla1, Δsla2, Δend3, Δmon1, and Δccz1. Sla1, Sla2, and End3 proteins form a complex on the lumenal side of the plasma membrane (Holtzman et al., 1993; Raths et al., 1993), where they regulate actin dynamics and proteins required for endocytosis (Toshima et al., 2006). The Ccz1p–Mon1p complex is required for nearly all membrane-trafficking pathways where the terminal acceptor compartment is the vacuole (Wang et al., 2002). Second, six deletion strains gave large (>100,000 units; P < 0.001) increases in ROS compared with the WT control (Fig. 6). These strains were: Δrvs161, Δsiw14, Δsnc1, Δtrx1, Δvta1, and Δvtc1. In general, these genes have roles in vesicle trafficking and actin filament organization. Third, eleven genes involved in the autophagy and Cvt pathways, when deleted, failed to increase ROS (Table S1). The broad picture that emerged from this screen was that YPP1 suppressed A30P toxicity via a pathway involving SLA1, SLA2, and END3, class E VPS genes, and MON1 and CCZ1. Given that these genes are involved in endocytosis and vesicle trafficking to the vacuole, such a finding, together with the fluorescence and TEM data (Figs. 3–5 ), indicate a role for YPP1 in the endocytic pathway.
Latrunculin A, a known inhibitor of actin (Ayscough et al., 1997), disrupts the trafficking of endosomal vesicles from the plasma membrane to the vacuole (Toshima et al., 2006). Latrunculin A (25 μM)–treated cells expressing A30P and overexpressing Ypp1p indeed exhibited a large increase in ROS signal (284,979 ± 10,816) compared with identically treated cells that lacked latrunculin A (25,818 ± 7005) (Fig. 6). Our interpretation of this result is that latrunculin A inhibits the formation of actin filaments which in turn blocks the budding of endocytic vesicles containing A30P and delivery of A30P to the vacuole.
Mechanism of YPP1 suppression
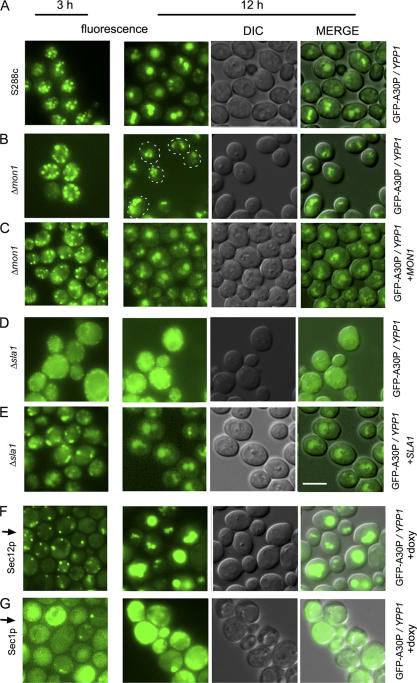
To verify results obtained from the ROS plate reader assay, the Δmon1 and Δsla1 strains were selected for further analysis. The S288c control strain expressing GFP-A30P and overexpressing Ypp1p exhibited 4 to 5 inclusions per cell at 3 h, and these inclusions had coalesced into 1 to 3 inclusions per cell at 12 h (Fig. 7 A). The Δmon1 strain expressing GFP-A30P and overexpressing Ypp1p also exhibited 4 to 5 inclusions per cell at 3 h (Fig. 7 B), but at 12 h multiple inclusions per cell were still evident. Given its role in catalyzing the fusion of vesicles with the vacuole (Wang et al., 2002), deletion of MON1 would be expected to hinder fusion of A30P-containing vesicles with the vacuole, and the microscopy images are consistent with such a defect. Notice that adding back MON1 on a plasmid rescued the inability of vesicles to coalesce at 12 h in the Δmon1 strain (Fig. 7 C). These experiments confirmed that MON1 is involved in the pathway by which YPP1 rids cells of A30P.
Figure 7.
Effects of gene deletions on YPP1-mediated A30P trafficking. The S288c strain and the two deletion strains transformed with pTF306 (GFP-A30P) and pTF602 (YPP1) were pregrown in noninducing media to mid-log phase, transferred to inducing media, induced for 3 or 12 h, and then visualized by fluorescence microscopy. The SEC1 and SEC12 Hughes strains transformed with pTF306 (GFP-A30P) and pTF602 (YPP1) were pregrown in noninducing media with doxycycline (20 μg/ml) for 7 h, transferred to inducing media with doxycycline, and then visualized at 3 and 12 h by fluorescence microscopy. (A) S288c; (B) Δmon1; (C) Δmon1 and pTF700 (MON1); (D) Δsla1; (E) Δsla1 and pTF701 (SLA1); (F) tet-SEC12 strain + doxycycline; and (G) tet-SEC1 strain + doxycycline. (A–E): Images labeled “MERGE” had brightness and contrast adjustments of −12 and +20, respectively. (F and G): images labeled MERGE had brightness and contrast adjustments of +40 and +20, respectively. Bar, 5 μm.
