Abstract
Sun1 and 2 are A-type lamin-binding proteins that, in association with nesprins, form a link between the inner nuclear membranes (INMs) and outer nuclear membranes of mammalian nuclear envelopes. Both immunofluorescence and immunoelectron microscopy reveal that Sun1 but not Sun2 is intimately associated with nuclear pore complexes (NPCs). Topological analyses indicate that Sun1 is a type II integral protein of the INM. Localization of Sun1 to the INM is defined by at least two discrete regions within its nucleoplasmic domain. However, association with NPCs is dependent on the synergy of both nucleoplasmic and lumenal domains. Cells that are either depleted of Sun1 by RNA interference or that overexpress dominant-negative Sun1 fragments exhibit clustering of NPCs. The implication is that Sun1 represents an important determinant of NPC distribution across the nuclear surface.
Introduction
The nuclear envelope (NE) is the selective barrier that defines the interface between the nucleus and the cytoplasm (Burke and Stewart, 2002; Gruenbaum et al., 2005). Because it mediates molecular trafficking between these two compartments, it plays an essential role in the maintenance of their biochemical identities. In addition to its transport function, the NE is also a key determinant of nuclear architecture, providing anchoring sites at the nuclear periphery for chromatin domains as well as for a variety of structural and regulatory molecules. A corresponding contribution to cytoplasmic structure has been described in which NE components may also influence cytoskeletal organization and mechanotransduction (Tzur et al., 2006; Worman and Gundersen, 2006).
The NE is composed of several structural elements, the most prominent of which are the inner nuclear membranes (INMs) and outer nuclear membranes (ONMs). These are separated by the perinuclear space (PNS), a gap of 30–50 nm. Annular junctions between the INM and ONM create aqueous channels that traverse the NE and that are occupied by nuclear pore complexes (NPCs). It is the NPCs that endow the NE with its selective transport properties (Tran and Wente, 2006).
The final major feature of the NE is the nuclear lamina, a thin (20–50 nm) protein layer that is associated with both the INM and chromatin. The lamina is composed primarily of A- and B-type lamins, which are members of the intermediate filament protein family (Gerace et al., 1978). The lamins interact with components of the INM and NPCs as well as with chromatin proteins and DNA (Zastrow et al., 2004). In this way, the lamina plays an important structural and organizational role at the nuclear periphery.
Despite their continuities, the INM and ONM are biochemically distinct. The ONM features numerous junctions with the peripheral ER, to which it is functionally and compositionally similar. In contrast, the INM contains its own unique selection of integral proteins. Clearly, the INM, ONM, and ER represent discrete domains within a single continuous membrane system. Accordingly, the PNS is a perinuclear extension of the ER lumen.
Localization of integral proteins to the INM involves a process of selective retention (Powell and Burke, 1990; Soullam and Worman, 1995; Ellenberg et al., 1997). Although proteins that are mobile within the ER and ONM may gain access to the INM via the continuities at each NPC, only proteins that interact with nuclear or other NE components are retained and concentrated. Recent studies suggest that additional mechanisms may overlie this basic scheme. Ohba et al. (2004) showed that movement of integral proteins through the NPC membrane domain is energy dependent. Other studies suggest a role for the nuclear transport receptor adaptor karyopherin/importin-α in the transit of proteins to the INM (King et al., 2006; Saksena et al., 2006).
Recognition of ONM-specific membrane proteins raises the question of what prevents these proteins from escaping to the peripheral ER. In Caenorhabditis elegans, the localization of Anc-1, an ONM protein involved in actin-based nuclear positioning, requires Unc-84, an INM protein whose retention is lamin dependent (Lee et al., 2002; Starr and Han, 2002). These observations led to a model in which Unc-84 and Anc-1 interact across the PNS via their lumenal domains, providing a mechanism for the tethering of ONM proteins.
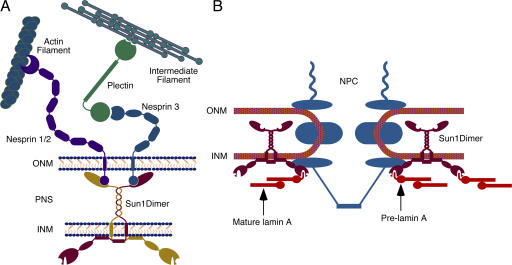
In mammals, two large actin-binding proteins, nesprin 1 Giant (nesp1G; 1,000 kD) and nesprin 2 Giant (nesp2G; 800 kD), reside in the ONM (Apel et al., 2000; Zhang et al., 2001; Mislow et al., 2002; Zhen et al., 2002). The nesprins (also known as Syne 1 and 2) are related to both Anc-1 and a Drosophila melanogaster ONM protein, Klarsicht (Welte et al., 1998; Mosley-Bishop et al., 1999), in that they contain an ∼50–amino acid C-terminal KASH (Klarsicht, Anc-1, Syne homology) domain consisting of a single transmembrane (TM) anchor and a short segment of ∼30–40 residues that resides within the PNS. A third ONM KASH domain–containing protein, nesprin 3, interacts with plectin, which is a large (466 kD) cytolinker (Wilhelmsen et al., 2005).
Unc-84 contains an ∼200–amino acid C-terminal region that shares homology with Sad1p, a Schizosaccharomyces pombe spindle pole body protein (Hagan and Yanagida, 1995). This sequence, which is known as the SUN (Sad1p, Unc-84) domain, resides within the PNS. The human genome encodes five SUN domain proteins. Two of these, Sun1 and 2, are lamin A–interacting proteins of the INM with topologies similar to that of Unc-84 (Hodzic et al., 2004; Crisp et al., 2006; Haque et al., 2006).
Both Sun1 and 2 cooperate in tethering nesp2G in the ONM (Padmakumar et al., 2005; Crisp et al., 2006; Haque et al., 2006; Hasan et al., 2006). This tethering involves interactions that span the PNS (Crisp et al., 2006), similar to that suggested for Unc84 and Anc-1. Unc84 also tethers Unc-83, another ONM KASH domain protein (McGee et al., 2006). Competition between nesprin 1 and 2 KASH domains (Zhang et al., 2007) suggests that nesp1G is similarly tethered. In this way, Sun1 and 2 function as links in a molecular chain that connects the actin cytoskeleton via nesprins to lamins and other nuclear components. We have termed this assembly the LINC (linker of nucleoskeleton and cytoskeleton) complex (Crisp et al., 2006). The fact that nesprin 3 binds plectin, a diverse cytolinker (Wilhelmsen et al., 2005), indicates that there may be multiple isoforms of the LINC complex responsible for integrating the nucleus with different components of the cytoskeleton.
As alluded to in the previous paragraphs, the NE can influence cytoplasmic mechanics and the responses of cells to mechanical stress. Cells depleted of either A-type lamins or emerin, an INM protein, exhibit reduced cytoplasmic resilience and an inability to activate mechanosensitive genes (Broers et al., 2004; Lammerding et al., 2004, 2005, 2006). In humans, the loss or mutation of either A-type lamins or emerin is associated with several diseases (Muchir and Worman, 2004), including Emery-Dreifuss muscular dystrophy. It is not hard to imagine that the LINC complex might be the mediator of these effects given its proposed role in nucleocytoplasmic coupling.
Less clear is the extent and nature of the interactions of LINC complex components and how these might affect LINC complex function. In the case of nesprins 1 and 2 versus nesprin 3, there are obvious differences in terms of actin versus plectin association. At the INM, the situation with the Sun proteins is more ambiguous. We know that there is some degree of functional redundancy between Sun1 and 2 with respect to nesprin 2 tethering. Furthermore, we know that Sun1 and 2 can associate with lamin A but that this interaction is not required for their localization. In this study, we further explore the interactions of SUN proteins at the nuclear periphery. In doing so, we have been able to describe discrete regions within Sun1 that function both in localization to the INM and in oligomerization. Most importantly, we are able to demonstrate that Sun1 and 2 are segregated within the INM. Although Sun2 displays a roughly uniform distribution across the NE, Sun1 is concentrated at NPCs. Elimination of Sun1 or overexpression of Sun1 mutants leads to NPC clustering. The inference is that Sun1 but not Sun2 functions in maintenance of the uniform distribution of NPCs. It also follows that certain LINC complex isoforms may mediate the differential association of cytoskeletal elements with NPCs versus NPC-free regions of the NE.
