Abstract
Human RNase H1 binds double-stranded RNA via its N-terminal domain and RNA–DNA hybrid via its C-terminal RNase H domain, the latter being closely related to Escherichia coli RNase HI. Using SELEX, we have generated a set of DNA sequences that can bind efficiently (Kd values ranging from 10 to 80 nM) to the human RNase H1. None of them could fold into a simple perfect double-stranded DNA hairpin confirming that double-stranded DNA does not constitute a trivial ligand for the enzyme. Only two of the 37 DNA aptamers selected were inhibitors of human RNase H1 activity. The two inhibitory oligomers, V-2 and VI-2, were quite different in structure with V-2 folding into a large, imperfect but stable hairpin loop. The VI-2 structure consists of a central region unimolecular quadruplex formed by stacking of two guanine quartets flanked by the 5′ and 3′ tails that form a stem of six base pairs. Base pairing between the 5′ and 3′ tails appears crucial for conferring the inhibitory properties to the aptamer. Finally, the inhibitory aptamers were capable of completely abolishing the action of an antisense oligonucleotide in a rabbit reticulocyte lysate supplemented with human RNase H1, with IC50 ranging from 50 to 100 nM.
INTRODUCTION
Ribonucleases H cleave specifically the RNA strand of RNA–DNA hybrids. This activity was first observed in calf thymus extracts (1) then recognized in all organisms examined to date in all three kingdoms (2). Moreover, RNase H constitutes an essential functional domain of the reverse transcriptases of retroviruses, hepadnaviruses (HBV RT) and transposons (3). The roles in the retroviral replication are firmly established and have been examined in increasingly great detail. Their participation in several key steps of nucleic acid processing such as DNA replication, repair and transcription is suspected on the ground of genetic studies in Escherichia coli (4) or from indirect indications derived from studies of cellular physiology (5,6). Recently, it has been found that embryonic development in RNase H1-deficient mice arrests about 8–9 days after fertilization due to the inability to amplify mitochondrial DNA (7). No lethal phenotype has been observed when the gene encoding RNase H1 of several unicellular eukaryotes has been deleted, and mitochondria persist in such mutant strains.
Both prokaryotic and eukaryotic cells usually contain two RNases H. The genes coding for RNase HI and HII in E.coli have been known for some time (8–10), but only recently have the human genes have been cloned (11–14). For the sake of uniformity, we will follow the nomenclature suggested elsewhere (15) and refer to RNase H1 and RNase H2 in eukaryotes as corresponding to RNase HI and RNase HII of prokaryotes. Recently, a third family of RNases H has been identified with amino acid sequence similar to RNase HII but with significantly different biochemical properties (2).
Human RNase H1, E.coli RNase HI and HIV-1 RT RNase H are closely related. The amino acid sequences of the C-terminal domain of the human RNase H1, of the C-terminal domain of the HIV-1 RT and of the RNase H1 of E.coli can be properly aligned showing strict conservation of all amino acid residues essential for the catalytic action of the enzyme (D10, E48, D70, H124 and D134 in the sequence of E.coli RNase HI) (11,13). Despite having only 24% sequence identity, the RNase H domain of the HIV-1 RT and the E.coli RNase HI both adopt a very similar 3D structure, a five-stranded mixed β-sheet surrounded by asymmetrically distributed α-helices (16). The major difference is the presence of a ‘basic protrusion’ region or ‘handle’ region in the E.coli enzyme which is absent in the HIV-1 RT RNase H domain. The ‘handle’ region is necessary for binding to the RNA–DNA hybrid and positioning the hydrolytic center for cleavage, a role fulfilled by the polymerase domain in the case of the HIV-1 RT. The 3D structure of the human enzyme is not known yet, but it is highly likely that its C-terminal RNase H domain adopts a fold similar to the one found in E.coli RNase HI and HIV-1 RT. Human RNase H1, as other known eukaryotic RNases H1, contains a N-terminal domain with a conserved dsRNA-binding motif which is highly similar to a region of caulimovirus ORF VI family of proteins (13).
Although both eukaryotic RNases H1 and H2 hydrolyze the RNA strand of a RNA–DNA hybrid they show distinct behavior towards hybrids of defined length and sequence. Distinct hydrolysis of these hybrids can be considered a signature of each class of enzyme (17). Besides their normal physiological role in the cell, RNases H have been identified as key players in antisense methodologies (18), acting both in a positive manner whereby oligodeoxynucleotides destroy the targeted RNA (19), and in a negative way by elimination of untargeted RNAs which contain a sequence to which the oligonucleotide can form an imperfect hybrid (20). The exact role played by each type of RNase H in antisense effects is still uncertain, although both potentially could participate in vivo. Their relative activities measured by liquid assays are very different, as is their relative contributions inside the cell. RNase H2 being essentially nuclear, whereas the RNase H1 is found to be more ubiquitous, including a mitochondrial localization (7,21). Assigning a role to these enzymes in the normal physiology of the cell, as well as their action in antisense-treated cells should be facilitated by the use of specific inhibitors for each class of RNases H. At present, only a few small molecules have been found to inhibit the RNase H activity of the retroviral reverse transcriptases (22–26) and E.coli RNase HI (25) but no information is available concerning inhibitors of eukaryotic RNases H.
One way to obtain specific inhibitors is to select aptamers by an in vitro systematic evolution of ligands by exponential amplification (SELEX) (27–29) that will bind with good affinity to the targeted protein, then to test them for possible inhibitory effect on the catalytic function of the enzyme. We have performed SELEX using cloned human RNase H1 as a target and found two inhibitory DNA aptamers V-2 and VI-2. They can completely and selectively abolish the antisense action of an oligonucleotide targeted to an mRNA in a rabbit reticulocyte lysate supplemented with human RNase H1. Whereas V-2 folds into a large, imperfect but stable, hairpin loop, VI-2 folds into a unimolecular quadruplex consisting of a stack of two guanine quartets flanked by a stem formed by base pairing of the 5′ and 3′ tails of the oligonucleotide.
MATERIALS AND METHODS
Nucleic acids
The initial DNA library consisted of a pool of oligonucleotides made of a continuous stretch of 40 randomized nucleotides flanked on both sides by fixed sequences used for the hybridization of PCR primers, P5 (24 nucleotides) and P3 (23 nucleotides), during subsequent rounds of selection amplification (Fig. 1A). P3 is connected at its 5′-end, via a linker made of two triethyleneglycol phosphate units, to an additional extra sequence of 20 nucleotides, so that the two strands of the PCR products could be easily separated from each other according to their size (87 and 107 nt) on a sequencing gel (30).
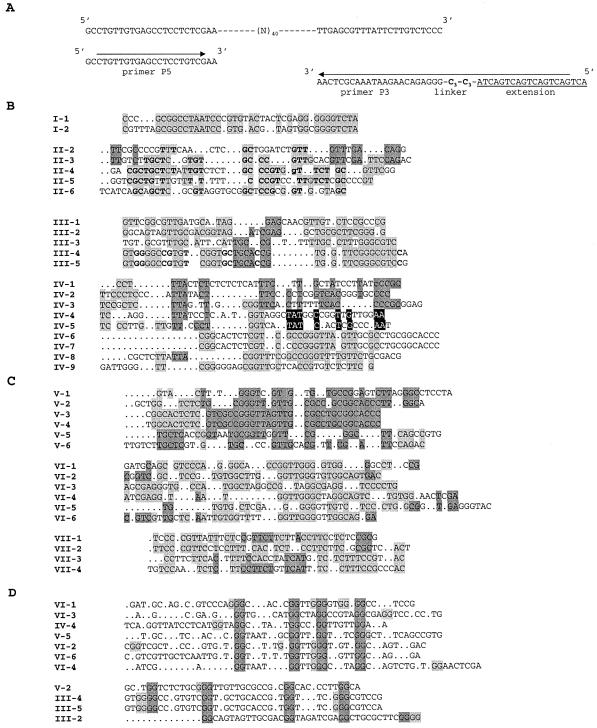
Figure 1.