The effect of the Δsla1 deletion on Ypp1p-mediated trafficking of GFP-A30P was also analyzed. After 3 or 12 h of induction Δsla1 cells expressing GFP-A30P and overexpressing Ypp1p displayed green fluorescence throughout the cell (Fig. 7 D). These images are very different from the images acquired from the S288c and Δmon1 strains. Our interpretation of the Δsla1 images is that deletion of SLA1 prevents the packaging of GFP-A30P into vesicles. Notice that adding back SLA1 on a plasmid resulted in the formation of ∼0–3 inclusions per cell after 3 h of induction; whereas, after 12 h of induction the inclusions had coalesced yielding 1–2 inclusions per cell (Fig. 7 E). These images from 12 h are remarkably similar to the images obtained at 12 h from the WT strain. Accordingly, we concluded that deletion of SLA1 resulted in a failure of cells to package A30P into vesicles.
An issue is whether A30P transits to the vacuole via the endocytic pathway, as proposed, or the secretory pathway. ER stress occurs when unfolded proteins accumulate in the ER, and such proteins are typically retrotranslocated out of the ER to the proteasome for degradation; this process is called ERAD (endoplasmic reticulum–associated degradation) (Meusser et al., 2005). YPP1 does not facilitate retrotranslocation and ERAD because A30P was still degraded when the proteasome was inhibited (Fig. 4, B and C). However, yeast have another pathway for the degradation of soluble, unfolded proteins that transit through the ER and Golgi, and this occurs via receptor-mediated forward transport of unfolded luminal proteins to the vacuole (Hong et al., 1996). The receptor for this process is coded for by VPS10. Because deletion of VPS10 caused intense ROS accumulation in cells expressing A30P (Fig. 6), perhaps YPP1 is an enhancer of the forward transport of luminal proteins to the vacuole. On the other hand, although WT α-syn and A53T use the secretory pathway and cause ER stress (Cooper et al., 2006), such findings have not been reported for A30P. Instead, evidence exists that A30P does not use the secretory pathway (Dixon et al., 2005).
We examined whether loss of function of one early-acting and two late-acting SEC genes, i.e., SEC12, SEC1, and SEC5, respectively, affected YPP1-mediated trafficking of A30P to the vacuole. Sec12p controls transport vesicle budding from the ER (Barlowe and Schekman, 1993). Sec1p controls the fusion of secretory vesicles with the plasma membrane (Wiederkehr et al., 2004), and when exocytosis is shut down actin regulation also becomes disrupted (Aronov and Gerst, 2004). The Sec5 protein is a component of the essential exocyst complex, which tethers secretory vesicles to the plasma membrane (TerBush et al., 1996). If YPP1 mediates A30P trafficking through the ER-Golgi to the vacuole, then this trafficking should be disrupted in the sec12 but not in the sec1 or sec5 strains. Conversely, if YPP1 mediates A30P trafficking to the vacuole via endocytosis, then this trafficking should be disrupted in the sec1 and sec5 but not in the sec12 strains. The Hughes collection of titratable promoter alleles (Mnaimneh et al., 2004) was used in these experiments.
Decreasing the level of the essential Sec12 protein had no appreciable effect on YPP1-mediated trafficking of A30P to the vacuole. Specifically, cells expressing GFP-A30P and overexpressing Ypp1p, but with a decreased level of Sec12p, exhibited 1–2 large GFP-A30P inclusions per cell, which coincided with the vacuole (Fig. 7 F). In contrast, decreasing the level of the essential Sec1 protein abolished YPP1-mediated trafficking of A30P to the vacuole. Specifically, cells expressing GFP-A30P and overexpressing Ypp1p, but with a decreased level of Sec1p, exhibited no GFP-A30P inclusions after 3 or 12 h of induction (Fig. 7 G). Similar results were obtained for cells with a decreased level of the Sec5 protein (unpublished data). Because decreasing the level of Sec1p, or Sec5p, or deleting SLA1 eliminated YPP1-mediated transport of A30P to the vacuole, we concluded that YPP1 mediates the trafficking of A30P to the vacuole via the endocytic pathway.
YPP1 participates in receptor-mediated endocytosis
Several reports have indicated that YPP1 functions in a MAPK pathway (Roberts et al., 2000; Hazbun et al., 2003; Mnaimneh et al., 2004). Because the yeast mating response is a MAPK pathway (Schwartz and Madhani, 2004) that involves receptor-mediated endocytosis (RME), we asked whether YPP1 functions in the yeast mating response (Engqvist-Goldstein and Drubin, 2003). In yeast, the response to pheromone is a well characterized example of RME: α-factor pheromone binds to Ste2p (on Mata cells) and triggers a sequence of events in which Ste2p and the α-factor are encapsulated into endosomes, which then transit along actin cables to the vacuole, where the pheromone and its receptor are proteolytically degraded (Dohlman, 2002). The hypothesis that YPP1 participates in RME was tested by incubating α-factor with a Mata haploid strain containing an integrated YPP1-GFP allele replacing the WT allele (Table I).