Results
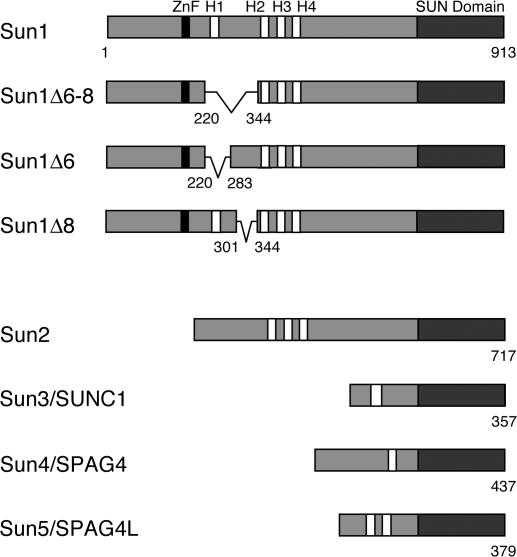
Mammalian SUN proteins are encoded by at least five genes (Fig. 1). Of these, only Sun1 and 2 are widely expressed in somatic cells (Hodzic et al., 2004; Crisp et al., 2006). Sun3 (Crisp et al., 2006; Haque et al., 2006; Tzur et al., 2006), Sun4 (SPAG4; Shao et al., 1999; Hasan et al., 2006), and Sun5 (SPAG4Like; unpublished data) seem to be restricted largely to the testis. Each of the SUN proteins conforms to the same basic structure featuring an N-terminal domain followed by a block of hydrophobic amino acid residues, likely representing a TM domain, and a C-terminal SUN domain. The relationships between these proteins are displayed in Fig. 1. In the case of Sun1, the largest of the mammalian SUN proteins, the nucleoplasmic N-terminal domain is composed of 350–400 amino acid residues (Crisp et al., 2006). All of the sequence variants of Sun1 that arise through alternative splicing involve changes in this segment of the molecule (Fig. 1).
Figure 1.
The mammalian SUN protein family. Sun1 features four hydrophobic sequences, H1–H4, each of roughly 20 amino acid residues. Its membrane-spanning domain is contained within the H2–H4 region. The Sun1 N terminus, including H1, is nucleoplasmic. The C-terminal SUN domain resides in the PNS. Murine but not primate Sun1 contains a predicted C2H2 zinc finger. Several splice isoforms of mouse Sun1 have been identified that feature the loss of sequences encoded by exons 6–8, including H1. Corresponding GenBank/EMBL/DDBJ accession nos. are BAB29445 (Sun1Δ6–8), AAT90501 (Sun1Δ6), and BAC29339 (Sun1Δ8). Four other mammalian SUN proteins are known. Sun2 is ubiquitously expressed and localizes to the INM. Sun3 (SUNC1), Sun4 (SPAG4), and Sun5 (SPAG4L; GenBank/EMBL/DDBJ accession no. NP_542406) appear to be expressed primarily in testis (unpublished data). When expressed in HeLa cells, Sun3 localizes to the NE (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200704108/DC1), whereas Sun4 (Hasan et al., 2006) and Sun5 (unpublished data) localize primarily to the ER.
A prominent feature of Sun1 is the presence of four hydrophobic sequences, H1–H4, any one of which could potentially function as a TM domain. Previously, we showed that H1 at least does not span the INM (Crisp et al., 2006). This conclusion was based on a naturally occurring splice isoform that is missing sequences encoded by exons 6, 7, and 8 (Sun1Δ221–344; Δexons 6–8). This isoform lacks H1 yet displays appropriate NE localization and has the same topology within the INM as full-length Sun1. Nevertheless, as will be expanded upon, although not a TM domain, H1 may still contribute to membrane association. Mouse Sun1 also contains a predicted C2H2 zinc finger. Because this is absent from primate Sun1, its significance remains unclear.
The C-terminal region of Sun1, like that of Sun2, consists of roughly 450 amino acid residues and resides within the PNS. It contains a membrane-proximal predicted coiled coil and a conserved distal SUN domain. The junction between these two features contains the KASH-binding site and is therefore essential for the tethering of ONM nesprins.
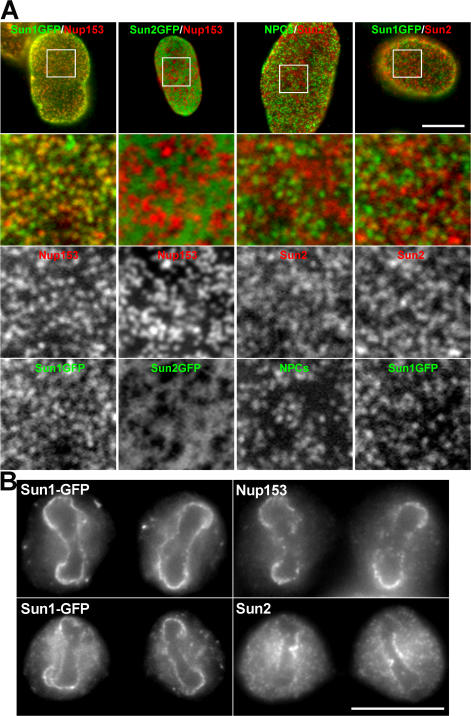
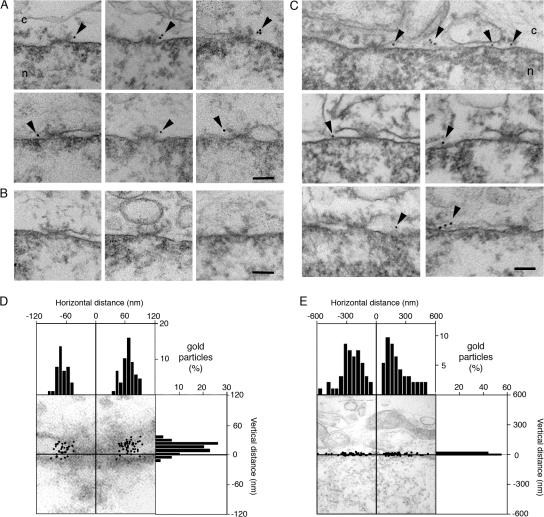
Although Sun1 and 2 have obvious similarities in terms of domain organization and topology, we noticed differences in their localizations within the NEs of multiple cell types. In particular, the distribution of Sun1 appears more punctate than that of Sun2 (Fig. 2 A). This was evident by both immunofluorescence microscopy using antibodies against the endogenous proteins as well as by observations on exogenous Sun1 or 2 carrying a C-terminal GFP tag. A direct comparison of the two proteins indicates that they are largely segregated within the plane of the NE. Double-label experiments using an antibody against Nup153, an NPC component, revealed that Sun1 but not Sun2 was concentrated at NPCs. This localization was confirmed by immunoelectron microscopy of HeLa cells expressing either Sun1- or Sun2-GFP (Fig. 3, A–C). Quantitative analysis revealed that in the case of Sun1, gold particles were all clustered within 120 nm of the NPC eightfold axis with a peak at 66 ± 20 nm (±SD; Fig. 3 D). Sun2 displayed a far broader distribution with a median distance from the NPC eightfold axis of 240 ± 120 nm (Fig. 3 E). Only 8% of gold particles were within 120 nm, and none were within 66 nm. Of necessity, the scale in Fig. 3 E is five times that in Fig. 3 D.
Figure 2.
Sun1 and 2 are segregated within the plane of the NE. Immunofluorescence microscopy of HeLa cells stably expressing mouse Sun1-GFP or human Sun2-GFP using antinucleoporin and anti-SUN protein antibodies. (A) Images of the nuclear surface reveal Sun1-GFP colocalization with Nup153. In contrast, the more diffuse Sun2-GFP is found in NPC-free regions. Endogenous Sun2 displays no colocalization with either NPCs labeled with the antinucleoporin antibody QE5 or with Sun1-GFP. The boxed areas in the top panels are magnified in the bottom three panels. (B) At late anaphase to early telophase, reforming nuclei exhibit a distinct distribution of NPC and NE components. Sun1-GFP localizes with Nup153 at the lateral margins of the mass of newly segregated chromatids and is absent from the Sun2-positive core region. These data suggest that Sun1 is closely associated with NPCs, a pattern that is established early in NE formation. Bars (A), 5 μm; (B) 4 μm.
Figure 3.
Sun1 but not Sun2 is closely associated with NPCs as revealed by immunoelectron microscopy. (A) Views of NPC cross sections from tetracycline-induced HeLa cells expressing Sun1-GFP that reveal anti-GFP–associated gold particles close to NPCs. (B) Sections of uninduced cells display little or no gold labeling. (C) Images of NE cross sections from HeLa cells expressing Sun2-GFP reveal gold particles in the PNS but with no preferential association with NPCs. c, cytoplasm; n, nucleus. Arrowheads indicate gold particles. (D and E) Quantitative analysis of the distribution of gold particles from NE cross sections of HeLa cells expressing Sun1- (D) or Sun2-GFP (E). The position of gold particles, which is defined by horizontal distance (from the NPC eightfold axis) and vertical distance (from the central plane of the NE), was measured in cross sectioned NEs (as in A and C) and plotted in a single dot graphic. Micrographs are provided as a visual reference for the position of the gold particles. Histograms for the distribution of gold particles for horizontal and vertical distances are shown on adjacent panels. For both Sun1 and 2, gold particles were scored within 200- and 600-nm windows on either side of each NPC. The larger, more conservative window size still reveals an absolute eightfold higher labeling density of Sun1 over Sun2 within 120 nm of the NPC center, with a peak density at 66 nm. A total of 88 and 92 gold particles were scored for D and E, respectively. Because of the far broader distribution of Sun2-GFP, the scale in E is five times that in D. Bars, 100 nm.