SELEX selection and sequences. (A) Randomized library and primers used for the selection. (B) Sequences obtained after round 9. (C) Sequences obtained after the ‘polishing’ step on Biacore. (D) Sequences with putative G-quartets ‘Group D’. Black, dark and light gray backgrounds of sequence letters in panels B and C highlight blocks of identical sequences in the alignments. In panel D, doublets of G putatively participating to G-quartets are highlighted in dark gray, and alternative G residues, which could also participate to G-tetrads are highlighted in light gray.
The RNA–DNA hybrid BD2 used as a test substrate for RNase H was a blunt-ended hybrid duplex of 20 bp previously identified as very discriminatory for the two major classes of cellular RNases H (17).
The aptamer ODN93 (5′-GGGGGTGGGAGGAGGGTAG GCCTTAGGTTTCTGA-3′) has been previously selected against the HIV-1 RT (31).
The 17mer antisense oligonucleotide 5′-CACCAACTTC TTCCACA-3′ is complementary of nucleotides 113–129 of the mRNA coding for the rabbit β-globin (19).
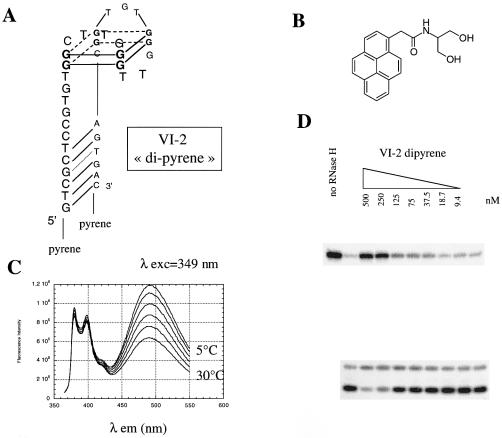
The chemically modified aptamer VI-2 ‘di-pyrene’, which 5′ and 3′ ends are linked to a pyrenyl group, was synthesized as described (32). Its fluorescence properties were recorded on a PTI spectrofluorimeter, as described (32).
DNA oligonucleotides were purchased from Genset SA (Paris, France) and the RNA oligonucleotide from Xeragon AG (Zurich, Switzerland).
Oligonucleotides were gel-purified and radiolabeled according to standard procedures (33). The sequences of aptamers and their variants which have been studied in great detail are listed in Table 1.
Table 1. Listing of sequences of aptamers and their variants studied in greater detail in this work.
| V-2 | GCTGGTCTCTGCGGGTTGTTGCGCCGCGGCACCCTTGGCA |
| V-2 ‘short’ | GGTCTCTGCGGGTTGTTGCGCCGCGGCACC |
| V-2 ‘mini’ | CTGCGGGTTGTTGCGCCGCGG |
| V-2 ‘linker’ | GCTGGTCTCTGCGG-C18 linker-CCGCGGCACCCTTGGCA |
| V-2 ‘perfect’ | GCTAAGGGTCTCTGCGGGTTGTTGCGCCGCGGAGACCCTTGGCA |
| VI-2 | CGGTCGCTCCGTGTGGCTTGGGTTGGGTGTGGCAGTGAC |
| VI-2 ‘minimum’ | GGCTTGGGTTGGGTGTGG |
| VI-2 Δ | GTGGCTTGGGTTGGGTGTGGCA |
| VI-2 dipyrene | Py-GTCGCTCCGTGTGGCTTGGGTTGGGTGTGGCAGTGAC-Py |
| VI-6 | GCTCGTTGCTCAATTGTGGTTTTGGTTGGGGTTGGCAGGA |
| VI-6 ‘extended’ | GCTCGTTGCTCAATTGTGGTTTTGGTTGGGGTTGGCAGGATTGAGCG |
Ribonucleases H
Human recombinant RNase H1 was prepared (H.Yamada, S.A.Gaidamakov and R.J.Crouch, manuscript in preparation). Details of the purification procedure can be obtained upon request (robert_crouch@nih.gov). The last step is a gel filtration and protein is at concentrations of 6–20 µM in 50 mM Tris–HCl pH 7.9, 1 M ammonium sulfate, 1 mM mercaptoethanol, 1 mM EDTA, 20 µg/ml PMSF, 10% glycerol. Dilutions for electrophoretic mobility shift assays (EMSAs) were done in 50 mM Tris–HCl pH 7.9, 40 mM DTT, 1 mg/ml BSA, 50% glycerol just prior to use. Partially purified human RNase H2 and beef heart mitochondrial RNase H have been obtained by phosphocellulose chromatography as described previously (17).
In vitro selection
200 pmol of the library (or selected sequences in the successive rounds of selection) were at first incubated in 100 µl buffer for 15 min at ambient temperature (23°C) with pieces of nitrocellulose filter (HAWP, Millipore). We used only filters which have been pretreated with alkali as described by McEntee et al. (34), in order to reduce non-specific adsorption of nucleic acids to the filters. Pieces of filter were collected after a brief centrifugation and the supernatant was divided into two identical parts, one being incubated with 0.1 µM human recombinant RNase H1, the other being incubated without protein for determination of background levels. Incubation was for 10 min at room temperature (∼23°C) in 100 µl of buffer A (30 mM Tris–HCl pH 8.5, 40 mM ammonium sulfate, 20 mM magnesium chloride, and 0.01% 2-mercaptoethanol), a buffer in which RNase H1 is very active. Each reaction mixture was then independently filtered through alkali-treated filters. Filters were washed with 3 ml of buffer A and the amount of retained sequences quantified by Cerenkôv counting. Nucleic acids retained on the filter were then extracted with 7 M urea and Tris-buffered phenol (pH 7.9) as described (35). The recovered sequences were amplified by PCR. In total, 5000–10 000 c.p.m. of 5′-end radiolabeled primers were included. PCR products were ethanol precipitated, resuspended in loading buffer and then electrophoresed on an 8% denaturing (7 M urea) polyacrylamide gel. The band corresponding to the shortest of the two strands was cut out, eluted, purified on a DEAE column, ethanol precipitated, counted and its concentration determined by UV absorption at 260 nm. Samples were heated for 2 min at 98°C, chilled in ice for 1 min, then incubated for 5 min at room temperature before being used in another round of selection. Seven rounds of selection were performed as described above. Afterwards the procedure was slightly modified, by the introduction of a second negative selection step (adsorption on pieces of nitrocellulose filter), by washing the filters after partitioning with 6 ml buffer instead of 3 ml, and by reducing the PCR to 15 cycles instead of 30, in a volume of 750 µl instead of 500 µl. These modifications were adopted to avoid the accumulation of exceedingly large amounts of non-specific nitrocellulose binding sequences. At the end of the 9th round of selection-amplification, DNA oligonucleotides were cloned using the topoTA cloning kit (InVitrogen, Carlsbad, CA), sequenced and analysed with the program Sequencing Analysis 3.0 (PE Applied Biosystems).