The Mata haploid strain expressing Ypp1p-GFP exhibited no change in the localization of Ypp1p-GFP upon incubation for 20 min, and no change in localization was detected even after hours of incubation. Weak, diffuse green fluorescence occurred throughout the cytosol in these cells, and somewhat more brightly fluorescent puncta appeared around the periphery of the cells (Fig. 8 A). The enhanced staining around the periphery was consistent with Ypp1p-GFP associating with actin cortical patches, which are thought to be the sites of exocytosis and endocytosis. In contrast, treatment of this strain with α-factor resulted in a rapid change in the localization of Ypp1p-GFP and a parallel increase in the fluorescence signal (Fig. 8 A). Cells exhibited numerous vesicles that appeared to coalesce into larger structures after 20 min. The green inclusions of Ypp1p-GFP merged with the vacuolar structures in the DIC images. These experiments showed that the endosomes formed in response to pheromone contained Ypp1p-GFP, and that such Ypp1p-GFP–containing endosomes then merged with the vacuole. This data strongly supported our conclusion that Ypp1p is involved in endocytosis.
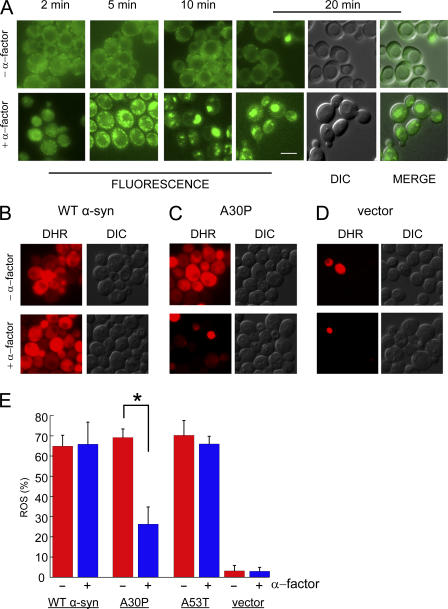
Figure 8.
Pheromone protects cells from A30P toxicity. (A) Effect of α-factor on cells expressing Ypp1p-GFP. Top (−α-factor): no change in the localization of Ypp1p-GFP occurred over 20 min. Bottom (+α-factor): 10 μM α-factor was added to cells and then images were acquired from 2 to 20 min. Strain: YPP1-GFP cultured in YPD. Identical instrumental conditions were used for all images. Bar, 5 μm. The ROS assay using the DHR 123 dye is shown in (B) WT α-syn, (C) A30P, and (D) vector control. The FY23 strain was transformed with pTF201 (WT α-syn), pTF202 (A30P), pTF203 (A53T), or pTF200 (empty vector) and pregrown in noninducing media to mid-log phase. 10 μM pheromone was added after shifting to inducing media; after 2 h in inducing media the DHR 123 dye was added to 5 μg/ml; and, before acquiring images at 3 h, cells were washed and resuspended in inducing media. The two images labeled “MERGE” (in A) had brightness and contrast adjustments of +10 and +40, respectively. (E) Plot of mean ROS (%) (± SD). Cells (n = 292–696) were counted in two independent experiments. *, P = 1.1 × 10−7. DIC, differential interference contrast; DHR, rhodamine 123.
Intrigued by the ability of pheromone to drive Ypp1p into vesicles, the hypothesis that pheromone itself could protect cells from the toxicity of the A30P protein—by driving A30P into endosomes, which then traffic to the vacuole, where A30P is degraded—was tested. The experiment was conducted by transforming the FY23 Mata haploid strain with plasmids for the various α-syns (WT, A30P, or A53T), pregrowing cells in noninducing media to mid-log phase, and shifting cells to inducing media for 3 h. The α-factor pheromone (5–10 μM) was added upon the shift into inducing media; the DHR 123 dye was added after 2 h in inducing media; and the images were acquired after 3 h in inducing media. The α-factor had no effect on ROS accumulation in cells expressing WT α-syn (or A53T), as judged by red fluorescence (Fig. 8, B and E). In contrast, in identically treated cells expressing A30P α-factor decreased the percentage of red staining A30P-expressing cells to 26.3 ± 4.2% from 69.2 ± 8.5% (P = 1.1 × 10−7) (Fig. 8, C and E). No appreciable amounts of ROS occurred in vector control cells with or without pheromone (Fig. 8 D). Our interpretation of these results was that pheromone triggered rapid RME, and because of Ypp1p's ability to bind A30P at the plasma membrane, A30P was encapsulated into the endocytic vesicles, which then trafficked to the vacuole.
Discussion
The biochemical, cell biological, and genetic experiments in this study have demonstrated a role for the essential gene YPP1 in endocytosis. We conducted a PSI-blast analysis of YPP1, using the Saccharomyces Genome Database, and found six potential functional homologues (orthologues) in humans. One of these, human TTC7B (Q86TV6), has 15% identical residues with YPP1. The TTC7B protein contains multiple tetratricopeptide repeat (TPR) protein–protein interaction domains (Lamb et al., 1995), and it is likely that the Ypp1 protein also contains such domains. Other than TPR domains, Ypp1p has no recognizable domains. Next, several issues are discussed relating to the role of Ypp1p in endocytosis and protection it affords against A30P.