The differential localization of Sun1 versus Sun2 is reflected in their behavior during mitosis. After NE breakdown, both proteins are dispersed throughout the cytoplasm, most likely in ER membranes. During late anaphase/early telophase, as NE reassembly commences, the resegregation of Sun1 and 2 occurs. Sun2 concentrates at a region of each newly separated chromatin mass adjacent to one of the spindle poles. This core region (Fig. 2 B) of chromatin is typically deficient in NPC reformation. The behavior of Sun2 mirrors that of another INM protein, emerin, which also concentrates at the chromatin core at roughly the same time (Maeshima et al., 2006). In contrast, Sun1 tends to be excluded from the core region and instead concentrates on the lateral margins of the chromatid masses where NPC assembly is initiated.
Localization of Sun1 to the INM involves determinants in both N- and C-terminal domains (Padmakumar et al., 2005; Crisp et al., 2006; Haque et al., 2006; Hasan et al., 2006; Wang et al., 2006), although their relative contribution to localization and role in NPC association remain unknown. In addition, although previous studies (Crisp et al., 2006; Haque et al., 2006) have demonstrated that the TM domains of Sun1 are contained within the H2–H4 region, they provide an ambiguous view of the targeting properties of this segment of the molecule. We and others (Padmakumar et al., 2005; Crisp et al., 2006) had proposed that Sun1 might be a multispanning protein with three TM sequences corresponding to H2, H3, and H4. However, direct evidence to support such a model is lacking.
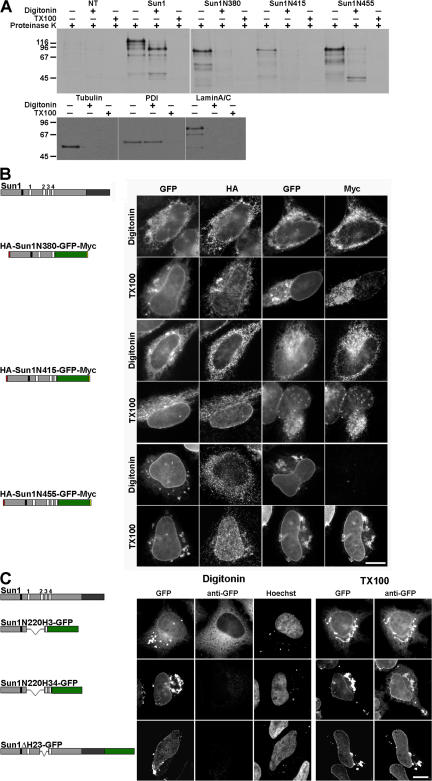
To better define Sun1 topology, we prepared a series of mutants containing different combinations of the H2, H3, and H4 hydrophobic sequences, all of which were found to confer some degree of membrane association. These mutants contained all or part of the N-terminal domain (residues 1–355) followed by one or more hydrophobic sequences and terminating with GFP (Fig. 4). At the N and C termini of certain chimeras, we placed HA and myc epitope tags, respectively. These constructs were expressed by transfection in HeLa cells, which were subsequently treated with low concentrations of digitonin. Under these conditions, the plasma membrane is permeabilized, but the ER and nuclear membranes remain intact. The permeabilized cells were then incubated with proteinase K. During the course of this incubation, Western blot analysis revealed that cytoplasmic (tubulin) and nuclear (lamin A/C and Nup153) proteins were degraded (proteinase K may enter the nucleus by degrading NPC proteins), whereas ER lumenal and PNS proteins such as protein disulfide isomerase (PDI) were protected (Fig. 4 A). Triton X-100 (TX-100) permeabilization permitted the digestion of all of these proteins. In the case of the Sun1 chimeras, we determined the latency of the HA and myc epitope tags by both SDS-PAGE analysis of immunoprecipitated, radiolabeled proteins and immunofluorescence microscopy (the latter on permeabilized but not proteinase-treated cells; Fig. 4, A and B, respectively). In some experiments, we used specific antibodies to monitor the latency of the GFP moiety (Fig. 4 C).
Figure 4.
Sun1 contains a single TM domain. (A) Three Sun1 mutants containing the N-terminal domain followed by H2, H2–H3, and H2–H4 were tagged with HA at the N terminus and GFP followed by a myc epitope at the C terminus. HeLa cells transfected (or not transfected [NT]) with these constructs were labeled with [35S]Met/Cys, permeabilized with digitonin, and incubated with proteinase K. After SDS lysis, immunoprecipitation with anti-myc, and SDS-PAGE analysis, only Sun1N455 retained a protected fragment of the predicted size for the GFP (∼30 kD). Permeabilization with TX-100 resulted in complete protein degradation. Nontransfected cells served as a negative control, whereas Sun1-GFP provided a positive control, with a 65–70-kD protected fragment. Western blot analysis was used to confirm the effectiveness of the digitonin permeabilization. Tubulin and lamins A/C were degraded after either digitonin or TX-100 permeabilization. In contrast, the ER lumenal protein PDI remained intact after digitonin permeabilization but was degraded after TX-100 treatment. (B) To further establish the orientation of these Sun1 constructs, 24 h after transfection, the HeLa cells were fixed and permeabilized with either digitonin or TX-100. Analyses focused on cells expressing sufficiently high levels of recombinant protein such that GFP fluorescence could be observed in both the NE and ER/cytoplasmic membranes. With digitonin permeabilization, both myc and HA epitopes were readily detected for HA-Sun1N380-GFP-myc and HA-Sun1N415-GFP-myc. In contrast, the HA but not C-terminal myc epitope tag was accessible for HA-Sun1N455-GFP-myc. In all cases, both myc and HA tags were accessible after TX-100 permeabilization. (C) Three more C-terminal GFP-tagged Sun1 constructs containing the first 220 residues of Sun1 fused to H3 or H3–H4 as well as full-length Sun1 lacking H2–H3 (Sun1N220H3-GFP, Sun1N220H34-GFP, and Sun1ΔH23-GFP, respectively) were permeabilized as described in A and labeled with anti-GFP antibodies. With digitonin permeabilization, the presence of the H4 domain rendered the GFP moiety inaccessible to antibody. Collectively, these results indicate that H4 serves as Sun1's sole TM domain. Bars, 4 μm.
The results reveal that the N-terminal HA tag is always exposed to the cytoplasm or nucleoplasm. In contrast, the myc tag or GFP becomes latent (i.e., it resides within the ER lumen/PNS) whenever the H4 sequence is present within the chimera. No combination of H2 and H3 (either singly or together) would confer such latency. Conversely, neither H2 nor H3 could affect the orientation of H4 and, therefore, the latency of the myc tag or GFP. The only reasonable conclusion is that although H2 and, to a lesser extent, H3 may confer membrane association (Fig. 4 C), they do not cross the bilayer. Therefore, rather than being a multispanning protein as previously suggested, Sun1 would appear to be a type II membrane protein with a single TM domain represented by H4 (see Fig. 9).
Figure 9.
Sun1 topology and interactions. (A) Sun1 is envisaged as forming homodimers via interactions involving the membrane-proximal coiled coil within its C-terminal lumenal domain. Nucleoplasmic domain interactions may also contribute to homodimer formation. Sun1 functions as a tether for ONM nesprin proteins. Nesprins 1 and 2 provide links to the actin cytoskeleton, whereas nesprin 3 binds plectin, a versatile cytolinker. (B) Sun1 is associated with NPCs, and, in addition, its nucleoplasmic domain displays preferential binding to newly synthesized pre–lamin A. Therefore, Sun1 may provide a link between NPCs and the A-type lamins. In this way, the Sun1-mediated nucleation of A-type lamina assembly may occur at NPCs.
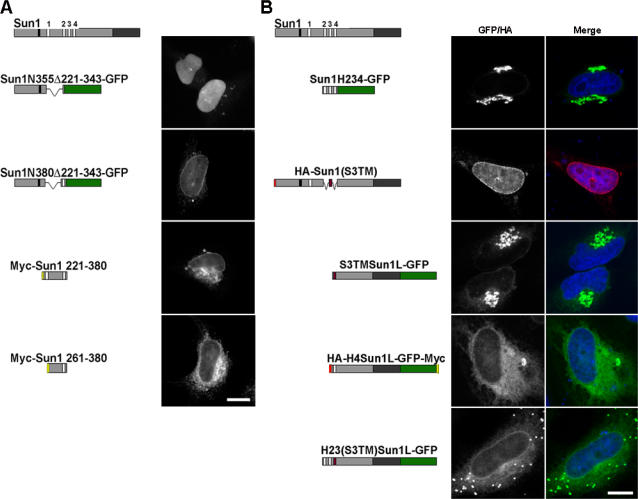
With a better understanding of Sun1 topology, we next wished to identify NE and NPC retention domains. To this end, we generated an extensive family of chimeric Sun1 proteins. Because H4 appears to represent the sole TM domain, we sought to clarify the role of the other hydrophobic sequences. Sun1N355, which contains the H1 sequence but lacks H2 and H3, was found to localize to the NE (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200704108/DC1; Haque et al., 2006). This is in contrast to the nucleoplasmic localization of Sun1N355Δ221–343 (this corresponds to the exon 6–8 deletion; Fig. 5 A) or of Sun1N220 (Crisp et al., 2006), both of which lack any hydrophobic motif. When Sun1N355Δ221–343 was extended to include the H2 domain (Sun1N380Δ221–343), NE association was rescued (Fig. 5 A). Taking all of this data together, we can conclude that H1, H2, and, to a certain extent, H3 are each sufficient to confer membrane association. However, because the Sun1N220 region itself will accumulate readily within the nucleoplasm (Crisp et al., 2006), these experiments do not reveal whether any of the hydrophobic sequences themselves have intrinsic INM-targeting activity.