BIAcore™ experiments were performed with RNase H1 immobilized on the surface of CM5 sensor chips via primary amino groups with the amine coupling kit from Pharmacia, following the procedure of Haruki et al. (36). Experiments were performed at 23°C in the buffer used for the selection, at a flow rate of 10 µl/min. The volume of sample of nucleic acids (either the initial library or the products of round 9) injected was 100 µl. Two minutes after the beginning of the dissociation phase, samples were collected for 8 min. DNA from these samples was cloned and sequenced as described above.
Electrophoretic Mobility Shift Assay (EMSA)
Binding of aptamers to the human RNase H1 was investigated by EMSA. Aptamers were heat-denatured for 1 min at 100°C, chilled in ice for 1 min, incubated for 2 h at 25°C and then stored for 18 h at 4°C. Aptamers (2 nM final concentration) were incubated for 10 min at room temperature with increasing amounts of RNase H1 (0–1 µM) in buffer A, with or without 40 µg/ml salmon sperm DNA in a final volume of 8 µl. At the end of the incubation, 2 µl of 5× loading buffer (20% glycerol, TB 2.5 X and xylene cyanol + bromophenol blue dyes), was added and the sample loaded, while the current was running, on a native 8% (19:1 acrylamide/bisacrylamide) gel made in 0.5× TB buffer. Electrophoresis was continued at 6 W constant power until the bromophenol blue had migrated half the distance of the gel. A quantitative analysis of the relative amounts of radioactivity present in the different bands was obtained from direct counting of the gel on a multiwire proportional counter (MWPC), Instant Imager (Packard Instruments).
Measurement of the RNase H activity
The activity of the RNase H1 in the presence of various concentrations (0–500 nM) of aptamers was tested in 10 µl of buffer A using BD2 hybrid (2 µm), whose RNA strand was radioactively 5′-end labeled. After the addition of 0.5 µl of a 1/10 dilution of the enzyme (made just prior to use), the reaction mixture was incubated for 10 min at 30°C. The reaction was stopped by addition of 10 µl of a stop/loading solution containing 7 M urea, 0.1× TBE, 30 mM EDTA, and bromophenol blue + xylene cyanol dyes). After heating for 2 min at 98°C, samples were loaded on a 20% acrylamide denaturing (7 M urea) gel. After migration the gel was analysed with Instant Imager and/or by autoradiography. Activity of E.coli RNase H was tested either in buffer A or in 20 mM Tris–HCl (pH 8.0), 100 mM KCl, 10 mM MgCl2, 1 mM DTT. RNase H activity of HIV-1 RT was measured as described in Andreola et al. (31).
Thermal denaturation
Thermal denaturation of aptamers V-2 and VI-2 was performed as described (37) using a Cary 1 UV/visible spectrophotometer interfaced with a Peltier effect device that controls temperature within ±0.1°C. The rate of temperature increase or decrease was 0.5°C/min from 5 to 90°C. Thermal denaturation were carried out in 10 mM sodium cacodylate buffer (pH 7.0) with added salts at the desired concentrations (140 mM for NaCl, KCl, LiCl and NH4Cl; 40 mM for (NH4)2SO4, and 3 or 20 mM for MgCl2).
Enzymatic probing of aptamer structures
Radiolabeled V-2 was incubated at 30°C in 100 µl of the RNase H1 activity buffer together with 0.5 µg of herring sperm DNA and 0.8 U of nuclease P1 (38). Aliquots were taken at various times and mixed with the same volume of stop/loading buffer, and analyzed on a 20% denaturing acrylamide gel. Sequencing ladders were obtained according to a simplified chemical sequencing procedure (39).
Chemical interference
The procedure is described in detail in the Supplementary Material. Briefly, each end-radiolabeled aptamer molecule had less than one chemical modification of either T with potassium permanganate (KMnO4) or G with dimethylsulfate (DMS), or a depurination (loss of an A or G residue and creation of an abasic site). The modified aptamers are then challenged with the RNase H in a gel shift experiment: retarded bands (B: bound aptamers) contain aptamers in which modification does not interfere with binding, whereas the non-retarded band (F: free aptamers) contain aptamers prevented from binding to the enzyme due to chemical modification. Bound and free bands are eluted, cleaved at the site of modification and finally loaded on a sequencing gel. For each modification reaction, bound (B) and free (F) aptamers are loaded side by side. After electrophoresis, the gel is autoradiographed. Bands present in lane F and absent in lane B correspond to modifications which interfere strongly in the interaction.
Hybrid-arrested translation in rabbit reticulocyte lysate
Untreated rabbit reticulocyte lysate was purchased from Ambion, Inc. and used according to the manufacturer’s instructions; 15 µl methionine master mix and 25 µl Tran 35S LABEL™ (ICN) (10.5 mCi/ml; 1175 Ci/mmol) and 0.5 µl human RNase H1 (6 µM) were added to 200 µl of lysate. Translation assays were performed in Eppendorf tubes containing 20 µl of complemented lysate and 5 µl containing either water or antisense oligonucleotide (5 µM) and aptamers at various concentrations. Samples were incubated for 60 min at 30°C; 2 µl aliquots were loaded on an acid urea polyacrylamide gel containing Triton X-100, suitable for convenient separation of the β and α chains of the globin (40). After overnight migration at 100 V, the gel was soaked in 20% methanol, 2% glycerol, dried and analysed with Instant Imager and/or autoradiography.
RESULTS
Selection
Starting from a library of 6 × 1013 different sequences of oligodeoxyribonucleotides, we selected aptamers binding to human RNase H1, using filter retention of protein–nucleic acid complexes (see Materials and Methods). We chose to select DNA rather than RNA sequences because the human RNase H1 contains a double-strand RNA binding domain (dsRBD) (13,14,41) that would select for any double-stranded RNA binding to the dsRBD.
Despite the systematic pretreatment of the filters and the negative selection step, the background increased dramatically after round 7. We therefore introduced a second pre-incubation step with nitrocellulose, extended the volume of washing and reduced the number of cycles in the PCR (see Materials and Methods). The selection was stopped after round 9 as we found no improvement by adding a tenth round.
This population of oligonucleotides was checked for binding to the RNase H1 immobilized on a BIAcore™ sensor chip. Sensorgrams obtained at various candidate concentrations gave dissociation constants ranging from 10 to 80 nM for the round 9 pool of sequences (see Supplementary Material), whereas under the same conditions, the initial library was observed to bind very poorly (Kd > 50 µM). By collecting DNA during the dissociation phase of the BIAcore™ analysis, we had an alternative to filter retention, and thus potentially eliminated sequences with a high affinity for nitrocellulose. We cloned and sequenced DNAs selected at round 9 and following the BIAcore™ experiment (Fig. 1B and C, respectively).
Analysis of selected sequences
Some oligonucleotides were selected several times being either strictly identical (IV-6 = V-3, II-3 = V-6, VI-6 has been found twice), with a single nucleotide difference (III-4 and III-5; IV-6 and IV-7; V-3 and V-4), or highly similar (VI-2 and VI-6; I-1 and I-2). In nearly all other cases, the oligonucleotides share several motifs, and can therefore tentatively be grouped in families. This resulted in the groups I to VII listed in Figure 1B and C. Except for the repeats and near repeats, no obvious primary consensus sequence was apparent. Therefore, to search for potential common secondary structures, we used the mfold program (42), with conditions set up at 0.14 M NaCl, 0.02 M MgCl2 and 23°C to see if a common structural pattern would emerge. Structures were computed for all the sequences with or without the flanking primer-binding sites, but no single structural pattern could be discerned.