Notice that A30P expression in the pep4Δ strain with YPP1 in high copy caused no appreciable toxicity, as judged by ROS accumulation (Fig. 6). We propose that YPP1 in high copy, whether in WT cells or pep4Δ cells, mediates the transport of A30P to vacuole. The lack of toxicity of A30P in pep4Δ cells with YPP1 in high copy is due to the removal of A30P from the plasma membrane and cytosol and sequestration in vacuoles. Suppression of A30P toxicity could be linked more to its sequestration in the vacuole rather than its degradation.
If Ypp1p functions in the endocytic pathway, why does Ypp1p select A30P, which appears to be a cytosolic protein, and target it to the vacuole instead of WT α-syn or A53T, which are membrane bound? Points addressing the selectivity of Ypp1p for A30P are: (1) A30P is not only a cytosolic protein; it also associates with membranes, albeit not as strongly as WT α-syn or A53T, and permeablizes the membranes to which it binds (Lashuel et al., 2002; Furukawa et al., 2006). Therefore, membrane-associated A30P should be able to associate with membrane-associated Ypp1p. (2) When overexpressed, Ypp1p rapidly packaged each of the α-syns (WT, A30P, and A53T) into inclusions/vesicles, albeit only A30P-containing vesicles then merged with the vacuole (Fig. 3, Fig. 4 A, and Fig. 5). The transport of A30P-containing vesicles to the vacuole is no doubt casually connected to the binding of A30P to Ypp1p. Such results suggest two separable functions for Ypp1p. The incomplete endocytosis of WT and A53T α-syn—which is proposed to be due to their inability to bind Ypp1p—results in the accumulation of Lewy body–like structures. (3) It is important to recognize that a spectrum of protein–protein equilibrium constants, from nanomolar to micromolar, occurs in cells. Ypp1p may weakly associate with WT α-syn or A53T, but the short-lived complexes cannot be pulled out of solution. The A→P substitution could increase the strength of the association between α-syn and Ypp1p for many reasons. We reason that, compared with WT α-syn and A53T, A30P may adopt a different conformation when membrane bound, it may possess a different phosphorylation pattern, and it may be ubiquitinylated at a different site. These unique features could foster a strong association between A30P and Ypp1p, resulting in kinetically stable complexes that can be pulled out of solution.
WT α-syn and the A53T mutant cause ER stress (Gosavi et al., 2002; Smith et al., 2005; Cooper et al., 2006), and, above a certain threshold concentration, each protein inhibits forward vesicle transport through the ER (Cooper et al., 2006). Alleles that suppress α-syn toxicity (WT and A53T) enhance forward ER-Golgi transport, and these suppressors are YPT1, YKT6, UBP3 and BRE5, and ERV29. YPP1 in high copy failed to suppress the toxicity of WT α-syn and A53T, thus YPP1 does not enhance forward ER-Golgi traffic vis-à-vis these two α-syns. Regarding A30P, A30P has not been shown to cause ER stress or inhibit ER-Golgi traffic, and our experiments gave no indication that YPP1 enhances the forward transport of A30P through the ER-Golgi (Fig. 7, F and G).
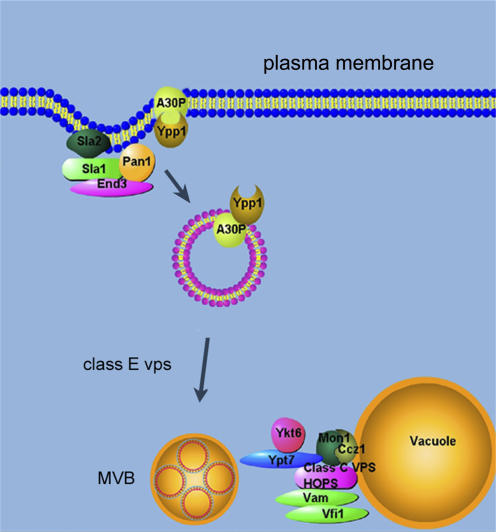
In summary, YPP1 interacts with the evolutionarily conserved genes SLA1, SLA2, END3, class E VPS, and CCZ1 and MON1 to target A30P α-syn to the vacuole for degradation via the endocytic pathway (Fig. 9). It will be of interest to characterize the step or steps that Ypp1p catalyzes in the endocytic pathway, correlate Ypp1p structure with function, and determine whether the potential human orthologues mediate the trafficking of A30P to the lysosome in human cells.
Figure 9.
Illustration of YPP1-mediated protection against A30P. Sla1p, Sla2p, and End3p form complexes at the actin cortical patches. Ypp1p binds to A30P and mediates the encapsulation of A30P into vesicles, which then bud off the membrane and transit to the vacuole. Class E Vps proteins are required for protein sorting into lumenal vesicles. The Mon1p–Ccz1p complex facilitates vesicle–vacuole fusion. MVB, multivesicular body.