Figure 5.
The Sun1 nucleoplasmic domain has overlapping NE localization motifs. Immunofluorescence microscopy of HeLa cells transiently expressing Sun1 deletion constructs. (A) Sun1N355, a region previously identified as conferring NE localization, was further mutated based on a natural splice isoform of Sun1 (Sun1N355Δ221–343-GFP). This protein, which lacks the H1 domain, is predominantly nucleoplasmic. Upon the inclusion of H2 (Sun1N380Δ221–343-GFP), it becomes NE associated. Deletion of the N-terminal 220 residues from the NE-localized Sun1N380 (myc-Sun1-221–380) fails to eliminate NE localization. However, extension of the deletion to include H1 (myc-Sun1-261–380) results in an ER localization with little concentration in the NE. (B) To examine the role of H2–H4 in the localization of Sun1, H234-GFP was expressed in HeLa cells, where it concentrates in the Golgi apparatus. This region (H2–H4) of Sun1 was replaced with the TM domain of Sun3 (HA-Sun1(S3TM)), resulting in localization that was indistinguishable from full-length Sun1. However, upon deletion of the nucleoplasmic domain (S3TMSun1L-GFP), the protein was found largely in the Golgi apparatus. Similarly, deletion of the H2 and H3 domain from H234Sun1L (H4Sun1L-GFP) led to the loss of NE association with a predominantly ER localization. NE localization of Golgi-associated S3TMSun1L-GFP could be partially rescued by addition of the H2–H3 sequence (H23(S3TM)Sun1L-GFP) to the N terminus. These data identify two overlapping regions of the Sun1 nucleoplasmic domain that are sufficient for NE targeting, H1–H2 and H2–H3. However, the latter is only functional in the context of a TM sequence and the Sun1 lumenal domain. Bars, 5 μm.
To address this question, we next examined the behavior of two additional H2-containing fusion proteins, which are both tagged at the N terminus with the myc epitope (Fig. 5 A). The first of these represented an N-terminal truncation lacking the initial 220 Sun1 residues but containing H1 and H2 (myc-Sun1 221–380), whereas the second was missing H1 in addition to the N-terminal 220 residues (myc-Sun1 261–380). The former localized to the NE and, to a lesser extent, to the peripheral ER. In contrast, the latter was primarily ER associated with little concentration in the NE. The implication of these results is that an NE localization motif is encoded by Sun1 residues 221–380. This region of the molecule must therefore share interactions with other nuclear or NE components.
We next examined whether the H2–H4 region alone has a role in NE targeting. When this sequence was fused to the N terminus of GFP, it localized predominantly to the Golgi apparatus and cell surface with little, if any, associated with the NE (Fig. 5 B). In contrast, a Sun1 N-terminal truncation consisting of the H2–H4 region followed by the Sun1 lumenal domain (H234Sun1L-GFP) localized efficiently to the NE (Fig S1; Padmakumar et al., 2005). However, we already know that a soluble form of the Sun1 lumenal domain that is appropriately localized to the ER lumen and PNS is itself insufficient for NE targeting (SS-HA-Sun1L-KDEL; Crisp et al., 2006). There are at least two explanations for these results. The first is that the lumenal domain does contain targeting information but that it is only functional when the domain is appropriately oriented or tethered to the ER or nuclear membranes. The second is that the H2–H4 hydrophobic region can direct localization to the INM but that this only occurs in the context of the Sun1 lumenal domain. In other words, the Sun1 lumenal domain can modify the behavior of the H2–H4 sequences. What we can rule out, however, is any suggestion that localization of H234Sun1L-GFP to the INM occurs by virtue of oligomerization with endogenous Sun1. Overexpression of H234Sun1L-GFP leads to the displacement of endogenous Sun1 from the NE while itself concentrating in the NE (Padmakumar et al., 2005; unpublished data). Additionally, depletion of Sun1 by RNAi has no effect on H234Sun1L-GFP localization (unpublished data).
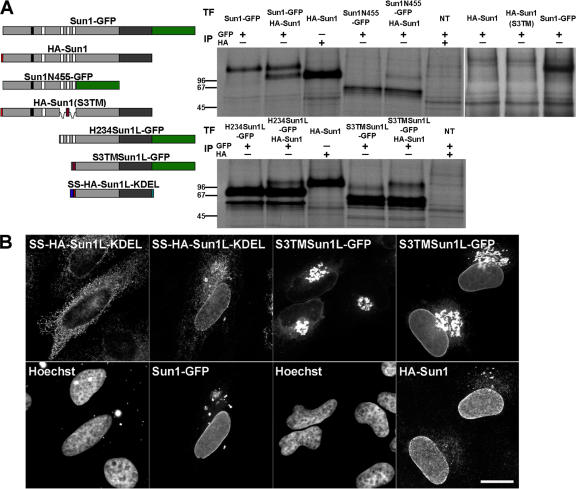
To further address these issues, we replaced the H2–H4 hydrophobic region of both full-length Sun1 and H234Sun1L-GFP with the unrelated TM domain of Sun3 (to yield HA-Sun1(S3TM) and S3TMSun1L-GFP, respectively; Fig. 5 B). As shown in Fig. S2 A (available at http://www.jcb.org/cgi/content/full/jcb.200704108/DC1), when expressed in HeLa cells, HA-tagged Sun3 localizes to the NE but does not associate with NPCs. Sun3 contains a single predicted TM domain that resides between residues 46 and 65. As will become evident below (Fig. 5 B), this sequence contains no intrinsic NE-targeting activity. Translation of Sun3 in vitro in the presence of microsomes confirms that this sequence must function as a TM domain with a type II orientation (Fig. S2 B).
HA-Sun1(S3TM) behaved exactly like full-length Sun1 in that it concentrated in the NE (Fig. 5 B) in association with NPCs (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200704108/DC1). In contrast, S3TMSun1L-GFP displayed little or no NE localization and instead was found in the Golgi apparatus and at the cell surface (Fig. 5 B). Evidently, it is not retained in the nuclear membrane/ER system. Deletion of H2–H3 from H234Sun1L (H4Sun1L-GFP) also resulted in the loss of NE association (Fig. 5 B). The implication, then, is that H2–H3 encodes an NE localization function. If this is the case, attaching H2–H3 to the N terminus of S3TMSun1L-GFP should lead to its accumulation at the NE. Indeed, we do observe a partial restoration of NE localization (Fig. 5 B).
It is evident from these results that although the H2–H3 sequence promotes localization to the NE, its activity is strongly influenced by the Sun1 lumenal domain. This is despite the fact that these regions of Sun1 reside on opposite sides of the INM. A possible explanation for this result is that the targeting activity of H2–H3 may be activated or enhanced by dimerization (or oligomerization), perhaps leading to increased avidity for nuclear or INM-associated binding partners. A prediction here is that the lumenal domain of Sun1 should mediate dimerization (or oligomerization). This is borne out in transfection experiments in which full-length Sun1 was coexpressed in HeLa cells with a variety of epitope-tagged Sun1 deletion mutants (Fig. 6 A). Immunoprecipitation analyses resulted in the efficient coprecipitation of full-length and mutant Sun1 only when the mutant form contained an intact lumenal domain. Further compelling evidence for lumenal domain–mediated oligomerization was provided by immunofluorescence observations of SS-HA-Sun1L-KDEL and S3TMSun1L. As described previously (Fig. 6 B), neither of these chimeric proteins concentrates to any great extent in the NE. However, the overexpression of full-length Sun1 will recruit both of these proteins to the NE. Collectively, these data clearly demonstrate that Sun1 homooligomerizes via lumenal domain interactions, most likely involving the predicted membrane proximal coiled coil.
Figure 6.
Sun1 forms homotypic oligomers in vivo. (A) To define the oligomerization state of Sun, HA-Sun1 was cotransfected into HeLa cells with Sun1-GFP, Sun1N455-GFP, H234Sun1L-GFP, and S3TMSun1L-GFP. All samples were labeled with [35S]Met/Cys and immunoprecipitated with anti-GFP antibodies. Full-length HA-Sun1 was most efficiently coprecipitated with Sun1-GFP followed by the lumenal then nucleoplasmic domain containing fusion proteins. NT, nontransfected. (B) In vivo evidence of Sun1 oligomerization mediated by the lumenal domain was provided by the cotransfection of Sun1-GFP or HA-Sun1 with SS-HA-Sun1L-KDEL or S3TMSun1L-GFP, respectively. Alone, these proteins localize predominantly to the ER or Golgi apparatus, respectively. Coexpression of full-length Sun1 recruited both proteins to the NE. Thus, Sun1 forms homotypic oligomers that independently involve the lumenal and, to a lesser extent, the nucleoplasmic domains. Bar, 6 μm.