It is common that SELEX-selected DNAs form G-quartets (43), a type of structure that cannot be predicted by mfold. Therefore, we examined the selected oligonucleotides for the presence of a minimum of four interspersed GG dinucleotides. Eleven sequences meeting this criterion were placed into group D (Fig. 1D).
Screening for aptamers inhibiting the RNase H activity
Faced with the absence of conserved secondary structures, we checked individual sequences, chemically synthesized without the primer binding sequences, for their potential to inhibit human RNase H1 activity at a concentration of 500 nM, using RNA–DNA hybrid BD2 (Fig. 2) as substrate. BD2 enables us to discriminate between RNase H1 and RNase H2 activities (17). Only two sequences were potent inhibitors of the human RNase H1. Hence, these two inhibitory aptamers, V-2 and VI-2, were selected for further studies (Fig. 2A).
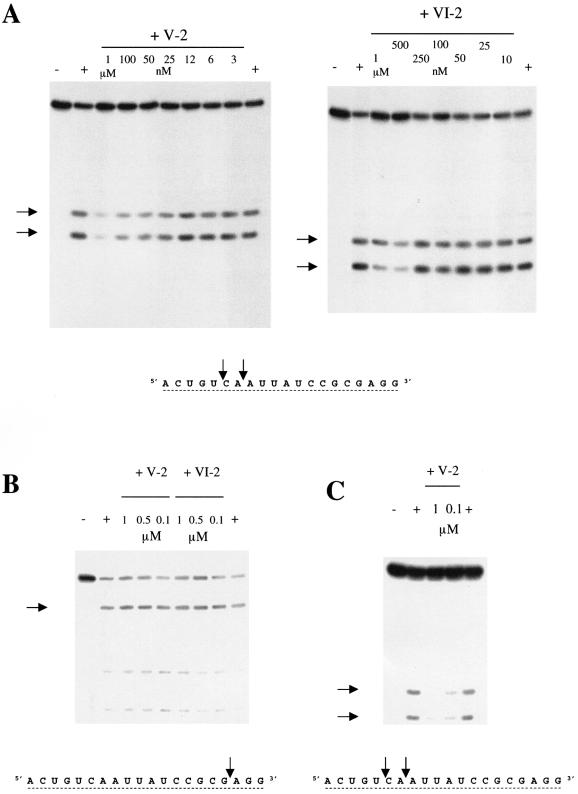
Figure 2.
Aptamer inhibition of RNase H. (A) Inhibition of the enzymatic activity of the human RNase H1 by aptamers V-2 and VI-2. (B) Test of aptamers V-2 and VI-2 for inhibition of the human RNase H2. The hybrid BD2 used is the same as used for the tests of the RNase H1, but the primary cut is located three nucleotides away from the 3′ end so that the major cleavage band does not migrate far from the initial intact hybrid. (C) Test of aptamer V-2 for inhibition of the mitochondrial RNase H from beef heart. Arrows to the left of the panels indicate cleavage products, which are indicated on the sequence of the RNA strand of the hybrid BD2 shown below panels B and C.
Selectivity of the aptamers for the human RNase H1
Aptamers V-2 and VI-2 were assayed for possible inhibitory effects on the activity of E.coli RNase HI and human RNase H2, partially purified from HeLa cell extracts by phosphocellulose chromatography (17). Activity was checked using the hybrid BD2 as a substrate. Aptamers V-2 and VI-2 had no effect at concentrations found to inhibit human RNase H1 (Fig. 2B) or on E.coli RNase HI (data not shown). These negative results were observed in assays using buffers for human RNase H1 or for E.coli enzyme (see Materials and Methods). Also, these aptamers did not inhibit the RNase H of HIV-1 RT even at concentrations as high as 4 µM (31). We have found that V-2 was also a good inhibitor of the RNase H partially purified from beef heart mitochondria, which appears to be very similar to the human RNase H1 (17). In conclusion, the aptamers appear to be selective for human RNase H1 or possibly a general inhibitor of the eukaryotic RNases H1 (Fig. 2C).
Binding of individual aptamers to the protein
Binding of the aptamers to human RNase H1 was investigated using an Electrophoretic Mobility Shift Assay (EMSA), as described in Materials and Methods. In the presence of denatured salmon sperm DNA at 40 µg/ml used as a nonspecific competitor, the unrelated ODN93 HIV-1 RT-specific aptamer failed to shift in the presence of RNase H1, even at the highest concentrations of protein used, whereas V-2 and VI-2 aptamers caused band-shifting (data not shown). Dissociation constants determined from the analysis of these gels (44) are 26.5 ± 2.5 nM for aptamer V-2 and 11 ± 0.8 nM for aptamer VI-2, in accordance with the Kd observed for the whole population of selected aptamers using surface plasmon resonance.
Structures of aptamers V-2 and VI-2
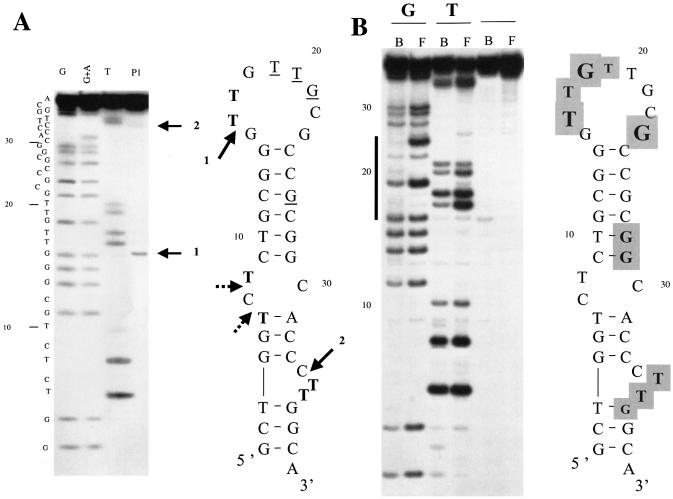
V-2. Although the V-2 aptamer has the potential to fold into four-stranded G-quartet structures, it appears to fold into a simpler structure. Melting curves obtained for V-2 with different monovalent cations show a single reversible cooperative transition (data not shown), and the deduced Tm (52°C) is in agreement with the Tm values predicted by mfold (53.1°C). Cations chosen for the experiment are known to either strongly favor the formation of G-quartets (K+) or to severely impair their formation (Li+). NH4+ was included in the comparison as it was used for both our SELEX procedure and for testing RNase H activity; this cation has been reported to support the formation of G-quartets (45). All these observations are in agreement with the large imperfect hairpin predicted by mfold and not with a G-quartet structure. Further confirmation of the structure was demonstrated by enzymatic mapping with the nuclease P1 which generates cuts at the proposed internal loop (C34), bulge (C7) and apical (G15) loops (Fig. 3A). Under very limited digestion conditions, a single strong cut is observed in the apical loop at G15, suggesting that the loop adopts an ordered structure. This is also deduced from the observed differential susceptibilities of G and T residues to modification by DMS and KMnO4 as assessed by chemical interference experiments (Fig. 3B). The modification procedure is performed on the folded native state of V-2 and the observed susceptibilities of T residues are in accordance with their single-stranded character: for example, T10 predicted to reside in a helix is particularly under-reactive. However, T20 in the apical loop is also under-reactive suggesting that it should be highly stacked with its neighbors. We observed that its immediate neighbor, G21, is anomalously under-reactive to the modification by DMS, suggesting it participates in an ordered structure in which the N7 of the guanine is engaged in hydrogen bonding, most probably with another base of the loop. Thus, this apical loop should be very constrained and therefore should exhibit very little purely single-stranded character in agreement with the data obtained with nuclease P1.