Materials and methods
Strains, plasmids, and media
The yeast strains used in this study were FY23 (Winston et al., 1995), S288c (Invitrogen), YPP1-GFP (Invitrogen), which contains an integrated copy of YPP1-GFP replacing the WT allele, and several strains from the Hughes collection (Open Biosystems) (Table I).Additionally, 116 strains from the ResGen deletion collection were also used; these strains were derived from the parental strain S288c. Cells transformed with various plasmids were pregrown in synthetic sucrose (2% wt/vol) drop out media to maintain selection for plasmids. Media containing sucrose is referred to as noninducing media. α-Syn expression, as well as Ypp1p overexpression, was induced in the same drop out media with galactose (2% wt/vol) replacing sucrose (Burke et al., 2000). Media containing galactose is referred to as inducing media. The various S288c deletion strains were cultured in noninducing media with selection for plasmids supplemented with 0.4 g/L G418 (Geneticin) or inducing media with selection for plasmids supplemented with 0.4 g/L G418. The Resgen deletion strains (Invitrogen) and the high copy yeast genomic library (Nasmyth and Tatchell, 1980) (YEp13 LEU2) were gifts from Dr. Kelly Tatchell (LSU Health Sciences Center, Shreveport, LA). In every experiment in this study, cells were pregrown in noninducing media (sucrose) with selection for plasmids to mid-log phase and then shifted to inducing media (galactose) with selection for plasmids and induced for 3 h. In some cases, 12-h inductions were used. Cells were grown at 30°C.
YPP1 was cloned from genomic DNA using the forward and reverse primers 5′-CCACTGGATCCATGCCTAACTCAAATGTTC-3′ and 5′-GAAGAAGAGCTCTTAGTAATTCGAATACCTTAG-3′, respectively. These primers contained the BamHI and SacI restriction sites. The PCR product was subcloned into the BamHI and SacI restriction sites on the 2-μm pRS326 plasmid. For all other YPP1 constructs, the YPP1 ORF was excised from the pRS326 plasmid at the BamHI and SacI sites and ligated into those same sites in the other plasmids. Preparation of the plasmids harboring untagged and GFP-tagged α-syns was described elsewhere (Flower et al., 2005). MON1 was cloned from genomic DNA using the forward and reverse primers 5′-CATAGACGGGCCGCATGGGTCTAGCTTAAATAC-3′ and 5′-AACCTCGAGCTCACAAGTTAAAACACGGCC-3′, respectively. These primers contained the NotI and SacI restriction sites; the PCR product was subcloned into the same restriction sites on the low copy pRS313 plasmid. SLA1 was cloned from genomic DNA using the forward and reverse primers 5′-CAGGTGACTAGTGAAGTAGGCCATTCACTGC-3′ and 5′-TCTTAACCGCGGTTAATCTAGAATCCAAACGGATTTTG-3′ These primers contained SpeI and SacII restriction sites; the PCR product was subcloned into the same restriction sites on the low copy pRS313 plasmid. Plasmids were propagated according to standard protocols using Escherichia coli DH5α (Sambrook et al., 1989).
Yeast α-factor pheromone (Zymo Research) was aliquoted into cells from a stock solution; the final concentration of α-factor was 5–10 μM. Latrunculin A, cycloheximide, and the proteasome inhibitor MG132 were purchased form Sigma-Aldrich and used at concentrations of 25, 10, and 50 μM, respectively. For the ROS experiments that used pheromone, FY23 cells transformed with the various plasmids for α-syn (pTF200-203) were pregrown in noninducing media to mid-log phase and then shifted into inducing media. Upon the shift into inducing media the α-factor pheromone was added to 5–10 μM. After 2 h in inducing media with pheromone the DHR 123 dye was added to a final concentration of 5 μg/ml, and cells were incubated 1 additional hour in this media. Before image acquisition (after 3 h in inducing media) cells were washed and resuspended in fresh media.
Hydrogen peroxide–based genetic screen
Cells transformed with pTF202 (A30P) and the high copy yeast genomic library and induced with galactose were grown to 1.5 × 107 cells/ml and then plated on SGal−Leu−Trp. A disk of sterile Whatman filter paper containing 10 μl of 8% H2O2 was placed in the center of each plate, and plates were incubated for 3 d. Colonies growing close to the peroxide disk were selected, cultured overnight, and the plasmid DNA was isolated and then amplified in DH5α cells. Plasmids were retested for protection in the hydrogen peroxide assay. Each strand of a protective plasmid was sequenced. YPP1 was discovered by this approach.
Western blot analysis
Westerns were conducted as described previously (Flower et al., 2005). The monoclonal antibody against human α-syn was purchased from Cell Signaling Technologies. The monoclonal antibody against Pgk1p was purchased from Invitrogen. The monoclonal antibody against myc was purchased from Sigma-Aldrich. Secondary antibodies were purchased from Sigma-Aldrich or Bio-Rad Laboratories. 20 μg of protein were loaded per well in each Western blot conducted (Fig. 1 B, Fig. 2 E, and Fig. 4 B).
Coimmunoprecipitation
The coimmunoprecipitation was performed using protein A–Sepharose beads saturated with the anti-myc antibody as described in Flower et al. (2005).