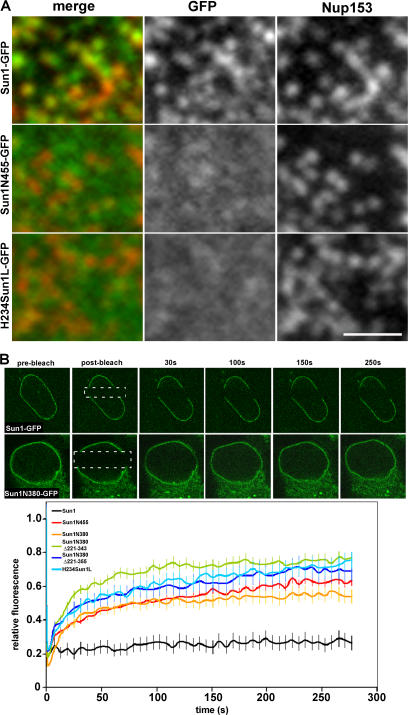
Which Sun1 sequence elements are required for association with NPCs? Analysis of all of the Sun1 constructs that we have prepared revealed that apart from wild-type Sun1, only Sun1Δ221–343 and Sun1(S3TM) associated with NPCs (Figs. 7 A and S3 A). Evidently, association with NPCs does not involve the H1 and H234 hydrophobic sequences acting in concert. To take these analyses further, we prepared a pair of chimeras in which we swapped the Sun1 and 2 lumenal domains. In neither case could we observe NPC association (Fig. S3 B). Instead, both recombinant proteins behaved like Sun2. Evidently, both nucleoplasmic and lumenal domains of Sun1 cooperate in conferring NPC association.
Figure 7.
Sun1 association with NPCs requires both the nucleoplasmic and lumenal domains. (A) To define regions of Sun1 involved in NPC association, Sun1 mutants lacking either the lumenal (Sun1N455-GFP) or nucleoplasmic domains (H234Sun1L-GFP) were transiently expressed in HeLa cells. Neither Sun1 deletion mutant displayed obvious colocalization with Nup153. (B) To further analyze the roles of various domains in Sun1 targeting and retention, FRAP analysis was performed on Sun1, Sun1N455, Sun1N380, Sun1N380Δ221–343, Sun1N380Δ221–355, and H234Sun1L, each bearing a GFP tag at the C terminus. In contrast to full-length Sun1-GFP, which is relatively immobile in the NE, fluorescence recovery occurred for all deletion proteins within a span of ∼1 min. Together, these data indicate that it is the combination of the nucleoplasmic and lumenal domains that stabilizes Sun1 in the NE, potentially involving an association with NPCs. Dotted boxes define the photobleached region. Bar, 1 μm.
So far, we have shown that there are multiple determinants within the Sun1 nucleoplasmic domain that can confer localization to the INM. Hasan et al. (2006) used FRAP analysis to show that wild-type Sun1 is relatively immobile within the INM. We performed similar analyses on a subset of our Sun1 deletion mutants that localize to the NE (Fig. 7 B). In all cases, these mutants display enhanced mobility relative to wild-type Sun1. Even deletion of the lumenal domain, which appears to contain no intrinsic targeting function but does promote oligomerization, leads to increased mobility within the INM. Thus, although Sun1 does contain multiple autonomous features involved in localization, stable localization to the NE requires that all be present. These findings are reminiscent of our conclusion that multiple features within the Sun1 molecule are required for NPC association.
What is the functional relevance of Sun1 association with NPCs? Proteomic studies provide no evidence that Sun1 is an intrinsic component of the NPC (Cronshaw et al., 2002). However, to determine whether Sun1 might contribute to NPC functionality, we examined nuclear transport in HeLa cells that had either been depleted of Sun1 by RNAi or that expressed Sun1 fragments, some of which resulted in a loss of endogenous NE-associated Sun1 (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200704108/DC1). To accomplish this, we took advantage of a GFP fusion protein bearing nuclear import and export signals (NLS-GFP–nuclear export sequence [NES]) and that shuttles between the nucleus and cytoplasm (Stade et al., 1997). We also used a hormone-inducible nuclear import substrate consisting of β-galactosidase fused to the glucocorticoid receptor (grβ; Bastos et al., 1996). Our results indicate that Sun1 has no substantial role in the nuclear transport of proteins, either import or export. Similarly, the distribution of poly A+ RNA revealed by in situ hybridization suggests that Sun1 makes little or no contribution to mRNA export (unpublished data).
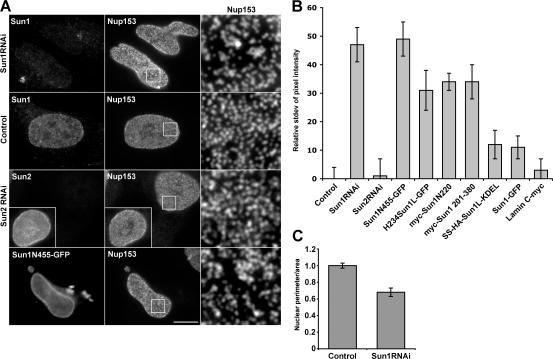
However, Sun1 depletion was not without effect on pore complexes. We noticed that the loss of Sun1 was always associated with an altered distribution of NPCs (Fig. 8 A) as well as altered nuclear shape (Fig. 8 C). In wild-type cells, NPCs tend to be uniformly distributed across the nuclear surface. After Sun1 depletion, NPC aggregates or clusters could be observed leaving NPC-free areas of varying sizes. This effect was Sun1 specific because the depletion of Sun2 left NPC distribution unchanged.
Figure 8.
Perturbation of Sun1 affects NPC distribution. (A) Immunofluorescence microscopy of HeLa cells 48 h after SUN protein depletion by RNAi. Cells were double labeled with antibodies against Sun1 or Sun2 and Nup153. The loss of Sun1 was associated with altered nuclear morphology and changes in the distribution of NPCs. Magnification of the nuclear surface reveals large pore-free tracts between clusters of NPCs. Nontransfected cells (control for Sun1 and insets for Sun2) or Sun2 RNAi had no such effect. The expression of Sun1N455-GFP altered NPC distribution in a manner similar to Sun1 RNAi. Boxed areas are magnified in the right panels. (B) To quantify the observed changes in NPC distribution, the relative SD of binned pixel intensity of anti-Nup153 fluorescence intensity across the projected nuclear surface was calculated. All measurements were standardized relative to the nontransfected control, which was set at 0%. Sun1 RNAi and Sun1N455-GFP were most effective at inducing NPC clustering followed by H234Sun1L-GFP, myc-Sun1N220, and myc-Sun1 220–380. SS-HA-Sun1L-KDEL and Sun1-GFP had a minimal effect on NPC distribution. Lamin C served as a transfection control and induced no obvious NPC clustering. n = 7–18. (C) To quantify the altered nuclear morphology induced by Sun1 RNAi, the ratio of the projected nuclear area to the perimeter was measured. RNAi of Sun1 led to a 35% reduction in this ratio over control cells, which were set to 100%. n = 10–11. Error bars represent SEM. Bar, 5 μm.
This effect of Sun1 depletion on NPC aggregation could be emulated by the overexpression of nucleoplasmic Sun1 deletion mutants in HeLa cells. The expression of these mutants often leads to a diminution in the amount of full-length Sun1 at the NE (and thus at NPCs). A quantitative analysis of NPC aggregation induced by both Sun1 depletion and Sun1 mutant expression is displayed in Fig. 8 B.
Because Sun1 may act as a tether for ONM nesprins, it is possible that NPC aggregation is a function of the loss of nesprins rather than a loss of Sun1. To determine whether this might be the case, we overexpressed a protein consisting of GFP fused to the KASH domain of either nesprin 1 or 2 (GFP-KASH1 or 2) in HeLa cells. Overexpression of GFP-KASH1 or 2 leads to the displacement of nesprins 1 and 2 from the NE (Zhang et al., 2007). Treatment of cells in this way was found to have no discernible effect on NPC distribution (unpublished data). These data suggest that Sun1 has a nesprin-independent role in maintenance of the uniform distribution of NPCs across the NE.
Discussion
Sun1 and 2 are of a pair of ubiquitous INM proteins that tether nesprins within the ONM. Nesp1G and nesp2G contain N-terminal actin-binding domains (Zhen et al., 2002; Padmakumar et al., 2004), whereas nesprin 3 binds plectin, a versatile cytolinker (Wilhelmsen et al., 2005). Thus, the SUN proteins represent links in a molecular chain that connects elements of the cytoskeleton to components within the nucleus. We have previously referred to translumenal Sun–nesprin pairs as LINC complexes (Crisp et al., 2006). Multiple LINC complex isoforms likely exist given the apparent redundancy of Sun1 and 2 in tethering nesprins. In addition, we can identify at least four or five splice isoforms of Sun1 alone, further increasing the LINC complex repertoire. The nesprins themselves (including nesprin 3) are also represented by dozens of splice isoforms. Aside from nesp1G and nesp2G, the number of these that may be tethered by Sun proteins at the ONM remains unknown.
