Abstract
Saccharomyces cerevisiae possesses two Escherichia coli endonuclease III homologs, NTG1 and NTG2, whose gene products function in the base excision repair pathway and initiate removal of a variety of oxidized pyrimidines from DNA. Although the glycosylase activity of these proteins has been well studied, the in vivo importance of the AP lyase activity has not been determined. Previous genetic studies have suggested that the AP lyase activities of Ntg1p and Ntg2p may be major contributors in the initial processing of abasic sites. We conducted a biochemical characterization of the AP lyase activities of Ntg1p and Ntg2p via a series of kinetic experiments. Such studies were designed to determine if Ntg1p and Ntg2p prefer specific bases located opposite abasic sites and whether these lesions are processed with a catalytic efficiency similar to Apn1p, the major hydrolytic AP endonuclease of yeast. Our results indicate that Ntg1p and Ntg2p are equally effective in processing four types of abasic site-containing substrates. Certain abasic site substrates were processed with greater catalytic efficiency than others, a situation similar to Apn1p processing of such substrates. These biochemical studies strongly support an important biological role for Ntg1p and Ntg2p in the initial processing of abasic sites and maintenance of genomic stability.
INTRODUCTION
Oxidative damage is one of the most common types of DNA base damage resulting from a variety of endogenous and exogenous exposures to reactive oxygen species (1–5). If left unrepaired, these base modifications may lead to mutations and cell death and have been implicated in a number of degenerative processes such as cancer and aging (2–5). The primary pathway for the removal of oxidative damage is thought to be base excision repair (BER). The first step of BER is the excision of the damaged base initiated by a DNA N-glycosylase. This creates an abasic site where the phosphodiester backbone is subsequently cleaved either by the hydrolytic activities of an apurinic/apyrimidinic (AP) endonuclease, creating a 3′ hydroxyl group and 5′ deoxyribose phosphate (dRP), or by an AP lyase activity associated with certain DNA N-glycosylases. The AP lyase activity produces a 3′ α,β-unsaturated aldehyde and a 5′ phosphate via a β-elimination reaction (6). The cleavage reactions mediated by hydrolytic AP endonucleases and β-elimination AP lyases are shown in Figure 1. Following the end-trimming activities of one or more of dRPase, 3′ phosphodiesterase, 3′ phosphatase or 3′ flap endonuclease, BER is completed when DNA polymerase fills in the gap and DNA ligase seals the ends together (7–10).
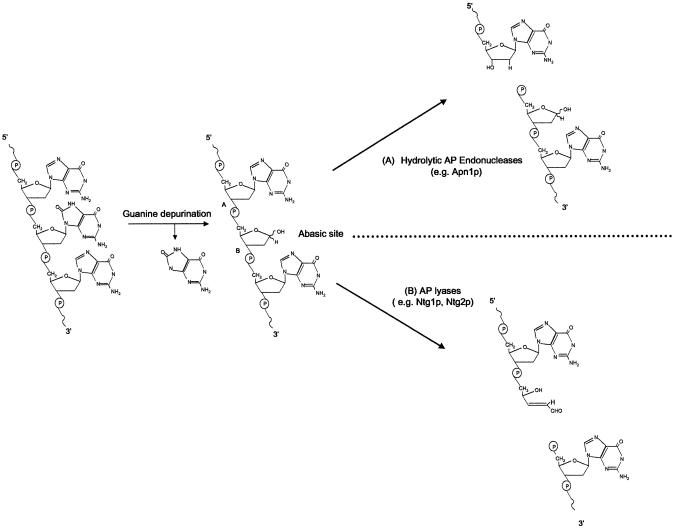
Figure 1.
Abasic site processing by AP endonucleases and AP lyases. Spontaneous hydrolysis of the N-glycosidic bond occurs frequently in genomic DNA guanine sites to produce abasic sites. Abasic sites are also produced by DNA N-glycosylases during the initial stops of BER (not shown). AP endonucleases (such as Apn1p) cleave DNA hydrolytically at (A) to generate a strand scission product containing a 5′ deoxyribose phosphate and a 3′ hydroxyl group. AP lyases cleave DNA by a β-elimination mechanism to generate a strand scission product containing a 5′ phosphate and a 3′ unsaturated aldehyde.
In Saccharomyces cerevisiae, Ntg1p and Ntg2p are N-glycosylase/AP lyases that remove oxidized pyrimidines from DNA. Both show significant sequence homology to each other (41% identity, 63% similarity) and exhibit a range of overlapping but not identical substrate specificities (11,12). As Escherichia coli endonuclease III homologs, both proteins contain a conserved hairpin–helix–hairpin motif thought to be involved in DNA binding (13). Despite their similarities, these proteins each display several notable differences. Ntg1p does not possess a consensus sequence for a C-terminal iron-sulfur center, typical of all other endonuclease III homologs, including Ntg2p (14). Ntg2p is localized exclusively to the nucleus whereas Ntg1p contains a 17 amino acid mitochondrial targeting sequence at the N-terminus and co-localizes to both the mitochondria and the nucleus (15,16). In addition, expression of NTG1 mRNA appears to be inducible in response to oxidizing agents while NTG2 mRNA expression seems to be unaffected (15,17). So far, S.cerevisiae is the only organism known to contain two such endonuclease III homologs.