Fluorescence microscopy
Fluorescent images of cells were obtained with a microscope (AX70; Olympus), including an Olympus UPlanFl 100×/1.35 NA objective with CoolSNAP HQ CCD camera (Roper Scientific). The acquisition software was IPLab v3.6 (Scanalytics). An Olympus U-MWG (510–550) filter set was used for detecting GFP (Figs. 3, 7, and 8), and a U-MNG filter (530–550 nm) was used to detect FM4-64, DHR 123, and FUN1 (Figs. 1, 2, 4, and 8; and Fig. S2). For the two-color experiment (Fig. 1 C and Fig. 4), a more restrictive Chroma JP1 (510 nm) was used to detect GFP; this filter set prevented bleed-through of GFP signal into the FM4-64 channel. All data were collected at room temperature.
The ROS assay, using the DHR 123 dye, was performed as described previously (Flower et al., 2005). The viability assay, using the FUN1 dye, was conducted according to Fannjiang et al. (2004). Vacuole staining, using the FM4-64 dye, was conducted according to a method adapted from Vida and Emr (1995). In brief, S288c cells transformed with a GFP-α-syn plasmid (pTF305-308) and with a YPP1 plasmid (pTF602) were pregrown in SSuc−Leu−Ura to mid-log phase. Cells were then washed and resuspended in SGal−Leu−Ura and induced for 12 h at 30°C. At various times aliquots were removed, washed, resuspended in YPD, and incubated with 40 μM FM4-64 (Invitrogen) for 10 min at 30°C. Cells were then washed twice, resuspended in YPD, incubated an additional 30 min at 30°C, and visualized by fluorescence microscopy. Adobe Photoshop 5.5 was used to prepare all figures. Adjustments in contrast were made to Westerns blots. Adjustments in brightness and contrast were made to some differential interference contrast (DIC) images and to some merged images (DIC merged with GFP fluorescence). Specific adjustments are given in the legends.
Transmission electron microscopy
TEM samples were prepared as described previously (Yang et al., 2006) and examined using an electron microscope (H-7000; Hitachi), equipped with a high-resolution digital camera (Gatan, Inc.). For immunogold labeling, cells were fixed with 4% formaldehyde and 0.25% glutaraldehyde in 40 mM phosphate buffer (pH 6.7), containing 1 mM MgCl2 and 1 mM EGTA for 1 h at room temperature. Cells were washed with phosphate buffer, incubated in 1% sodium metaperiodate for 15 min, then in 50 mM ammonium chloride for 15 min. Cells were dehydrated with graded ethanol, embedded in LR White resin (Electron Microscopy Sciences). 60-nm-thin sections were cut and collected on nickel grids. Grids were incubated in 0.1 M glycine in PBS containing 0.1% (vol/vol) Triton X-100 for 15 min to inactivate residual aldehydes. Sections were blocked with blocking buffer (PBS containing 5% normal goat serum, 0.2% Tween 20, and 0.2% BSA) for 40 min. Grids were then incubated in the primary rabbit anti-A30P antibody, diluted 1:50 in blocking buffer for 2 h. Grids were washed then incubated in 12-nm gold–conjugated goat anti–rabbit IgG (Jackson ImmunoResearch Laboratories) diluted 1:30 in blocking buffer for 1.5 h. Labeled sections were stained with 2% uranyl acetate for 30 min and lead citrate for 30 s, examined as described above. The control was performed by substituting primary antibody with blocking buffer.
ROS plate reader assay
A Perkin Elmer Wallac Victor3 1420 multilabel counter was used to screen deletion strains for ROS accumulation. Cells transformed with the pTF302 (A30P) and pTF602 (YPP1) plasmids were pregrown in noninducing media until mid-log phase and then shifted into galactose inducing media and induced for 3 h. After the second hour of induction, the DHR 123 dye was added to a final concentration of 5 μg/ml. Before adding the cells to the 96-well plates, cells were washed twice using PBS and the assay was conducted in PBS. The excitation and emission wavelengths were 485 and 535 nm, respectively. At minimum, deletion strains were analyzed in two independent experiments in triplicate. Thirty of the deletion strains were analyzed in three to four independent experiments in triplicate. ROS readings from each deletion strain were compared with the ROS readings from the S288c strain (transformed with pTF302 [A30P] and pTF602 [YPP1]). Acetic acid treatment has been shown to induce apoptosis and ROS in yeast cells (Ludovico et al., 2002), and acetic acid–treated cells were prepared and incubated with DHR 123 as described previously (Flower et al., 2005) as a control.
Deletion strains exhibiting greater than fourfold increases in ROS signal compared with the control (S288c transformed with pTF202 and pTF602) were selected for further evaluation. One two-plasmid control consisted of the various deletion strains expressing Ypp1p (pTF602 (YPP1) plus pTF300 (empty vector)). The other two-plasmid controls consisted of the various deletion strains harboring plasmids with no inserts (pTF300 and pTF604). ROS accumulation was measured after 3 h of induction. Ypp1p overexpression did not appreciably increase the ROS signal, and the two-plasmid control cells yielded a signal indicative of zero fluorescence.