Figure 3.
Susceptibility of V-2 to enzymatic and chemical probes. (A) Sequencing gel identifying primary cuts generated by nuclease P1 on aptamer V-2, and structure of V-2 predicted by mfold. Arrows indicate cleavages generated by nuclease P1 after 1 min at 30°C. Thymines highly sensitive to KMnO4 are indicated in bold. Thymines poorly reactive to modification by KMnO4 and guanines minimally modified by DMS are underlined. (B) Identification of thymines and guanines in free and human RNase H1-bound V-2. The bold line on the left of the gel marks the sequences of the apical loop. Bold letters on a gray background indicate residues with decreased reactivities. Amplitudes of the observed changes are reflected by the relative letter sizes.
Study variants of V-2. Four variants of V-2 were prepared and evaluated for RNase H1 binding: (i) one (V-2 ‘short’) encompassing nucleotides from G4 to C33 comprising the internal and apical loops, (ii) one (V-2 ‘mini’) restricted to the upper part of the stem and the apical loop from C9 to G29, (iii) an oligomer in which the entire apical loop of V-2 was replaced by a C18 spacer (V-2 ‘linker’), and (iv) one (V-2 ‘perfect’) in which a perfect double-stranded stem was generated by inserting the sequence AAG in front of the CTT bulge and by replacing the C residue on the 3′ side of the internal loop by the dinucleotide AG. All of these V-2 variants failed to bind to human RNase H1 as assessed by EMSA (data not shown). Chemical interference was performed to identify critical residues of the aptamer to bind to the RNase H1 as illustrated in Figure 3B. Depurination at almost all sites decreases or eliminates binding whereas chemical modifications of T and G residues are restricted to a few residues mainly located in the apical loop, adjacent to the internal loop and at residues in and next to the bulge.
Therefore, experimental evidence obtained with the V-2 variants and the chemical interference data point to the fact that the whole sequence of V-2 is involved in binding and that all the particular structural features of the hairpin, the apical loop, the internal loop and the bulge are crucial for recognition by the RNase H1.
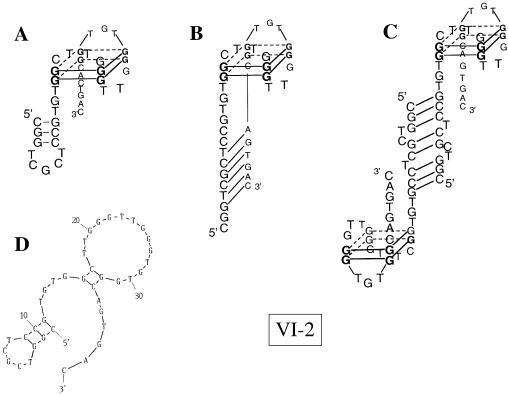
VI-2. In contrast to V-2, VI-2 does form a G-quartet structure. Its Tm is dependent on the monovalent cation used for the experiment and the presence of a hypochromic cooperative transition at 295 nm (46) is indicative of G-quartet structures. The synthetic oligonucleotide VI-2 ‘minimum’ 5′ GGCTT GGGTTGGGTGTGG 3′, representing the minimum sequence required for folding into an intramolecular quadruplex, gave perfectly monophasic, fully reversible melting curves at both 275 and 295 nm for each monovalent ion tested. However, the melting transition at 275 nm was hyperchromic, and the transition at 295 nm was hypochromic. Tm values determined at both wavelengths were 40°C in LiCl, 50°C in NH4Cl and 67°C in KCl, the expected ranking in stability for G-quartets (47). The same experiments performed with VI-2 also gave a hypochromic cooperative transition at 295 nm, with a Tm of 40°C in both LiCl and NH4Cl, and 64°C in KCl, demonstrating the presence of the postulated quadruplex (see Supplementary Material). Transitions observed at both 275 and 295 nm were coincident in LiCl and NH4Cl. We observed two transitions in KCl at 275 nm, one at 64°C coincident with the transition observed at 295 nm, therefore representing the melting of the G-quartets, and a second at 40°C without concomitant transition at 295 nm. This transition should correspond to the independent melting of canonical Watson–Crick base pairs outside the G-quartet. As illustrated in Figure 4, several different base pairings can be considered, one involving a short hairpin at the 5′ end of the aptamer, as predicted by mfold, another with pairing between the 5′ and the 3′ tails outside the G-quartet, and finally the possibility of head-to-tail dimerization.
Figure 4.
G-quartet structure of VI-2 aptamer. Data obtained by DMS reactivity of VI-2 indicate the G-quartet structure shown as (A). (B) and (C) are alternative structures revealing multiple base pairing possibilities for the 5′ end. The structure computed by mfold is shown for comparison (D).
Other features of the VI-2 aptamer in addition to the G-quartet are important for binding and inhibition. A shortened version of VI-2, VI-2Δ (5′-GTGGCTTGGGTTGGGTGTGG CA-3′), restricted to the G-quartet forming domain plus two adjacent nucleotides at each end did not bind to the RNase H1 by EMSA (data not shown). This result suggests the putative stem formed by the 5′ and 3′ extensions (Fig. 4B) are critical for binding and inhibition by VI-2. To support this assertion, a modified oligonucleotide, VI-2 dipyrene (Fig. 5), in which the last three nucleotides at the 5′ end have been removed and the resulting terminal 5′ and 3′ ends have been linked to a pyrene moiety (31), was generated and found to be an inhibitor of the RNase H1 activity (Fig. 5D). This oligonucleotide is locked in a conformation where the 5′ and 3′ tails pair, as demonstrated by the observation of the excimer fluorescence indicating a precise stacking overlap of the two pyrenyl rings at the end of the small DNA helix resulting from the pairing of the last six bases of the 5′ tail with the last six bases of the 3′ tail (Fig. 5C). As expected (32), the end-stacked pyrenyl rings provide an additional stabilization of this helix. Thermal denaturation of VI-2 dipyrene results in a major transition at 65°C at both 275 and 295 nm corresponding to the melting of the G-quartet. A slight shoulder observed at 275 nm around 52°C corresponds to the melting of the helix, indicating an increase in Tm of 12°C for this short helix as compared to the unmodified VI-2 (data not shown). Because both ends were linked to pyrenyl moieties, it could not be radiolabeled and used in gel-shift assays. We therefore attempted to determine Kd in BIAcore experiments with immobilized RNase H. Kd values obtained were in the 10–30 nM range (see Supplementary Material).
Figure 5.
VI-2 ‘dipyrene’. (A) Proposed structure of the aptamer ‘dipyrene’. (B) Pyrenyl compound used for linking to the 5′ or 3′ ends of oligonucleotides via one of the two hydroxyl groups (31). (C) Fluorescence emission spectra of aptamer VI-2 ‘dipyrene’ excited at 349 nm, in 10 mM sodium cacodylate buffer, pH 7.0 with 40 mM ammonium sulfate recorded at 5, 10, 15, 20, 25 and 30°C. Peaks at 379 and 395 nm represent monomer emission bands whereas the broad band centered at 485 nm corresponds to the excimer. (D) Inhibition of human RNase H1 activity by the aptamer VI-2 ‘dipyrene’, using RNA–DNA hybrid BD2 as substrate.