Abasic sites are the intermediate step of BER and can also be generated by chemical induction or spontaneous base hydrolysis (1). It has been generally assumed that hydrolytic AP endonucleases are the primary proteins involved in the initial processing of abasic sites (18). Yeast strains lacking the major hydrolytic AP endonuclease, Apn1p, have been reported to be moderately sensitive to oxidizing agents and alkylating agents (19). Further, an increase in the level of spontaneous mutations and sensitivity to methyl methane sulfonate has been reported in apn1Δ apn2Δ strains (20). A series of BER yeast mutants lacking Ntg1p, Ntg2p and Apn1p singly and in combination were assessed for their respective contributions in protecting the cell from various oxidizing agents. Surprisingly, no single or combination of double mutants showed a decrease in cell survival over the wild-type strain. Furthermore, the BER-deficient triple mutant, ntg1 ntg2 apn1, also did not exhibit substantial sensitivity to oxidizing agents over the wild-type but did exhibit a hyper-recombination and mutator phenotype. When this BER defect was combined with a deficiency in either nucleotide excision repair (NER), translesion synthesis (TLS) or recombination (REC), then oxidizing agent sensitivity was observed (21). These findings indicated that overlap exists for the processing of a lesion(s), probably abasic sites, between the BER, NER, TLS and REC pathways. This genetic analysis also led to the conclusion that Ntg1p, Ntg2p and Apn1p proteins were most likely competing for a common substrate (abasic sites) and suggested that AP lyase activity might be as effective as Apn1p in the initial processing of abasic sites in yeast (21,22). Therefore, the AP lyase activities of Ntg1p and Ntg2p may play an important biological role for the repair of abasic sites.
To address the above possibility, we have conducted biochemical analyses of Ntg1p, Ntg2p and Apn1p on a series of duplex oligonucleotides containing abasic sites opposite to G, A, T or C for the purpose of revealing any potential differences in substrate specificity and catalytic efficiency among these proteins. A comparison of the kinetic parameters was carried out. Our biochemical results provide strong support for the notion that the AP lyase activities of Ntg1p and Ntg2p are biologically relevant functions of these proteins for eliminating the potential mutagenic and cytotoxic effect of abasic sites in yeast.
MATERIALS AND METHODS
Enzymes and chemicals
Escherichia coli endonuclease III was a generous gift from Dr Yoke Wah Kow (Emory University). T4 polynucleotide kinase was purchased from Promega and uracil DNA-glycosylase (UDG) was obtained from New England Biolabs. All other chemicals were of molecular biology grade or better.
Expression and purification of Ntg1p, Ntg2p and Apn1p
The NTG2 coding region was amplified by PCR using pRSETA-Ntg2 as template and primers d(pCATGCCATGGGAGAGGAAAGTAGG) and d(pCGGGATCCTTAGTGATGGTGATGGTGATGTTTTTTCTTGTGTCTTTCTG). The PCR product was cloned into the NcoI and BamHI restriction sites of pET15b (Novagen).
Ntg1p and Ntg2p were purified as previously described (11,14). Briefly, E.coli strain BL21 (DE3) containing the expression vector, either pGEX2T for GST–Ntg1p or pET15b for Ntg2p–His6, were grown in LB broth with ampicillin (100 µg/µl) to an OD600 of 0.5–1.0. Expression of GST–Ntg1p was induced by the addition of 1 mM isopropyl β-d-thiogalactoside (IPTG) for 2 h at 37°C. Cells were centrifuged at 12 000 g for 30 min and then lysed via sonication. The supernatant was applied to glutathione–Sepharose affinity beads (Pharmacia). Bound GST–Ntg1p was eluted with 10 mM reduced glutathione and then further purified to apparent homogeneity by Mono S FPLC (Pharmacia). Expression of Ntg2p–His6 was attained by the addition of 1 mM IPTG for 3 h at 25°C. The fractions that contained Ntg2p were further purified by a combination of Ni+ affinity chromatography (Qiagen) and Mono S FPLC (Pharmacia), resulting in a near-homogeneous protein preparation.
For expression and purification of Apn1p, an expression plasmid, pGEX-4T-1-Apn1, was obtained from Dr Dindial Ramotar (Montreal, Canada). BL21(DE3) cells containing this plasmid were grown as described above for Ntg1p and Ntg2p. Protein expression was induced by the addition of 1 mM IPTG at 25°C for 4 h. Cells were lysed via sonication and harvested by centrifugation at 12 000 g for 30 min at 4°C. The supernatant was applied to glutathione–Sephorase beads and GST–Apn1p was eluted with 10 mM reduced glutathione and further purified by Mono S FPLC. Protein concentrations were determined by the Bradford assay (23).