Statistical analysis
For the ROS (and FUN1 viability assays) (Figs. 1 and 8, and Fig. S2), P values were determined using a two-tailed t test (heteroscedastic) that compared red cell counts from various +α-syn/+Ypp1 cultures to red cell counts from various +α-syn/−Ypp1 cultures. Cells were counted in two to three independent experiments. The program Excel was used for this analysis. For the ROS plate reader assay (Fig. 6), a paired t test was used to compare the ROS signals from deletion strains to matched controls (S288c +A30P/+YPP1). To adjust for multiple comparisons, the Bonferroni correction method was used, with a family-wise error rate of 0.05 or 0.001. For example, because 116 gene deletion strains were tested, the highest accepted P value was 4.3 × 10−4 (= 0.05/116) for a family error rate of 0.05. The statistical software used for this analysis was SAS v9.13 (SAS Institute, Inc.). Table S1 gives exact P values (available at http://www.jcb.org/cgi/content/full/jcb.200610071/DC1).
Online supplemental material
Fig. S1 shows that in high copy YPP1 permitted normal growth of cells expressing A30P, but not of cells expressing WT or A53T. Fig. S2 shows that YPP1 in high copy fails to protect cells from hydrogen peroxide–induced ROS. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200610071/DC1.
Supplementary Material
Acknowledgments
We thank Kelly Tatchell for the yeast genomic library and deletion strains; Jennifer Larson for streaking out the deletion strains; Lucy Robinson and Michelle Kearney for retesting some library plasmids found in the hydrogen peroxide assay; and Brent Reed for help in figure preparation.
This work was supported in part by grants from the Parkinson's Resource of Northwest Louisiana and from the National Institutes of Health (R21NS053678) to S.N. Witt.
Abbreviations used in this paper: α-syn, α-synuclein; RME, receptor-mediated endocytosis; ROS, reactive oxygen species; WT, wild-type.
References
- Aronov, S., and J.E. Gerst. 2004. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J. Biol. Chem. 279:36962–36971. [DOI] [PubMed] [Google Scholar]
- Ayscough, K.R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D.G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C., and R. Schekman. 1993. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 365:347–349. [DOI] [PubMed] [Google Scholar]
- Bowers, K., and T.H. Stevens. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1744:438–454. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson, and T. Stearns. 2000. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 205 pp.
- Christianson, T.W., R.S. Sikorski, M. Dante, J.H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene. 110:119–122. [DOI] [PubMed] [Google Scholar]
- Cooper, A.A., A.D. Gitler, A. Cashikar, C.M. Haynes, K.J. Hill, B. Bhullar, K. Liu, K. Xu, K.E. Strathearn, F. Liu, et al. 2006. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 313:324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davierwala, A.P., J. Haynes, Z. Li, R.L. Brost, M.D. Robinson, L. Yu, S. Mnaimneh, H. Ding, H. Zhu, Y. Chen, et al. 2005. The synthetic genetic interaction spectrum of essential genes. Nat. Genet. 37:1147–1152. [DOI] [PubMed] [Google Scholar]
- Dawson, T.M., and V.L. Dawson. 2003. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 302:819–822. [DOI] [PubMed] [Google Scholar]
- Dixon, C., N. Mathias, R.M. Zweig, D.A. Davis, and D.S. Gross. 2005. alpha-Synuclein targets the plasma membrane via the secretory pathway and induces toxicity in yeast. Genetics. 170:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman, H.G. 2002. G proteins and pheromone signaling. Annu. Rev. Physiol. 64:129–152. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A.E., and D.G. Drubin. 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19:287–332. [DOI] [PubMed] [Google Scholar]
- Fannjiang, Y., W.C. Cheng, S.J. Lee, B. Qi, J. Pevsner, J.M. McCaffery, R.B. Hill, G. Basanez, and J.M. Hardwick. 2004. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 18:2785–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower, T.R., L.S. Chesnokova, C.A. Froelich, C. Dixon, and S.N. Witt. 2005. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson's disease. J. Mol. Biol. 351:1081–1100. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., M. Matsuzaki-Kobayashi, T. Hasegawa, A. Kikuchi, N. Sugeno, Y. Itoyama, Y. Wang, P.J. Yao, I. Bushlin, and A. Takeda. 2006. Plasma membrane ion permeability induced by mutant alpha-synuclein contributes to the degeneration of neural cells. J. Neurochem. 97:1071–1077. [DOI] [PubMed] [Google Scholar]
- Gosavi, N., H.J. Lee, J.S. Lee, S. Patel, and S.J. Lee. 2002. Golgi fragmentation occurs in the cells with prefibrillar alpha-synuclein aggregates and precedes the formation of fibrillar inclusion. J. Biol. Chem. 277:48984–48992. [DOI] [PubMed] [Google Scholar]
- Hazbun, T.R., L. Malmstrom, S. Anderson, B.J. Graczyk, B. Fox, M. Riffle, B.A. Sundin, J.D. Aranda, W.H. McDonald, C.H. Chiu, et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 12:1353–1365. [DOI] [PubMed] [Google Scholar]
- Holtzman, D.A., S. Yang, and D.G. Drubin. 1993. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122:635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, E., A.R. Davidson, and C.A. Kaiser. 1996. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 135:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., J.V. Falvo, L.C. Gerke, A.S. Carroll, R.W. Howson, J.S. Weissman, and E.K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. [DOI] [PubMed] [Google Scholar]