Previous studies indicated that KASH domain proteins play an important role in nuclear positioning in certain cell types (Mosley-Bishop et al., 1999; Starr et al., 2001; Starr and Han, 2002; Malone et al., 2003; Grady et al., 2005; Yu et al., 2006). However, the existence of links spanning the NE have far broader implications than mere nuclear location and present us with a mode (or modes) of nucleocytoplasmic coupling that may bypass NPCs. This notion is highlighted by biomechanical studies on Lmna-null fibroblasts, which exhibit impaired mechanotransduction and decreased viability under mechanical strain (Broers et al., 2004; Lammerding et al., 2004, 2005, 2006). Induction of the mechanosensitive genes iex-1 and egr-1 is attenuated, as is nuclear factor κB–regulated transcription in response to either cytokine or mechanical stimulation. Although nuclei in Lmna-null cells are both mechanically fragile and highly deformable, a surprising finding of Lammerding et al. (2004) is that these cells also feature reduced cytoplasmic resilience. Given that both Sun1 and 2 interact with A-type lamins, it is possible that the LINC complex might mediate mechanotransduction and the lamin-dependent changes in cytoplasmic organization.
Retention of Sun1 and 2 in the INM is independent of A-type lamins in some cell types (Padmakumar et al., 2005; Crisp et al., 2006; Haque et al., 2006; Hasan et al., 2006). This implies that there have to be other nuclear or NE components that interact with and retain the SUN proteins. Logically, based on our studies here, there have to be at least two discrete regions within Sun1 that are sufficient for INM localization. Evidence for this can be seen in the differential effect of exogenous Sun1 and 2 on each other. Sun2 will not substantially displace Sun1 in HeLa cells. However, Sun1 can efficiently displace Sun2 from the INM (Crisp et al., 2006), presumably by competition for a common binding partner or anchor . Therefore, Sun1 likely has an additional binding partner that is not shared with Sun2. This is perhaps most obvious when considering that both of these proteins are segregated within the plane of the NE. Although Sun2 predominates in NPC-free regions, Sun1 is concentrated in the vicinity of NPCs, possibly forming a halo around each NPC. The mechanisms of interaction with NPCs remain unknown. However, it clearly requires contributions from both the nucleoplasmic and lumenal domains.
Our analyses suggest that there are at least two separate INM-targeting regions within the Sun1 nucleoplasmic domain. The first lies between residues 1–260 and includes the H1 hydrophobic sequence. The second is immediately downstream of the H1 sequence and includes the H2 and H3 hydrophobic sequences. The bulk of this second targeting region is absent in the Sun1Δ221–343 (i.e., the Δexon 6–8) splice isoform, although the H2 and H3 sequences are retained. Because Sun1Δ221–343 still colocalizes with NPCs, the bulk of this second targeting region cannot have an essential role in NPC association. The same is also true of the entire H234 region, which can be substituted by the Sun3 TM domain without affecting NPC association.
Although the H2–H3 hydrophobic sequence exhibits INM-targeting activity, it is only functional in the context of molecules containing the lumenal domain. The lumenal domain has no intrinsic targeting properties but does promote oligomerization, most likely based upon coiled-coil homodimers. We would suggest that manifestation of the INM localization function of H2–H3 requires dimerization/oligomerization, perhaps leading to increased avidity for an NE or nuclear binding partner.
The Sun1 TM domain is contained within the region of the molecule defined by the H2–H4 hydrophobic sequences. We and others had suggested that these might represent three TM domains (Padmakumar et al., 2005; Crisp et al., 2006). Our current studies suggest that H2 and H3 do not, in fact, span the INM, leaving H4 as the only TM sequence within the Sun1 molecule (Fig. 9 A). This view is reinforced by the existence of an apparent human Sun1 splice isoform (GenBank/EMBL/DDBJ accession no. NM_25154) lacking sequences encoded by exons 6–9 and missing H2. Our conclusion is that Sun1 has the topology of a type II membrane protein.
Although Sun1 has only a single membrane-spanning domain (H4), the three other hydrophobic sequences, H1, H2, and H3, can confer membrane association. Nucleoplasmic domain constructs that contain at least one of the three all become associated with the INM, whereas their absence leads to nucleoplasmic localization. It remains unclear whether these hydrophobic sequences interact directly with the INM lipid bilayer or whether association is mediated by other INM proteins. The former would appear more likely because regardless of expression level, H1-, H2-, or H3-containing proteins always appear membrane associated. The interaction of extended hydrophobic sequences such as H2–H3 with the lipid bilayer is not without precedent. For instance, the tubular ER protein reticulon 4 contains a 30–40-residue hydrophobic sequence that forms a hairpin, which dips into the cytoplasmic face of the ER lipid bilayer without crossing it (Voeltz et al., 2006).
The segregation of Sun1 and 2 within the plane of the NE and the association of Sun1 with NPCs is quite striking. Could Sun1 be an NPC component? The complement of mammalian NPC subunits identified by Cronshaw et al. (2002) using proteomic approaches does not include Sun1. However, the same is also true of the authentic vertebrate NPC membrane protein Ndc1 (Stavru et al., 2006). Perhaps Sun1's additional associations with the nuclear lamina and possibly chromatin limits its coextraction with NPC proteins. Regardless, we can find no evidence that Sun1 contributes to nucleocytoplasmic transport, and, consequently, we feel that Sun1 is unlikely to represent an intrinsic NPC component. Instead, a more reasonable scenario is that Sun1 is associated with the NPC periphery and may define a novel microdomain within the nuclear membranes, which, in turn, could blur the boundary between NPCs and the bulk of the NE (Fig. 9 B). The presence of Sun1 and, by implication, nesprins at NPCs could provide a basis for older ultrastructural observations that cytoskeletal elements, particularly intermediate filaments, frequently seem to contact pore complexes (Goldman et al., 1985).
The distribution of NPCs across the NE is not random. Rather, they are arrayed in a uniform (although not regular) fashion that is constrained by a minimum NPC separation (Maul, 1977). We have observed that the depletion of Sun1 (but not Sun2) or overexpression of truncated forms of Sun1 lead to the formation of NPC aggregates or clusters. This suggests that Sun1 has a role in the maintenance of uniform NPC distribution across the nuclear surface. In mammalian cells, NPCs are largely immobile and maintain their relative positions over many hours (Daigle et al., 2001; Rabut et al., 2004). The implication is that NPC clusters in Sun1-depleted cells may arise during de novo NPC assembly as well as during postmitotic reassembly.
A-type lamins are also determinants of NPC distribution because Lmna-null mouse embryonic fibroblasts (MEFs) frequently feature clustered or aggregated NPCs (Sullivan et al., 1999). Furthermore, Maeshima et al. (2006) have shown that A-type lamins strongly influence the distribution of NPCs and pore-free regions of the NE. It is important to bear in mind, however, that cells that normally lack A-type lamins (early embryonic cells, for instance) do not display obviously clustered NPCs. It follows that there must be additional mechanisms to define NPC distribution that predominate in certain cell types. Such mechanisms might potentially involve B-type lamins (Maeshima et al., 2006).
Because Sun1 interacts with lamin A via the N220 region of its N-terminal domain (Crisp et al., 2006), it could function as an adaptor between the nuclear lamina and the NPC (Fig. 9). Of more significance is our observation that Sun1 has a preference for farnesylated pre–lamin A. Given that pre–lamin A exists only transiently in normal cells, this raises the possibility that Sun1 might function in the targeting and assembly of newly synthesized lamin A at the nuclear face of the INM. If this is the case, given the localization of Sun1, NPCs could actually function as nucleation sites of A-type lamina assembly. Ultimately, this may help define the distribution of NPCs. Our next goal will be to test this notion by determining whether there is a spatial relationship between A-type lamina assembly and NPCs. Regardless of the outcome of these studies, it is becoming increasingly clear that there are complex networks of interactions at the nuclear periphery involving NPCs, INM and ONM proteins, and nuclear lamins. These interactions appear to define not only the organization of the NE but also determine cytoskeletal mechanics and perhaps mediate signaling between the nucleus and cytoplasm.
Materials and methods
Cell culture and transfections
HeLa cells and MEFs, both Lmna+/+ and Lmna−/− (Sullivan et al., 1999), were maintained in 7.5% CO2 and at 37°C in DME (Invitrogen) plus 10% FBS (Hyclone), 10% penicillin/streptomycin (Invitrogen), and 2 mM glutamine. Plasmid DNA was introduced into HeLa cells and MEFs by using the Polyfect reagent as described previously (Crisp et al., 2006) or with the LipofectAMINE 2000 reagent (Invitrogen). To transfect a 3.5-cm2 tissue culture dish of cells with LipofectAMINE 2000, 6 μl of transfection reagent or 2 μg of plasmid DNA were each added to separate 100-μl vol of Optimem (Invitrogen) and were combined and incubated at RT for 20 min. Subsequently, the cell medium was replaced with serum-free DME and the 200-μl transfection mix added dropwise and was incubated at 37°C for 1 h, after which time the medium was replaced with DME/10% FCS. Cells were analyzed 1–2 d later.
Generation of tetracycline-inducible stable cell lines
A HeLa cell line stably expressing a tetracycline repressor protein from a pcDNA6/TR plasmid (TRex-HeLa; Invitrogen) was transiently transfected with pcDNA4/TO plasmid (Invitrogen) containing murine Sun1GFP or human Sun2GFP. After transfection, cells were selected with 200 μg/ml zeocin, and stably expressing subclones were isolated. Stable cells were analyzed 24 h after the addition of 1 μg/ml tetracycline.