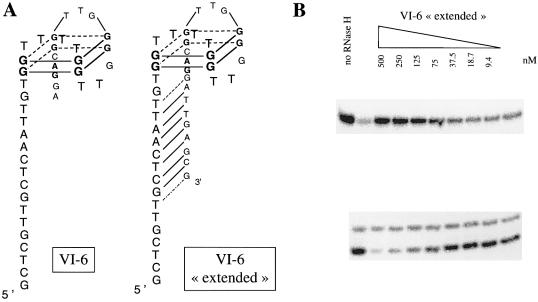
A variant of VI-2. Additional evidence of the importance of the base pairing between the 5′ and 3′ tails for these aptamers to be inhibitors comes from the observation that the VI-6 was not inhibitory despite a G-quartet fold (confirmed by melting curves—data not shown). The 40 nt aptamer, VI-6, has a long 5′ tail and a short 3′ tail with no significant base pairing possible between these regions (Fig. 6). However, when the additional oligonucleotides present as fixed sequences necessary for SELEX are included, seven consecutive nucleotides in the 3′ portion have potential base pairing with the 5′ tail of the 40 nt VI-6. Therefore, we synthesized an ‘extended’ version of VI-6 containing these seven additional nucleotides. This VI-6 ‘extended’ aptamer was capable of inhibiting human RNase H1 activity (Fig. 6) and the Kd determined by EMSA was 6.9 ± 0.8 nM (data not shown).
Figure 6.
VI-6 ‘extended’. (A) Proposed structure of aptamers VI-6 and VI-6 ‘extended’. G residues involved in G-quartet formation were identified by their low reactivity to modification by DMS (see text). (B) Inhibition by the aptamer VI-6 ‘extended’ of cleavage of the hybrid BD2 by human RNase H1.
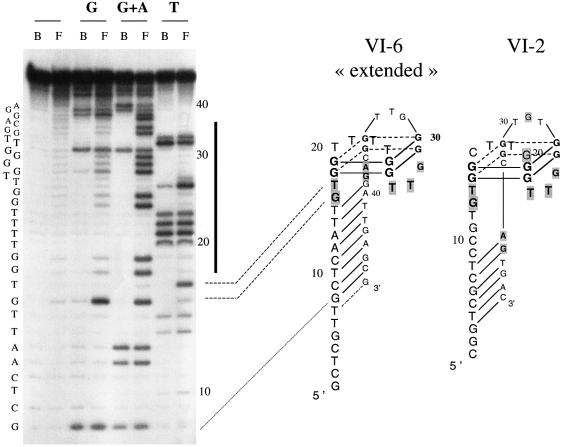
VI-2 and VI-6 have significant differences in nucleotide sequence yet both form G-quartet structures. To determine which G residues in VI-2 and VI-6 are engaged in G-quartets, we treated VI-2 and VI-6 with DMS in TE buffer, where the oligonucleotide is denatured, and in TE + 80 mM KCl where it is folded. Identification of the protected G residues in both VI-2 and VI-6 ‘extended’ aptamers led us to conclude that both aptamers adopt an identical fold. Although the two aptamers have different upper loop sequences, they are identical in length. The lower three-membered loop is conserved, as are the three contiguous bases immediately 5′ and 3′ of the G-quartet. Interestingly, these highly conserved bases are all located on the lower face of the G-quartet, pointing toward the stem, and are the same as those which chemical modification prevent binding to the enzyme (Fig. 7).
Figure 7.
Results of the chemical interference experiments obtained with aptamers VI-2 and VI-6 ‘extended’. End labeled aptamers were modified at T, G or G + A residues as described in Materials and Methods. Identical amounts of free (F) or protein-bound aptamers (B) were cleaved at the site of modification by treatment with pyrrolidine and loaded side by side on a sequencing gel. A representative experiment is illustrated by the gel shown on the left and put in register with the corresponding structure by dotted lines. Nucleotides belonging to the intramolecular quadruplex are located by the black bold line on the right side of the gel. Residues whose modification interferes with the binding to the human RNase H1 are indicated by a gray background. Major interferences are highlighted by bold letters.
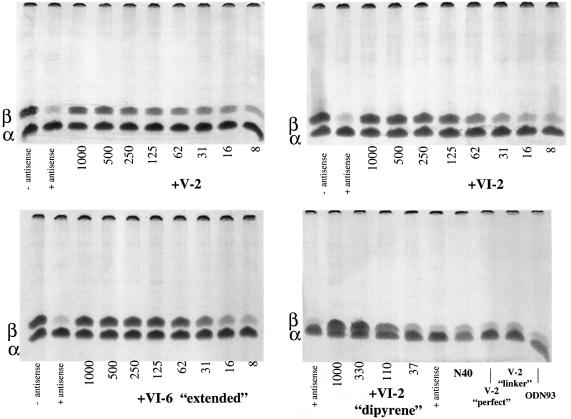
Abolition of antisense inhibition of translation in rabbit reticulocyte lysate
Antisense inhibition of translation observed in cell-free systems upon addition of an oligodeoxynucleotide complementary to the coding region of a targeted messenger RNA relies on the presence of an active RNase H in the medium. Endogenous RNase H activity in the wheat germ extract was sufficient to observe efficient antisense effects, but exogenous E.coli RNase HI has to be added to the rabbit reticulocyte lysate for oligonucleotide-targeted inhibition of translation. Addition of an inhibitor of the RNase H should therefore abolish antisense action and restore full translation of the targeted mRNA. We have used it as a test of the efficiency of our aptamers to inhibit the human RNase H1 in complex rabbit reticulocyte lysate translation system. These lysates retain their endogenous messenger RNAs encoding the α- and β-globin polypeptides. We have used an antisense oligodeoxynucleotide complementary to the nucleotides 113–129 of the mRNA coding for the β-globin chain. This antisense oligonucleotide efficiently inhibits the translation of this mRNA, provided that RNase H activity is present to cleave the mRNA at the site of hybridization to the antisense oligodeoxynucleotide. After preliminary experiments to adjust the concentrations of antisense oligonucleotides and human RNase H1 to reduce translation of the β-globin chain to less than 6%, we tested the aptamers at several concentrations to reverse the antisense effect of the oligonucleotide. As shown in Figure 8, V-2, VI-2, VI-6 ‘extended’ and VI-2 dipyrene efficiently inhibited human RNase H1, thereby permitting synthesis of the β-globin. Several aptamers used as controls had no effect even at the highest concentration (1 µM) tested in these experiments. Quantification of the radioactive bands corresponding to the α- and β-globin chains and plotting the percent inhibition of antisense effect versus the concentration of the aptamer tested indicate an IC50 of about 100 nM for V-2 and VI-2 and about 50 nM for VI-6 ‘extended’. No gross toxic or unspecific effect on translation was noticed when using these aptamers, as translation of the α-globin was the same as in the control reaction without added aptamer in all cases. These inhibitory aptamers had no effect on antisense inhibition of translation when the lysates were supplemented with E.coli RNase HI (data not shown).
Figure 8.
Reversal of antisense-mediated inhibition of translation by aptamers. Electrophoresis of translation products from a rabbit reticulocyte lysate supplemented with 10 nM human RNase H1, with (antisense +) or without (antisense –) 0.5 µM antisense oligo SC[113–129] targeted to β-globin mRNA and with antisense oligo together with various concentrations (nM) of aptamers as indicated below the lanes of the gel. Positions of the β-globin and α-globin chains are indicated as β and α bold letters on the left of each gel. Aptamers used as negative controls are N40, a population of 40mers of random sequences, V-2 ‘linker’ a variant of V-2 with a C18 spacer in place of the apical loop, V-2 ‘perfect’ a variant of V-2 where the bulge and internal loop have been converted into regular double stranded DNA, and ODN93 an inhibitory aptamer of the HIV-1 RT RNase H.