Preparation of DNA repair enzyme oligonucleotide substrates
An oligonucleotide (37mer) containing uracil at position 19 (U-37mer) was purchased from Integrated DNA Technologies (Coralville, IA) with the sequence d(pCTTGGACTGGATGTCGGCCXAGCGGATACAGGAGCA) (X = U). Complementary strands to the U-37mer containing either G, A, T or C opposite to the U were also obtained from Integrated DNA Technologies. All oligomers were gel purified prior to use. The U-37mer was 5′-end-labeled with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Promega) prior to annealing to the complementary strand (11). Unincorporated radioactivity was removed via purification on a G-30 spin column (Bio-Rad). Single-stranded U-37mer was annealed in a 1:2 molar ratio to the appropriate complementary strand and heated to 80°C for 10 min, then cooled slowly to room temperature. All duplex oligos (U/G-, U/A-, U/T- and U/C-37mer) were gel purified on a 15% non-denaturing gel (11). For the generation of abasic sites, each duplex oligonucleotide was incubated with UDG (4 U) at 37°C for 45 min in UDG buffer (30 mM HEPES–KOH, pH 7.5, 1 mM EDTA and 50 mM NaCl) followed by phenol/chloroform extraction and ethanol precipitation. All AP-37mer oligos were quantified by UV spectroscopy then evaluated for abasic site content by quantitative cleavage with 0.1 M piperidine at 90°C for 10 min followed by analysis on a 20% denaturing (7 M urea) polyacrylamide gel.
DNA strand scission assays
DNA strand scission assays were performed in either 10 or 20 µl reaction volumes containing buffer B (15 mM KH2PO4, pH 6.8, 10 mM EDTA and 10 mM β-mercaptoethanol) plus 40 mM KCl. To measure AP lyase activity on AP/G, AP/A, AP/T or AP/C substrates, reaction mixtures contained 20 nM duplex abasic site-containing oligos and were incubated with a final protein concentration of 10 nM Ntg1p, Ntg2p or Apn1p. Three replicates for each substrate type were incubated at 37°C for various time intervals (0–8 min). All reactions were terminated by the addition of 5 µl formamide dye loading solution and held on ice. Prior to gel loading, samples were heated at 65°C for 45 s then placed back on ice.
Kinetic assays were performed in triplicate using a range of substrate concentrations (5–100 nM) and the protein concentrations were 2 nM for Ntg1p and Apn1p and 10 nM for Ntg2p. Reactions were incubated at 37°C for various times (0–4 min). Reaction products were electrophoresed on a 20% denaturing (7 M urea) polyacrylamide gel. Following electrophoresis, DNA strand scission product formation was determined by phosphorimager (Molecular Dynamics) analysis of the gel using ImageQuant (Molecular Dynamics). Data were taken from the 4 min time point and used in Origin 5.0 (Origin Lap) statistical analysis software and Excel (Microsoft) to compute kinetic parameters.
RESULTS
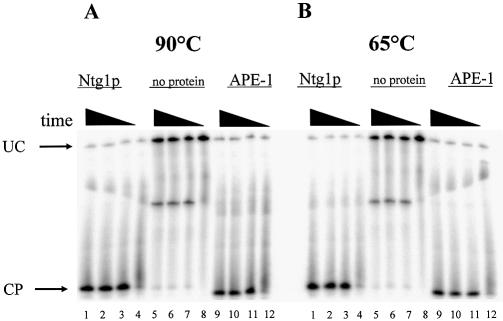
Abasic sites in DNA are generally regarded as thermolabile and susceptible to strand cleavage when exposed to heating. Many studies employ procedures that avoid exposure of samples containing non-modified abasic sites to elevated temperatures prior to gel loading or use modified abasic site analogs such as tetrahydrofuran to determine enzymatic processing of abasic site substrates (24–27). For methodological/analytical purposes, we examined the effect of heating samples containing unmodified, natural abasic site substrates for DNA strand scission at different times and temperatures following incubation with either yeast Ntg1p or human APE-1. In non-heated samples, the strand scission product was difficult to quantify due to significant smearing of the corresponding band in the gel (Fig. 2). In contrast, heating the samples at 65 or 90°C eliminated the band smearing, significantly improved band resolution and improved quantification of DNA strand scission. In addition, direct comparisons of the product bands at 65 or 90°C for short periods of time (0–2 min) produced negligible quantitative differences, indicating that unmodified, natural abasic sites are stable under these analytical conditions. In the negative control lanes (no protein), the background DNA strand scission observed between heated and non-heated samples varied from 1 to 4% at both 65 and 90°C regardless of time of incubation. Thus, under our experimental conditions, briefly heating the samples improved quantification of DNA strand scission products and did not introduce significant background into the analysis of enzyme-mediated DNA cleavage at abasic sites. Conse quently, the unmodified abasic site appears to be relatively stable under appropriate pH, temperature and chemical conditions. Therefore, the biochemical characterization of abasic site processing using oligonucleotides containing an unmodified, natural abasic site within a random DNA sequence may reflect a more realistic environment for enzyme-mediated processing.