Antibodies
The following antibodies were used in this study: the monoclonal antibodies 9E10 and 12CA5 against the myc, HA, and GFP epitope tags were obtained from the American Type Culture Collection, Covance, and Abcam, respectively. Rabbit antibodies against the same epitopes were obtained from Abcam. Rabbit antibodies against Sun1 and 2 were previously described (Hodzic et al., 2004). Mouse monoclonal anti-nup153 (clone SA1) and anti-nucleoporin (clone QE5) were described previously (Pante et al., 1994; Bodoor et al., 1999). Rabbit anti-emerin was a gift from G. Morris (Robert Jones and Agnes Hunt Orthopaedic Hospital, Oswestry, UK). Mouse anti–β-galactosidase was purchased from Promega. Mouse anti-PDI and antitubulin were obtained from Abcam. Goat anti– lamin A/C was obtained from Santa Cruz Biotechnology, Inc. Secondary antibodies conjugated with AlexaFluor dyes were obtained from Invitrogen. Peroxidase-conjugated secondary antibodies were obtained from Biosource International.
Immunofluorescence microscopy
For immunofluorescence microscopy, cells were grown on glass coverslips and fixed in 3% formaldehyde (prepared in PBS from PFA powder) for 10 min followed by a 5-min permeabilization with 0.2% TX-100. Cells labeled with anti-Sun1 were fixed for 6 min in 3% formaldehyde and permeabilized for 15 min in 0.4% TX-100 in PBS. The cells were then labeled with the appropriate antibodies plus the DNA-specific Hoechst dye 33258. For experiments involving selective permeabilization, the cells were first fixed in 3% formaldehyde. This was followed by permeabilization in 0.001% digitonin in PBS on ice for 15 min (Adam et al., 1990). The cells were then labeled with appropriate primary and secondary antibodies. Specimens were observed using a fluorescence microscope (DMRB; Leica). Images were collected using a CCD camera (CoolSNAP HQ; Roper Scientific) linked to a computer (G4; Macintosh) running IPLab Spectrum software (Scanalytics). Image quantification was performed using IPLab software.
FRAP analysis
FRAP experiments were performed on a confocal microscope (LSM 510; Carl Zeiss MicroImaging, Inc.) with a 63× 1.4 NA oil objective (Carl Zeiss MicroImaging, Inc.). GFP was excited with the 488-nm line of an Ar laser, and GFP emission was monitored using a 505-nm longpath filter. Cells were maintained at 37°C using an incubator (ASI Air Stream; Nevtek). In transfected cells, a rectangular region typically 2–4 μm in height was bleached in three iterations using a 488-nm laser line at 100% laser power. Cells were monitored at 5-s intervals for up to 300 s. Data were normalized as described previously (Dundr et al., 2002; Phair et al., 2004) to take into account bleaching during the imaging phase. Recovery values are means ± SD from at least five cells from at least two independent experiments.
Immunoelectron microscopy
Immunoelectron microscopy of Sun1 was performed by preembedding and labeling of the tetracycline-inducible cell line expressing Sun1-GFP. After fluorescence examination to verify GFP expression, cells were trypsinized and pelleted by centrifugation. The pellet was permeabilized with 0.1% TX-100 for 1 min in PBS and fixed with 3% PFA in PBS for 10 min followed by three washes with PBS. The fixed cells were then incubated with 2% BSA in PBS for 10 min followed by incubation with the primary polyclonal anti-GFP antibody ab290 (Abcam) for 1 h. After washing three times with 0.1% BSA in PBS, the cells were incubated with a secondary anti–rabbit IgG antibody conjugated to 10-nm gold particles (Ted Pella Inc.) for 1 h followed by washing three times in PBS. Cells were then fixed and prepared for embedding/thin section electron microcopy (Rollenhagen et al., 2003).
In situ proteinase K digestions
HeLa cells were transfected in triplicate for each construct. After transfection (24 h), the cells were incubated in Met/Cys-free media for 45 min followed by incubation in medium containing 50 μCi [35S]Met/Cys (MP Biomedicals) for 1 h. After two rinses with ice-cold PBS, one well was incubated in 4 μg/ml proteinase K (Sigma-Aldrich) in KHM buffer (110 mM KOAc, 20 mM Hepes, pH 7.4, and 2 mM MgCl2) for 45 min. Another well was permeabilized with 24 μM of ice-cold digitonin in KHM for 15 min followed by 4 μg/ml proteinase K digestion in KHM for 45 min. The third well was incubated with 4 μg/ml proteinase K for 45 min in 0.5% TX-100/KHM. Subsequently, PMSF was added to all wells to a final concentration of 40 μg/ml. The first two wells were gently washed in KHM buffer with 40 μg/ml PMSF again to remove excess proteinase K. Cells were lysed in 0.4% SDS, 2% TX-100, 400 mM NaCl, 50 mM Tris-HCl, pH 7.4, 40 μg/ml PMSF, 1 mM DTT, plus 2 μg/ml pepstatin A and 1 μg/ml leupeptin and passed through a 23-gauge needle five times before centrifugation for 10 min at 16,000 g. Soluble proteins were immunoprecipitated with rabbit anti-myc or GFP by protein A–Sepharose. After three washes, proteins were incubated with sample buffer before separation by SDS-PAGE. Gels were stained with Coomassie brilliant blue and incubated with Amplify (GE Healthcare) for 20 min before drying. Autoradiographs were obtained from dried gels. In parallel, a nontransfected cell lysate was analyzed by Western blotting to validate the permeabilization and digestion conditions.
In vitro translations
In vitro translations and proteinase K digestions were performed as described previously (Crisp et al., 2006).
Nuclear transport assays
To observe nuclear export, an NES-GFP-NLS (NES-MGNELALKLAGLDI and NLS-PKKKRKV) construct was transfected into HeLa cells either alone or with other expression vectors. CRM1-dependent nuclear export was inhibited by a 2-h incubation with 10 ng/ml leptomycin B. To assay nuclear import, HeLa cells were transfected with a glucocorticoid receptor β-galactosidase fusion protein (grβ) 24 h before a 30-min incubation with 10 μg/ml dexamethasone to induce nuclear import of the fusion protein (Bastos et al., 1996).
Plasmids
Full-length murine Sun1 and human Sun2 were cloned as previously described (Hodzic et al., 2004; Crisp et al., 2006) and were used as a template for PCR mutagenesis to generate the Sun1 constructs described in this study. Sun1- and Sun2-GFP were cloned into the tetracycline-responsive pcDNA4/TO (Invitrogen). All other GFP-tagged Sun1 constructs were cloned into pEGFP-N1 (BD Biosciences). HA-Sun1, HA-Sun1(Sun3TM), HA-Sun1(1–221), myc-Sun1(221–380), and myc-Sun1(261–380) were all generated in pcDNA3.1−. NES-GFP-NLS was created by PCR mutagenesis and inserted into pEGFP-N1. Sun2-RNAi plasmids were obtained from Open Biosystems. Sun1 RNAi was accomplished by cloning the sequences 5′-GATCCGACCGGGATGGTGGACTTTCTCAAGAGAAAAGTCCACCATCCCGGTCTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGACCGGGATGGTGGACTTTTCTCTTGAGAAAGTCCACCATCCCGGTCG-3′ into pSilencer 3.1-H1 neo (Ambion), which were derived from the open reading frame of human Sun1 using the pSilencer expression vectors insert design tool (Ambion).
Online supplemental material
Fig. S1 shows that large regions of Sun1 are sufficient for NE targeting. Fig. S2 shows that human Sun3 is a type II membrane protein that localizes to the NE but not NPCs when expressed in HeLa cells. Fig. S3 shows the NPC association of various Sun1 isoforms and chimeras. Fig. S4 shows that all Sun1 mutants containing the H4 TM domain exhibit the topology of a type II membrane protein. Fig. S5 shows that Sun1 does not contribute to the functionality of the NPC. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704108/DC1.
Supplementary Material
Acknowledgments
We would like to thank Dr. David Wilson (Department of Mathematics, University of Florida, Gainesville, FL) for advice on the quantification of NPC clustering and Dr. Manfred Lohka for insightful discussions at the Southern Alberta Nuclear Envelope meeting.
This work was supported by grants from the National Institutes of Health to B. Burke and grants from the Muscular Dystrophy Association to D. Hodzic.
Abbreviations used in this paper: INM, inner nuclear membrane; LINC, linker of nucleoskeleton and cytoskeleton; MEF, mouse embryonic fibroblast; NE, nuclear envelope; NES, nuclear export sequence; PC, nuclear pore complex; ONM, outer nuclear membrane; PDI, protein disulfide isomerase; PNS, perinuclear space; TM, transmembrane; TX-100, Triton X-100.