DISCUSSION
SELEX experiments targeting the human RNase H1 allowed us to identify DNA oligonucleotides with high affinity for the protein but lacking a consensus sequence, a situation reminiscent of that encountered by Schneider et al. for DNA aptamers directed against HIV-1 RT (48). Because all DNAs have identical 5′- and 3′-ends necessary for amplification in the SELEX procedure and in most cases can be eliminated (49), we generated aptamers of the internal 40 nt, screening for inhibition of RNase H1. Two of the aptamers, V-2 and VI-2, were found to be inhibitory. Although they share some similarities at the primary sequence level (about the same nucleotide composition and the presence of 4 or 5 doublets of G residues), they adopt quite different secondary structures.
Whereas both aptamers have the theoretical possibility of forming a G-quartet structure, a motif which has been observed many times for DNA aptamers (43), only VI-2 does so. V-2 is a hairpin structure with bulges within the stem as was predicted by a thermodynamic mfold algorithm. The aptamer VI-2 folds into a G-quartet structure involving 18 nucleotides close to the center of the sequence. Comparing V-2 and VI-2, we notice that they can be described as a stem loop structure in which the loop itself adopts an ordered structure: for V-2 this structure results probably from stacking interactions and non-canonical hydrogen bonding as suggested by enzymatic and chemical probing, and for VI-2 the structure is a unimolecular quadruplex. It is the unique combination of the stem and the ordered loop which confers binding and inhibitory properties to these aptamers as indicated by the lack of inhibition of shortened variants. Also the presence of regular double-stranded helical regions of DNA is not by itself sufficient. These helical regions are at most 6 bp in length in either V-2 or VI-2, and converting the stem of V-2 into a longer regular DNA helix abolished binding. Also the major chemical interferences identified in these aptamers are principally located outside the regular structures in both aptamers, essentially in the apical loop and bulge for V-2 and a set of conserved nucleotides present between the stem and the stack of G-quartets for VI-2 and VI-6. We can imagine that these nucleotides located between two very structured domains, i.e. the stack of G-quartets and the stem, would be forced to adopt a very peculiar structure which is recognized by the enzyme. Presence of G-quartets is not by itself sufficient for promoting binding. Shortened versions of VI-2 restricted to the G-quartet motifs do not bind the enzyme. Also ODN93, an oligonucleotide inhibitory of the RNase H of the HIV-1 reverse transcriptase, has poor affinity for the human enzyme and exhibits no inhibition of the cellular enzyme (31). ODN93 generated by the SELEX procedure was found to be the most inhibitory oligonucleotide exhibiting an IC50 of 500 nM and a Kd of 70–80 nM compared with the IC50 of 50–100 nM and Kd values of 10–30 nM for aptamers V-2 and VI-2. Because the 5′ half of ODN93 is G rich, it has the potential characteristics necessary for intra- or inter-molecular G-quartet formation.
We should stress that the selected aptamers are very specific at least for the eucaryotic RNase H1 as they are ineffective against the E.coli RNase H and the HIV RT RNase H despite the common structural properties of these three enzymes. In gel shift experiments performed in conditions identical to those used for the human enzyme, we observed no retarded bands with either HIV RT or E.coli enzyme up to 3 and 0.5 µM, respectively indicating that Kd values for these enzymes are much higher (data not shown). The fact that they inhibit both the human and the bovine enzyme indicates the recognition of a common structural motif shared by the eucaryotic RNases H1 but absent from procaryotic and viral RNases H. Future work aimed at determining this structural motif on the protein should address this issue. The inhibition by the aptamers of the human RNase H1 and not RNase H2, and their ability to function in the reticulocyte lysate opens the way for their use as specific inhibitory tools to examine the roles of the two classes of RNase H in the general physiology of the cell and their relative participation in the success of antisense oligonucleotides. In addition, these aptamers appear well suited for examination of the role of human RNase H1 in replication, transcription or repair in either complex biological media such as nuclear extracts or isolated mitochondria. Also, they could be introduced in intact cells where they could be employed to unravel the specific implication of the RNase H1 in the efficiency of antisense oligodeoxyribonucleotides.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Stein H. and Hausen,P. (1969) Enzyme from calf thymus degrading the RNA moiety of DNA–RNA hybrids: effect on DNA-dependent RNA polymerase. Science, 166, 393–395. [DOI] [PubMed] [Google Scholar]
- 2.Ohtani N., Haruki,M., Morikawa,M., Crouch,R.J., Itaya,M. and Kanaya,S. (1999) Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry, 38, 605–618. [DOI] [PubMed] [Google Scholar]
- 3.Wintersberger U. (1990) Ribonucleases H of retroviral and cellular origin. Pharmacol. Ther., 48, 259–280. [DOI] [PubMed] [Google Scholar]
- 4.Kogoma T. and Foster,P.L. (1998) Physiological functions of E. coli RNase H. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. John Libbey, Paris, pp. 39–66. [Google Scholar]
- 5.Crouch R.J. and Toulmé,J.J. (eds), Ribonucleases H. John Libbey, Paris. [Google Scholar]
- 6.Büsen W. and Frank,P. (1998) Bovine RNases H. In Crouch,J.J. and Toulmé,J.J. (eds), Ribonucleases H. John Libbey, Paris, pp. 113–146. [Google Scholar]
- 7.Cerritelli S.M., Frolova,E.G., Feng,C., Grinberg,A., Love,P.E. and Crouch,R.J. (2003) Failure to produce mitochondrial DNA results in embryonic lethality in RNaseh1 null mice. Mol. Cell, 11, 807–815. [DOI] [PubMed] [Google Scholar]
- 8.Kanaya S. (1998) Enzymatic activity and protein stability of E. coli ribonuclease H. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. John Libbey, Paris, pp. 1–38. [Google Scholar]
- 9.Kanaya S. and Crouch,R.J. (1983) DNA sequence of the gene coding for Escherichia coli ribonuclease H. J. Biol. Chem., 258, 1276–1281. [PubMed] [Google Scholar]
- 10.Itaya M. (1990) Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc. Natl Acad. Sci. USA, 87, 8587–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank P., Braunshofer-Reiter,C., Poltl,A. and Holzmann,K. (1998) Cloning, subcellular localization and functional expression of human RNase HII. Biol. Chem., 379, 1407–1412. [DOI] [PubMed] [Google Scholar]
- 12.Frank P., Braunshofer-Reiter,C., Wintersberger,U., Grimm,R. and Büsen,W. (1998) Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII. Proc. Natl Acad. Sci. USA, 95, 12872–12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerritelli S.M. and Crouch,R.J. (1998) Cloning, expression and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics, 53, 300–307. [DOI] [PubMed] [Google Scholar]