Figure 2.
Effect of post-enzyme incubation sample heating on DNA strand scission analysis. The abasic site-containing strand was 5′-end-labeled with 32P and annealed to the complementary strand containing G opposite the abasic site. Oligonucleotide cleavage products were electrophoresed and subjected to phosphorimager analysis as described in Materials and Methods. (A) AP/G-37mer (70 nM) was incubated with 160 ng Ntg1p (lanes 1–4), no protein (lanes 5–8) or 160 ng APE-1 (lanes 9–12) for 20 min, then heated to 90°C for 2 min, 1 min, 20 s or 0 s, respectively. (B) AP/G-37mer (70 nM) was incubated with 160 ng Ntg1p (lanes 1–4), no protein (lanes 5–8) or 160 ng APE-1 (lanes 9–12) for 20 min then heated to 65°C for the increasing time intervals described above. UC designates uncleaved duplex oligonucleotide and CP designates the single-stranded cleaved product.
Ntg1p and Ntg2p efficiently cleave DNA at abasic sites
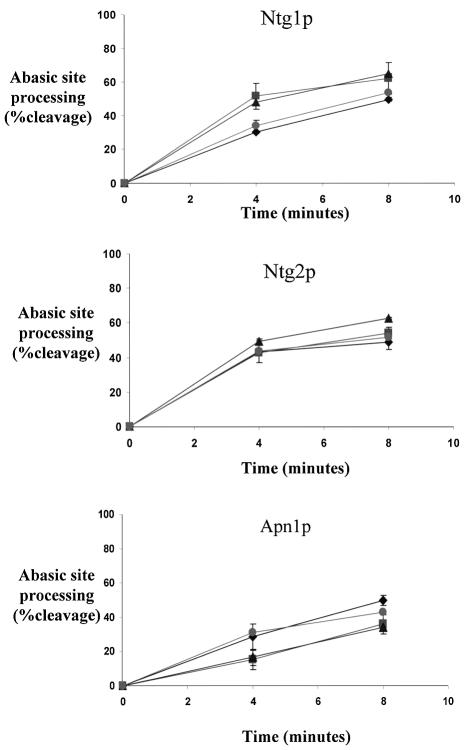
Genetic evidence suggests that the AP lyase activity of Ntg1p and Ntg2p may play an important biological role in the initial processing of abasic sites (21). Due to the high frequency of spontaneous depurination at guanine sites, we hypothesized that AP/C might be a preferred substrate, as well as apurinic sites generated by the glycosylase activities of Ntg1p and Ntg2p on oxidized pyrimidines. To assess whether these proteins demonstrated preferences for abasic sites located opposite different DNA bases, we prepared a series of duplex oligonucleotides containing abasic sites in all four opposite base contexts (AP/G-37mer, AP/A-37mer, AP/T-37mer and AP/C-37mer). Figure 3 shows the enzymatic activities of Ntg1p and Ntg2p on this series of AP-37mer substrates. Quantification of the DNA strand scission products shows that Ntg1p and Ntg2p exhibit very similar abasic site substrate specificities and are approximately equal in their abilities to process all four types of abasic site-containing substrates. These results also indicate that both Ntg1p and Ntg2p exhibit a moderate preference for AP/T sites. These findings contrast with the human endonuclease III homolog, hNTH1, which shows a distinct preference for AP/G substrates compared to other opposite base contexts (28). We conclude that both Ntg1p and Ntg2p efficiently recognize and process abasic sites in duplex DNA regardless of the opposite base context and that their abilities to catalyze strand cleavage are comparable to that of Apn1p.
Figure 3.
Excision efficiencies of Ntg1p, Ntg2p and Apn1p processing abasic sites in different opposite base contexts. The abasic site-containing oligonucleotide duplex contained either guanine (diamonds), adenine (squares), thymine (triangles) or cytosine (circles) opposite to the abasic site. A 2:1 molar ratio of substrate to protein was subjected to incubation for 0–8 min at 37°C. Experiments were performed in triplicate. The percent cleavage was quantified by phosphorimager analysis. Error bars represent the standard error.