- Hurley, J.H., and S.D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E.W., G.S. Zubenko, and R.R. Parker. 1982. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics. 102:665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontopoulos, E., J.D. Parvin, and M.B. Feany. 2006. {alpha}-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum. Mol. Genet. 15:3012–3023. [DOI] [PubMed] [Google Scholar]
- Kruger, R., W. Kuhn, T. Muller, D. Woitalla, M. Graeber, S. Kosel, H. Przuntek, J.T. Epplen, L. Schols, and O. Riess. 1998. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat. Genet. 18:106–108. [DOI] [PubMed] [Google Scholar]
- Lamb, J.R., S. Tugendreich, and P. Hieter. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20:257–259. [DOI] [PubMed] [Google Scholar]
- Lashuel, H.A., B.M. Petre, J. Wall, M. Simon, R.J. Nowak, T. Walz, and P.T. Lansbury Jr. 2002. Alpha-synuclein, especially the Parkinson's disease-associated mutants, forms pore-like annular and tubular protofibrils. J. Mol. Biol. 322:1089–1102. [DOI] [PubMed] [Google Scholar]
- Ludovico, P., F. Rodrigues, A. Almeida, M.T. Silva, A. Barrientos, and M. Corte-Real. 2002. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, L.J., Y.H. Pan, A.C. Price, W. Sterling, N.G. Copeland, N.A. Jenkins, D.L. Price, and M.K. Lee. 2006. Parkinson's disease alpha-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J. Neurosci. 26:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser, B., C. Hirsch, E. Jarosch, and T. Sommer. 2005. ERAD: the long road to destruction. Nat. Cell Biol. 7:766–772. [DOI] [PubMed] [Google Scholar]
- Mnaimneh, S., A.P. Davierwala, J. Haynes, J. Moffat, W.T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, et al. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell. 118:31–44. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K.A., and K. Tatchell. 1980. The structure of transposable yeast mating type loci. Cell. 19:753–764. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos, M.H., C. Lavedan, E. Leroy, S.E. Ide, A. Dehejia, A. Dutra, B. Pike, H. Root, J. Rubenstein, R. Boyer, et al. 1997. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 276:2045–2047. [DOI] [PubMed] [Google Scholar]
- Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J., B. Nelson, M.J. Marton, R. Stoughton, M.R. Meyer, H.A. Bennett, Y.D. He, H. Dai, W.L. Walker, T.R. Hughes, et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 287:873–880. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pena, J.M., V.J. Cid, M. Sanchez, M. Molina, J. Arroyo, and C. Nombela. 1998. The deletion of six ORFs of unknown function from Saccharomyces cerevisiae chromosome VII reveals two essential genes: YGR195w and YGR198w. Yeast. 14:853–860. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Schulz, J.B., M. Weller, and T. Klockgether. 1996. Potassium deprivation-induced apoptosis of cerebellar granule neurons: a sequential requirement for new mRNA and protein synthesis, ICE-like protease activity, and reactive oxygen species. J. Neurosci. 16:4696–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M.A., and H.D. Madhani. 2004. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38:725–748. [DOI] [PubMed] [Google Scholar]
- Smith, W.W., H. Jiang, Z. Pei, Y. Tanaka, H. Morita, A. Sawa, V.L. Dawson, T.M. Dawson, and C.A. Ross. 2005. Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant alpha-synuclein-induced toxicity. Hum. Mol. Genet. 14:3801–3811. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., S. Engelender, S. Igarashi, R.K. Rao, T. Wanner, R.E. Tanzi, A. Sawa, V.L. Dawson, T.M. Dawson, and C.A. Ross. 2001. Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum. Mol. Genet. 10:919–926. [DOI] [PubMed] [Google Scholar]
- TerBush, D.R., T. Maurice, D. Roth, and P. Novick. 1996. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Toshima, J.Y., J. Toshima, M. Kaksonen, A.C. Martin, D.S. King, and D.G. Drubin. 2006. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc. Natl. Acad. Sci. USA. 103:5793–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky, V.N., J.R. Gillespie, and A.L. Fink. 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 41:415–427. [DOI] [PubMed] [Google Scholar]
- Vida, T.A., and S.D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.W., P.E. Stromhaug, J. Shima, and D.J. Klionsky. 2002. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 277:47917–47927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb, P.H., W. Zhen, A.W. Poon, K.A. Conway, and P.T. Lansbury Jr. 1996. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 35:13709–13715. [DOI] [PubMed] [Google Scholar]
- Wiederkehr, A., J.O. De Craene, S. Ferro-Novick, and P. Novick. 2004. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J. Cell Biol. 167:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., C. Dollard, and S.L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 11:53–55. [DOI] [PubMed] [Google Scholar]
- Xu, J., S.Y. Kao, F.J. Lee, W. Song, L.W. Jin, and B.A. Yankner. 2002. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat. Med. 8:600–606. [DOI] [PubMed] [Google Scholar]
- Yang, H., Q. Ren, and Z. Zhang. 2006. Chromosome or chromatin condensation leads to meiosis or apoptosis in stationary yeast (Saccharomyces cerevisiae) cells. FEMS Yeast Res. 6:1254–1263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.