References
- Adam, S.A., R.S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, E.D., R.M. Lewis, R.M. Grady, and J.R. Sanes. 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275:31986–31995. [DOI] [PubMed] [Google Scholar]
- Bastos, R., A. Lin, M. Enarson, and B. Burke. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor, K., S. Shaikh, D. Salina, W.H. Raharjo, R. Bastos, M. Lohka, and B. Burke. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264. [DOI] [PubMed] [Google Scholar]
- Broers, J.L., E.A. Peeters, H.J. Kuijpers, J. Endert, C.V. Bouten, C.W. Oomens, F.P. Baaijens, and F.C. Ramaekers. 2004. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 13:2567–2580. [DOI] [PubMed] [Google Scholar]
- Burke, B., and C.L. Stewart. 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3:575–585. [DOI] [PubMed] [Google Scholar]
- Crisp, M., Q. Liu, K. Roux, J.B. Rattner, C. Shanahan, B. Burke, P.D. Stahl, and D. Hodzic. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw, J.M., A.N. Krutchinsky, W. Zhang, B.T. Chait, and M.J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L.I. Rothblum, R.D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 298:1623–1626. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., E.D. Siggia, J.E. Moreira, C.L. Smith, J.F. Presley, H.J. Worman, and J. Lippincott-Schwartz. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner Nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace, L., A. Blum, and G. Blobel. 1978. Immunocytochemical localization of the major polypeptides of the nuclear complex-lamina fraction: interphase and mitotic distribution. J. Cell Biol. 79:546–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, R., A. Goldman, K. Green, J. Jones, N. Lieska, and H.Y. Yang. 1985. Intermediate filaments: possible functions as cytoskeletal connecting links between the nucleus and the cell surface. Ann. NY Acad. Sci. 455:1–17. [DOI] [PubMed] [Google Scholar]
- Grady, R.M., D.A. Starr, G.L. Ackerman, J.R. Sanes, and M. Han. 2005. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc. Natl. Acad. Sci. USA. 102:4359–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., A. Margalit, R.D. Goldman, D.K. Shumaker, and K.L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31. [DOI] [PubMed] [Google Scholar]
- Hagan, I., and M. Yanagida. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque, F., D.J. Lloyd, D.T. Smallwood, C.L. Dent, C.M. Shanahan, A.M. Fry, R.C. Trembath, and S. Shackleton. 2006. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 26:3738–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, S., S. Guttinger, P. Muhlhausser, F. Anderegg, S. Burgler, and U. Kutay. 2006. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 580:1263–1268. [DOI] [PubMed] [Google Scholar]
- Hodzic, D.M., D.B. Yeater, L. Bengtsson, H. Otto, and P.D. Stahl. 2004. Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 279:25805–25812. [DOI] [PubMed] [Google Scholar]
- King, M.C., C.P. Lusk, and G. Blobel. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007. [DOI] [PubMed] [Google Scholar]
- Lammerding, J., P.C. Schulze, T. Takahashi, S. Kozlov, T. Sullivan, R.D. Kamm, C.L. Stewart, and R.T. Lee. 2004. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding, J., J. Hsiao, P.C. Schulze, S. Kozlov, C.L. Stewart, and R.T. Lee. 2005. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 170:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding, J., L.G. Fong, J.Y. Ji, K. Reue, C.L. Stewart, S.G. Young, and R.T. Lee. 2006. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281:25768–25780. [DOI] [PubMed] [Google Scholar]
- Lee, K.K., D. Starr, M. Cohen, J. Liu, M. Han, K.L. Wilson, and Y. Gruenbaum. 2002. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol. Biol. Cell. 13:892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima, K., K. Yahata, Y. Sasaki, R. Nakatomi, T. Tachibana, T. Hashikawa, F. Imamoto, and N. Imamoto. 2006. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J. Cell Sci. 119:4442–4451. [DOI] [PubMed] [Google Scholar]
- Malone, C.J., L. Misner, N. Le Bot, M.C. Tsai, J.M. Campbell, J. Ahringer, and J.G. White. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836. [DOI] [PubMed] [Google Scholar]
- Maul, G. 1977. Nuclear pore complexes: elimination and reconstruction during mitosis. J. Cell Biol. 74:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee, M.D., R. Rillo, A.S. Anderson, and D.A. Starr. 2006. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell. 17:1790–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mislow, J.M., M.S. Kim, D.B. Davis, and E.M. McNally. 2002. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J. Cell Sci. 115:61–70. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop, K.L., Q. Li, L. Patterson, and J.A. Fischer. 1999. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 9:1211–1220. [DOI] [PubMed] [Google Scholar]
- Muchir, A., and H.J. Worman. 2004. The nuclear envelope and human disease. Physiology (Bethesda). 19:309–314. [DOI] [PubMed] [Google Scholar]
- Ohba, T., E.C. Schirmer, T. Nishimoto, and L. Gerace. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar, V.C., S. Abraham, S. Braune, A.A. Noegel, B. Tunggal, I. Karakesisoglou, and E. Korenbaum. 2004. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 295:330–339. [DOI] [PubMed] [Google Scholar]
- Padmakumar, V.C., T. Libotte, W. Lu, H. Zaim, S. Abraham, A.A. Noegel, J. Gotzmann, R. Foisner, and I. Karakesisoglou. 2005. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118:3419–3430. [DOI] [PubMed] [Google Scholar]
- Pante, N., R. Bastos, I. McMorrow, B. Burke, and U. Aebi. 1994. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R.D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D.T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, L., and B. Burke. 1990. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the association of p55 with the nuclear lamina. J. Cell Biol. 111:2225–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut, G., V. Doye, and J. Ellenberg. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. [DOI] [PubMed] [Google Scholar]
- Rollenhagen, C., P. Muhlhausser, U. Kutay, and N. Pante. 2003. Importin beta-depending nuclear import pathways: role of the adapter proteins in the docking and releasing steps. Mol. Biol. Cell. 14:2104–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena, S., M.D. Summers, J.K. Burks, A.E. Johnson, and S.C. Braunagel. 2006. Importin-alpha-16 is a translocon-associated protein involved in sorting membrane proteins to the nuclear envelope. Nat. Struct. Mol. Biol. 13:500–508. [DOI] [PubMed] [Google Scholar]
- Shao, X., H.A. Tarnasky, J.P. Lee, R. Oko, and F.A. van der Hoorn. 1999. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 211:109–123. [DOI] [PubMed] [Google Scholar]
- Soullam, B., and H.J. Worman. 1995. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 130:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade, K., C.S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 90:1041–1050. [DOI] [PubMed] [Google Scholar]
- Starr, D.A., and M. Han. 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 298:406–409. [DOI] [PubMed] [Google Scholar]
- Starr, D.A., G.J. Hermann, C.J. Malone, W. Fixsen, J.R. Priess, H.R. Horvitz, and M. Han. 2001. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 128:5039–5050. [DOI] [PubMed] [Google Scholar]
- Stavru, F., B.B. Hulsmann, A. Spang, E. Hartmann, V.C. Cordes, and D. Gorlich. 2006. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173:509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, T., D. Escalante-Alcalde, H. Bhatt, M. Anver, N. Bhat, K. Nagashima, C.L. Stewart, and B. Burke. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147:913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E.J., and S.R. Wente. 2006. Dynamic nuclear pore complexes: life on the edge. Cell. 125:1041–1053. [DOI] [PubMed] [Google Scholar]
- Tzur, Y.B., K.L. Wilson, and Y. Gruenbaum. 2006. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 7:782–788. [DOI] [PubMed] [Google Scholar]
- Voeltz, G.K., W.A. Prinz, Y. Shibata, J.M. Rist, and T.A. Rapoport. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. [DOI] [PubMed] [Google Scholar]
- Wang, Q., X. Du, Z. Cai, and M.I. Greene. 2006. Characterization of the structures involved in localization of the SUN proteins to the nuclear envelope and the centrosome. DNA Cell Biol. 25:554–562. [DOI] [PubMed] [Google Scholar]
- Welte, M.A., S.P. Gross, M. Postner, S.M. Block, and E.F. Wieschaus. 1998. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 92:547–557. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen, K., S.H. Litjens, I. Kuikman, N. Tshimbalanga, H. Janssen, I. van den Bout, K. Raymond, and A. Sonnenberg. 2005. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171:799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman, H.J., and G.G. Gundersen. 2006. Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 16:67–69. [DOI] [PubMed] [Google Scholar]
- Yu, J., D.A. Starr, X. Wu, S.M. Parkhurst, Y. Zhuang, T. Xu, R. Xu, and M. Han. 2006. The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev. Biol. 289:336–345. [DOI] [PubMed] [Google Scholar]
- Zastrow, M.S., S. Vlcek, and K.L. Wilson. 2004. Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 117:979–987. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., J.N. Skepper, F. Yang, J.D. Davies, L. Hegyi, R.G. Roberts, P.L. Weissberg, J.A. Ellis, and C.M. Shanahan. 2001. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114:4485–4498. [DOI] [PubMed] [Google Scholar]
- Zhang, X., R. Xu, B. Zhu, X. Yang, X. Ding, S. Duan, T. Xu, Y. Zhuang, and M. Han. 2007. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 134:901–908. [DOI] [PubMed] [Google Scholar]
- Zhen, Y.Y., T. Libotte, M. Munck, A.A. Noegel, and E. Korenbaum. 2002. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115:3207–3222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.