The kinetic parameters of Ntg1p and Ntg2p for processing abasic sites are similar to Apn1p
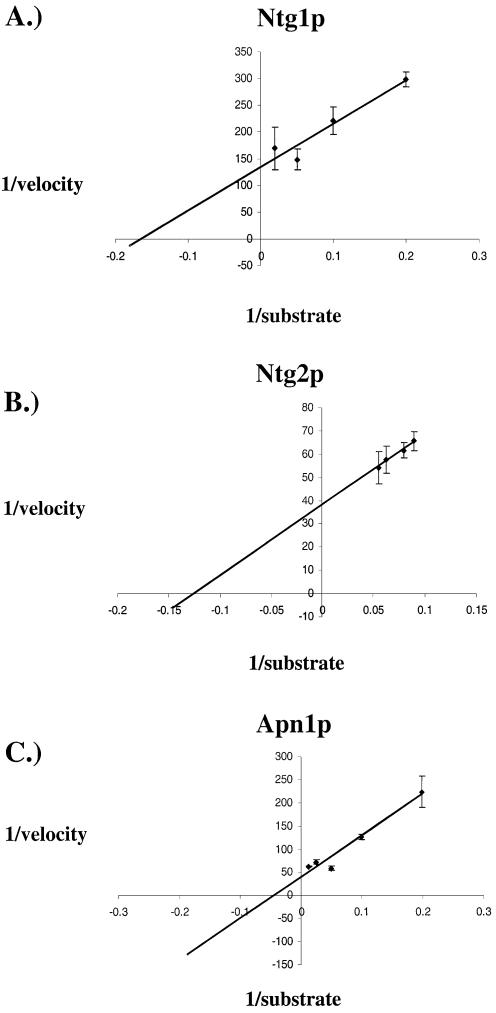
The kinetic parameters for Ntg1p, Ntg2p and Apn1p were determined using all four base contexts opposite an abasic site. Figure 4 shows Lineweaver–Burk plots for Ntg1p, Ntg2p and Apn1p for the AP/T-37mer substrate. The calculated kinetic parameters for the processing of all abasic site substrates used in this study are given in Table 1. The Km value for Apn1p obtained here was in the range of that reported from previously published studies on depurinated poly[d(A-T)] and abasic site plasmid substrates at 5.5 and 20 nM, respectively (29). However, other kinetic parameters (kcat or turnover number and kcat/Km or catalytic efficiency) were lower for the AP-37mer substrates used in the present study (29). Such differences may be attributed to the use of recombinant protein (here) versus the purified, endogenous cellular protein used in the previous studies (29). Further, it is also likely that specific flanking sequences surrounding the abasic site may have contributed to the differences in our kinetic parameters for Apn1p. Our results show that the AP lyase activities of Ntg1p and Ntg2p exhibit enzymatic efficiencies for processing AP/T and AP/C that are comparable to Apn1p. Interestingly, both Ntg1p and Ntg2p, but not Apn1p, had significantly lower enzymatic efficiencies when processing apurinic (abasic sites opposite pyrimidines) sites when compared to apyrimidinic (abasic sites opposite purines) sites.
Figure 4.
Kinetic analysis of AP/T 37mer DNA strand cleavage by Ntg1p (A), Ntg2p (B) and Apn1p (C). Kinetic assays were performed in triplicate using a range of substrate concentrations (5–100 nM). Protein concentrations were 2 nM for Ntg1p and Apn1p and 10 nM for Ntg2p. Strand scission was determined by phosphorimager analysis. The panels are plots of 1/reaction rate (pmol/min) versus 1/substrate concentration (nM). The average values were used to obtain a best fit Lineweaver–Burk plot. Error bars represent the standard error.
Table 1. Kinetic parameters for incision at abasic sites by Ntg1p, Ntg2p and Apn1p.
| Repair enzyme | Substrate | kcat (min–1) | Km (nM) | kcat/Km (min–1 nM) | |
|---|---|---|---|---|---|
| Ntg1p | AP/G | 0.056 | 24.86 | 0.002 | |
| AP/A | 0.042 | 11.37 | 0.003 | ||
| Ntg1p | AP/T | 0.42 | 6.67 | 0.063 | |
| Ntg1p | AP/C | 1.35 | 36.58 | 0.037 | |
| Ntg2p | AP/G | 0.194 | 6.43 | 0.031 | |
| AP/A | 0.307 | 21.8 | 0.014 | ||
| AP/T | 1.40 | 9.40 | 0.149 | ||
| AP/C | 0.90 | 46.0 | 0.190 | ||
| Apn1p | AP/G | 2.26 | 39.80 | 0.057 | |
| AP/A | 0.061 | 10.05 | 0.061 | ||
| AP/T | 1.54 | 11.67 | 0.133 | ||
| AP/C | 2.14 | 28.79 | 0.074 |
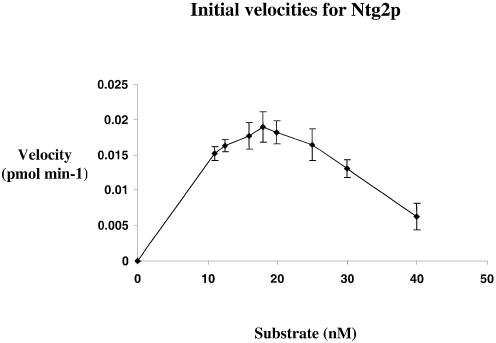
A striking difference between Ntg1p and Ntg2p is that Ntg2p exhibited apparent substrate inhibition. Figure 5 shows that the initial velocity of Ntg2p decreases with increasing substrate concentration. As a result, whenever the substrate concentration exceeded the protein concentration more than 2-fold, the initial velocities decreased. Consequently, the initial velocities for Ntg2p were determined for a relatively narrow range of substrate concentrations.
Figure 5.