- 14.Wu H., Lima,W.F. and Crooke,S.T. (1998) Molecular cloning and expression of cDNA for human RNase H. Antisense Nucleic Acid Drug Dev., 8, 53–61. [DOI] [PubMed] [Google Scholar]
- 15.Crouch R.J., Arudchandran,A. and Cerritelli,S.M. (2001) RNase H1 of Saccharomyces cerevisiae: Methods and nomenclature. Methods Enzymol., 341, 395–413. [DOI] [PubMed] [Google Scholar]
- 16.Hughes S.H., Arnold,E. and Hostomsky,Z. (1998) RNase H of retroviral reverse transcriptases. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. John Libbey, Paris, pp. 195–224. [Google Scholar]
- 17.Pileur F., Toulmé,J.J. and Cazenave,C. (2000) Eukaryotic ribonucleases HI and HII generate characteristic hydrolytic patterns on DNA–RNA hybrids: further evidence that mitochondrial RNase H is an RNase HII. Nucleic Acids Res., 28, 3674–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toulmé J.J. and Tidd,D. (1998) Role of RNase H in antisense oligonucleotide-mediated effects. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. John Libbey, Paris, pp. 225–250. [Google Scholar]
- 19.Cazenave C., Loreau,N., Thuong,N.T., Toulmé,J.J. and Hélène,C. (1987) Enzymatic amplification of translation inhibition of rabbit beta-globin mRNA mediated by anti-messenger oligodeoxynucleotides covalently linked to intercalating agents. Nucleic Acids Res., 15, 4717–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larrouy B., Blonski,C., Boiziau,C., Stuer,M., Moreau,S., Shire,D. and Toulmé,J.J. (1992) RNase H-mediated inhibition of translation by antisense oligodeoxyribonucleotides: use of backbone modification to improve specificity. Gene, 121, 189–194. [DOI] [PubMed] [Google Scholar]
- 21.Cazenave C. (1998) Ribonucleases H of oocytes from the South-African clawed toad Xenopus laevis. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. John Libbey, Paris, pp. 101–112. [Google Scholar]
- 22.Loya S. and Hizi,A. (1993) The interaction of illimaquinone, a selective inhibitor of the RNase H activity, with the reverse transcriptases of human immunodeficiency and murine leukemia retroviruses. J. Biol. Chem., 268, 9323–9328. [PubMed] [Google Scholar]
- 23.Hnatyszyn O., Broussalis,A., Herrera,G., Muschietti,L., Coussio,J., Martino,V., Ferraro,G., Font,M., Monge,A., Martinez-Irujo,J.J., Sanroman,M., Cuevas,M.T., Santiago,E. and Lasarte,J.J. (1999) Argentine plant extracts active against polymerase and ribonuclease H activities of HIV-1 reverse transcriptase. Phytother. Res., 13, 206–209. [DOI] [PubMed] [Google Scholar]
- 24.Min B.S., Nakamura,N., Miyashiro,H., Kim,Y.H. and Hattori,M. (2000) Inhibition of human immunodeficiency virus type 1 reverse transcriptase and ribonuclease H activities by constituents of Juglans mandshurica. Chem. Pharm. Bull. (Tokyo), 48, 194–200. [DOI] [PubMed] [Google Scholar]
- 25.Borkow G., Fletcher,R.S., Barnard,J., Arion,D., Motakis,D., Dmitrienko,G.I. and Parniak,M.A. (1997) Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry, 36, 3179–3185. [DOI] [PubMed] [Google Scholar]
- 26.Shaw-Reid C.A., Munshi,V., Graham,P., Wolfe,A., Witmer,M., Danzeisen,R., Olsen,D.B., Carroll,S.S., Embrey,M., Wai,J.S., Miller,M.D., Cole,J.L. and Hazuda,D.J. (2003) Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem., 278, 2777–2780. [DOI] [PubMed] [Google Scholar]
- 27.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 28.Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 29.Toulmé J.J. (2000) Aptamers: selected oligonucleotides for therapy. Curr. Opin. Mol. Ther., 2, 318–324. [PubMed] [Google Scholar]
- 30.Williams K.P. and Bartel,D.P. (1995) PCR product with strands of unequal length. Nucleic Acids Res., 23, 4220–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreola M.L., Pileur,F., Calmels,C., Ventura,M., Tarrago-Litvak,L., Toulmé,J.J. and Litvak,S. (2001) DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry, 40, 10087–10094. [DOI] [PubMed] [Google Scholar]
- 32.Michel J., Bathany,K., Schmitter,J.M., Monti,J.-P. and Moreau,S. (2002) New ligand combinations for the efficient stabilization of short nucleic acid hairpins. Tetrahedron, 58, 7975–7982. [Google Scholar]
- 33.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.McEntee K., Weinstock,G.M. and Lehman,I.R. (1980) recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc. Natl Acad. Sci. USA, 77, 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzwater T. and Polisky,B. (1996) A SELEX primer. Methods Enzymol., 267, 275–301. [DOI] [PubMed] [Google Scholar]
- 36.Haruki M., Noguchi,E., Kanaya,S. and Crouch,R.J. (1997) Kinetic and stoichiometric analysis for the binding of Escherichia coli ribonuclease HI to RNA–DNA hybrids using surface plasmon resonance. J. Biol. Chem., 275, 22015–22022. [DOI] [PubMed] [Google Scholar]
- 37.Duconge F., Di Primo,C. and Toulmé,J.J. (2000) Is a closing GA pair a rule for stable loop–loop RNA complexes? J. Biol. Chem., 275, 21287–21294. [DOI] [PubMed] [Google Scholar]
- 38.Wohlrab F. (1992) Enzyme probes in vitro. Methods Enzymol., 212, 294–301. [DOI] [PubMed] [Google Scholar]
- 39.Williamson J.R. and Celander,D.W. (1989) Rapid procedure for chemical sequencing of small oligonucleotides without ethanol precipitation. Nucleic Acids Res., 18, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rovera G., Magarian,C. and Borun,T.W. (1978) Resolution of hemoglobin subunits by electrophoresis in acid urea polyacrylamide gels containing Triton X-100. Anal. Biochem., 85, 506–518. [DOI] [PubMed] [Google Scholar]
- 41.Cerritelli S.M. and Crouch,R.J. (1995) The non-RNase H domain of Saccharomyces cerevisiae RNase H1 binds double-stranded RNA: magnesium modulates the switch between double-stranded RNA binding and RNase H activity. RNA, 1, 246–259. [PMC free article] [PubMed] [Google Scholar]
- 42.SantaLucia J. Jr (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breaker R.R. (1997) DNA aptamers and DNA enzymes. Curr. Opin. Chem. Biol., 1, 26–31. [DOI] [PubMed] [Google Scholar]
- 44.Long K.S. and Crothers,D.M. (1995) Interaction of human immunodeficiency virus type 1 Tat-derived peptides with TAR RNA. Biochemistry, 34, 8885–8895. [DOI] [PubMed] [Google Scholar]
- 45.Hud N.V., Schultze,P. and Feigon,J. (1998) Ammonium ion as an NMR probe for monovalent cation coordination sites of DNA quadruplexes. J. Am. Chem. Soc., 120, 6403–6404. [Google Scholar]
- 46.Mergny J.L., Phan,A.T. and Lacroix,L. (1998) Following G-quartet formation by UV-spectroscopy. FEBS Lett., 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 47.Guschlbauer W., Chantot,J.-F. and Thiele,D. (1990) Four-stranded nucleic acid structures 25 years later: from guanosine gels to telomer DNA. J. Biomol. Struct. Dynam., 8, 491–511. [DOI] [PubMed] [Google Scholar]
- 48.Schneider D.J., Feigon,J., Hostomsky,Z. and Gold,L. (1995) High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry, 34, 9599–9610. [DOI] [PubMed] [Google Scholar]
- 49.Jayasena S.D. (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem., 45, 1628–1650. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.