Ntg2p exhibits substrate inhibition. Initial velocities were determined by incubating 10 nM Ntg2p with increasing concentrations of AP/T-37mer (11–40 nM) at different time intervals (0–4 min). Reaction products were analyzed as described in Materials and Methods. Initial velocities were calculated at the 4 min time point and plotted against increasing substrate concentration. Error bars represent the standard error.
DISCUSSION
Abasic sites are one of the most frequently occurring spontaneous lesions in cellular DNA (30). They arise through hydrolysis of the N-glycosidic bond between the base and the deoxyribose moiety, from free radical attack via oxidizing agents and ionizing radiation or as an intermediate product during the process of BER (10,18). If abasic sites are not repaired, they may cause DNA strand breaks leading to genetic instability or cell death. Therefore, it is critical that the cell has multiple repair proteins to counter such potentially deleterious effects and restore genomic fidelity. It has been generally assumed that in S.cerevisiae, Apn1p is the major repair protein responsible for the initial processing of abasic sites. This notion is due in part to the fact that the biological importance of AP lyase activity in processing abasic sites in vivo is not well understood. However, genetic studies by Swanson et al. provide biological evidence that the AP lyase activities of Ntg1p and Ntg2p are major contributors to the removal of abasic sites (21). In this study, we have directly compared the activities of Ntg1p and Ntg2p to Apn1p on a variety of abasic sites substrates. Since Ntg1p and Ntg2p recognize a wide spectrum of oxidized pyrimidines, it might be reasonable to expect robust AP lyase activities on AP/A and AP/G substrates and perhaps less substantial activites on AP/T and AP/C substrates. In this regard, our results are somewhat unexpected. We have presented the first direct biochemical evidence that both Ntg1p and Ntg2p possess robust AP lyase activities directed against abasic site-containing substrates in all opposite base contexts and support the idea that this AP lyase activity mediates an important physiological role in yeast. A comparison of the kinetic parameters in Table 1 suggests that for Apn1p there are minimal catalytic differences between the four types of abasic site-containing substrates. In addition, all three proteins seem to process AP/C and AP/T sites with equal efficiencies. However, Ntg1p and Ntg2p exhibit significant differences when the abasic site is opposite purines or pyrimidines. Furthermore, the catalytic efficiencies of Ntg1p and Ntg2p AP lyase activities are much lower when compared with the catalytic efficiencies of their DNA N-glycosylase activities on substrates such as dihydrouracil (Ntg1p 21.5 min–1 nM, Ntg2p 0.833 min–1 nM) (11). Therefore, the chemical nature of the initial substrate is important for determining the enzymatic activity of Ntg1p and Ntg2p and is consistent with another DNA N-glycosylase/AP lyase in S.cerevisiae, yOGG1p, whose AP lyase activity is approximately 5-fold lower than its glycosylase activity (31). These findings could also be related to the type of base pairing generated by DNA N-glycosylases that do not possess AP lyase activity and are directed against base lesions produced by cytosine deamination or alkylation. For example, 3-methyladenine DNA glycoslyase removes toxic 3-methyladenine adducts producing an abasic site opposite to T. If Ntg1p and Ntg2p show a preference for handling AP/T sites, perhaps these proteins are just as likely to process the AP/T site as Apn1p. Spontaneous base hydrolysis occurs an estimated 10 000 times per mammalian cell per day and is thought to involve purines more frequently than pyrimidines. It is conceivable that a similar level of spontaneous base hydrolysis occurs in S.cerevisiae and concurrent repair events to minimize abasic sites may require the collective, simultaneous actions of Apn1p, Ntg1p and Ntg2p. Although the precise cellular levels for Ntg1p and Ntg2p are unknown, if these proteins are expressed at levels comparable to Apn1p, all three enzymes would be potentially available for efficient processing of abasic sites.
Our finding that Ntg2p consistently exhibited a relatively low turnover for abasic sites reveals an important difference between Ntg1p and Ntg2p and could be indicative of either substrate or product inhibition. The inhibition observed occurred at relatively low substrate concentrations and somewhat limits the interpretation of the kinetics measurements for Ntg2p. This result was somewhat surprising, as the kinetic parameters for Ntg2p glycosylase activity for substrates containing a variety of other base damage products were determined and did not show substrate concentration dependence (11,12). While the crystal structures for Ntg1p and Ntg2p have not been determined, they are assumed to be similar to E.coli endonuclease III. It is possible that some unknown, subtle structural differences could help explain the biochemical differences between Ntg1p and Ntg2p.
It is recognized that the steps of BER are highly coordinated as a mechanism to protect the exposure of the abasic site to the cellular environment (32,33). Another potential explanation for Ntg2p low turnover seen in these in vitro studies could be that Ntg2p has an increased association time for abasic sites as a means to protect the abasic site until Apn1p uses its 3′ diesterase activity to remove the potentially toxic 3′ α,β-unsaturated aldehyde to produce a 3′ hydroxyl group, thus creating a substrate for DNA polymerase. It was recently reported that human thymine-DNA glycosylase (TDG) was covalently modified through a family of ubiquitin-like proteins, SUMO-1 and SUMO-2/3 (34). SUMO conjugation dramatically changed the enzymatic properties of TDG, reducing DNA binding affinity and modulating its AP endonuclease-mediated turnover number. To date, although no other BER proteins, yeast or human, have been shown to exhibit post-translational sumoylation, it is conceivable that similar modifications of N-glycosylases such as Ntg1p and Ntg2p could significantly modify their kinetic parameters. In this regard, there are several examples of other types of yeast proteins that undergo sumoylation and subsequent activity modification (35–37).
In conclusion, our findings provide a biochemical basis for the notion that the AP lyase activities of Ntg1p and Ntg2p are important for the initial processing of abasic sites in S.cerevisiae. It should be emphasized that ntg1 ntg2 apn1 triple mutants exhibit an 18-fold increase in recombination rates while neither ntg1 ntg2 double nor apn1 single mutants exhibit recombination rates significantly above wild-type (21). These genetic data together with our in vitro kinetic studies suggest that both the hydrolytic AP endonuclease activity of Apn1p and the AP lyase activities of Ntg1p and Ntg2p are important for the processing of abasic sites in S.cerevisiae. It should also be emphasized that the role of the AP lyase activity may be a yeast-specific function. For human proteins, it has been reported that when APE-1, the major hydrolytic AP endonuclease, is present at certain concentrations, it stimulates the glycosylase activity of hOGG1 and hNth, resulting in a bypass of their associated AP lyase activities (25,38,39). Conversely, a recent report characterizing mutants defective in Nth1, the Ntg1/2p homolog in Schizosaccharomyces pombe, demonstrated that the AP lyase activity of this protein and not the S.pombe Apn1p homolog, Apn1p (possessing no AP endonuclease activity), is the major cellular enzyme for the repair of abasic sites (40,41). Therefore, the contributions of hydrolytic AP endonucleases and/or AP lyases, either individually or collectively, to initiating the repair of abasic sites seems to be species dependent. We propose that in addition to Apn1p, Ntg1p and Ntg2p are major contributors to the repair of abasic sites in S.cerevisiae.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the members of the Doetsch laboratory for their helpful discussions and Dr Yoke Wah Kow for critical reading of the manuscript. This work was supported by grants ES11163 and CA073041 from the National Institutes of Health.
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 2.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 3.Marnett L.J. (2000) Oxyradicals and DNA damage. Carcinogenesis, 21, 361–370. [DOI] [PubMed] [Google Scholar]
- 4.Ames B.N., Shigenaga,M.K. and Hagen,T.M. (1993) Oxidants, antioxidants and the degenerative diseases of aging. Proc. Natl Acad. Sci. USA, 90, 7915–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckman K.B. and Ames,B.N. (1997) Oxidative decay of DNA. J. Biol. Chem., 272, 19633–19636. [DOI] [PubMed] [Google Scholar]
- 6.Doetsch P.W. and Cunningham,R.P. (1990) The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res., 236, 173–201. [DOI] [PubMed] [Google Scholar]
- 7.Krokan H.E., Nilsen,H., Skorpen,F., Otterlei,M. and Slupphaug,G. (2000) Base excision repair of DNA in mammalian cells. FEBS Lett., 476, 73–77. [DOI] [PubMed] [Google Scholar]
- 8.Karumbati A.S., Deshpande,R.A., Jilani,A., Vance J.R., Ramotar,D. and Wilson,T.E. (2003) The role of yeast DNA 3′ phosphatase Tpp1 and Rad1/Rad10 endonuclease in processing spontaneous and induced base lesions. J. Biol. Chem., 278, 31434–31443. [DOI] [PubMed] [Google Scholar]
- 9.Guillet M. and Boiteux,S. (2002) Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J., 21, 2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memisoglu A. and Samson,L. (2000) Base excision repair in yeast and mammals. Mutat. Res., 451, 39–51. [DOI] [PubMed] [Google Scholar]
- 11.You H.J., Swanson,R.L. and Doetsch,P.W. (1998) Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry, 37, 6033–6040. [DOI] [PubMed] [Google Scholar]
- 12.Senturker S., Auffret van der Kemp,P., You,H.J., Doetsch,P.W., Dizdaroglu,M. and Boiteux,S. (1998) Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res., 26, 5270–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo C.F., McRee,D.E., Fisher,C.L., O’Handley,S.F., Cunningham,R.P. and Tainer,J.A. (1992) Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science, 258, 434–440. [DOI] [PubMed] [Google Scholar]
- 14.Augeri L., Lee,Y.M., Barton,A.B. and Doetsch,P.W. (1997) Purification, characterization, gene cloning and expression of Saccharomyces cerevisiae redoxyendonuclease, a homolog of Escherichia coli endonuclease III. Biochemistry, 36, 721–729. [DOI] [PubMed] [Google Scholar]
- 15.You H.J., Swanson,R.L., Harrington,C., Corbett,A.H., Jinks-Robertson,S., Senturker,S., Wallace,S.S., Boiteux,S., Dizdaroglu,M. and Doetsch,P.W. (1999) Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]
- 16.Alseth I., Eide,L., Pirovano,M., Rognes,T., Seeberg,E. and Bjoras,M. (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol., 19, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eide L., Bjoras,M., Pirovano,M., Alseth,I., Berdal,K.G. and Seeberg,E. (1996) Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson D.M. III and Barsky,D. (2001) The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res., 485, 283–307. [DOI] [PubMed] [Google Scholar]
- 19.Ramotar D., Popoff,S.C., Gralla,E.B. and Demple,B. (1991) Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol., 11, 4537–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson R.E., Torres-Ramos,C.A., Izumi,T., Mitra,S., Prakash,S. and Prakash,L. (1998) Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1 and its role in the repair of abasic sites. Genes Dev., 12, 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson R.L., Morey,N.J., Doetsch,P.W. and Jinks-Robertson,S. (1999) Overlapping specificities of base excision repair, nucleotide excision repair, recombination and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao W., Chow,B.L., Hanna,M. and Doetsch,P.W. (2001) Deletion of the MAG1 DNA glycosylase gene suppresses alkylation-induced killing and mutagenesis in yeast cells lacking AP endonucleases. Mutat. Res., 487, 137–147. [DOI] [PubMed] [Google Scholar]
- 23.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 24.Vidal A.E., Boiteux,S., Hickson,I.D. and Radicella,J.P. (2001) XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J., 20, 6530–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill J.W., Hazra,T.K., Izumi,T. and Mitra,S. (2001) Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res., 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unk I., Haracska,L., Johnson,R.E., Prakash,S. and Prakash,L. (2000) Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem., 275, 22427–22434. [DOI] [PubMed] [Google Scholar]
- 27.Wilson D.M. III, Takeshita,M., Grollman,A.P. and Demple,B. (1995) Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem., 270, 16002–16007. [DOI] [PubMed] [Google Scholar]
- 28.Eide L., Luna,L., Gustad,E.C., Henderson,P.T., Essigmann,J.M., Demple,B. and Seeberg,E. (2001) Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry, 40, 6653–6659. [DOI] [PubMed] [Google Scholar]
- 29.Johnson A.W. and Demple,B. (1988) Yeast DNA 3′-repair diesterase is the major cellular apurinic/apyrimidinic endonuclease: substrate specificity and kinetics. J. Biol. Chem., 263, 18017–18022. [PubMed] [Google Scholar]
- 30.Nakamura J. and Swenberg,J.A. (1999) Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res., 59, 2522–2526. [PubMed] [Google Scholar]
- 31.Girard P.M., D’Ham,C., Cadet,J. and Boiteux,S. (1998) Opposite base-dependent excision of 7,8-dihydro-8-oxoadenine by the Ogg1 protein of Saccharomyces cerevisiae. Carcinogenesis, 19, 1299–1305. [DOI] [PubMed] [Google Scholar]
- 32.Nilsen H. and Krokan,H.E. (2001) Base excision repair in a network of defence and tolerance. Carcinogenesis, 22, 987–998. [DOI] [PubMed] [Google Scholar]
- 33.Wilson S.H. and Kunkel,T.A. (2000) Passing the baton in base excision repair. Nature Struct. Biol., 7, 176–178. [DOI] [PubMed] [Google Scholar]
- 34.Hardeland U., Steinacher,R., Jiricny,J. and Schar,P. (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J., 21, 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachant J., Alcasabas,A., Blat,Y., Kleckner,N. and Elledge,S.J. (2002) The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell, 9, 1169–1182. [DOI] [PubMed] [Google Scholar]
- 36.Hoege C., Pfander,B., Moldovan,G.L., Pyrowolakis,G. and Jentsch,S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO [comment]. Nature, 419, 135–141. [DOI] [PubMed] [Google Scholar]
- 37.Johnson E.S. and Blobel,G. (1999) Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol., 147, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marenstein D.R., Ocampo,M.T., Chan,M.K., Altamirano,A., Basu,A.K., Boorstein,R.J., Cunningham,R.P. and Teebor,G.W. (2001) Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J. Biol. Chem., 276, 21242–21249. [DOI] [PubMed] [Google Scholar]
- 40.Osman F., Bjoras,M., Alseth,I., Morland,I., McCready,S., Seeberg,E. and Tsaneva,I. (2003) A new Schizosaccharomyces pombe base excision repair mutant, nth1, reveals overlapping pathways for repair of DNA base damage. Mol. Microbiol., 48, 465–480. [DOI] [PubMed] [Google Scholar]
- 41.Ramotar D., Vadnais,J., Masson,J.Y. and Tremblay,S. (1998) Schizosaccharomyces pombe apn1 encodes a homologue of the Escherichia coli endonuclease IV family of DNA repair proteins. Biochim. Biophys. Acta, 1396, 15–20. [DOI] [PubMed] [Google Scholar]