Abstract
To quantitatively understand the binding affinity and target selectivity of small-molecule RNA interactions, it is useful to have a rapid, highly reproducible binding assay that can be readily generalized to different RNA targets. To that end, an assay has been developed and validated for measuring the binding of low-molecular weight ligands to RNA by monitoring the fluorescence of a covalently incorporated fluorophore. As a test system, the fluorescence of a pyrene-derivatized HIV-1 TAR (transactivating response element) RNA was measured upon titration with aminoglycoside antibiotics. The binding isotherms thus obtained fit well with a model for a 1:1 interaction and yield an accurate measure of the equilibrium dissociation constant. Among a series of natural aminoglycosides, the binding affinity correlates with the number of amines, supporting an electrostatic compensation model for binding. Furthermore, the ionic strength dependence confirms that much of the binding energy is electrostatic. Finally, by measuring the binding affinity in the presence of nucleic acid competitors, we confirm that although aminoglycosides show high RNA to DNA selectivity, their selectivity among different RNA targets is sub- optimal. We conclude that this newly developed assay can be generalized to measure the binding affinities and selectivities of a variety of small molecules to a specific RNA target.
INTRODUCTION
As a pivotal player in numerous eukaryotic, prokaryotic and viral life functions, RNA is an attractive target for small-molecule therapeutics (1,2). Yet, several challenges still hinder the common use of RNA as a therapeutic target. Perhaps foremost among these challenges is the lack of selectivity of the small molecule for a given RNA target to the exclusion of a myriad of other cellular RNAs. Often the population of the desired RNA target (mRNAs or viral RNAs) is very small relative to the total cellular RNA, demanding selectivity by many orders of magnitude to achieve the desired binding effect without numerous spurious interactions. To realize the goal of making RNA-directed small molecules, it is critical to thoroughly understand, and thereby modulate, target selectivity.
From a biochemical perspective, two approaches can be used to evaluate target selectivity. First, a precise quantitative comparison of the binding affinities of a series of small molecules to multiple RNA targets would reveal characteristics of the small molecules that facilitate selection among the different targets. A complementary approach is to measure the affinity with which the small molecule binds to a single RNA target in the presence of other competing nucleic acids. To facilitate both approaches, it is desirable to have a real-time equilibrium assay, which is quantitative, rapid and can easily be generalized to several different RNA targets. Here we describe the development and validation of such an assay and use it to examine the binding affinity and specificity of aminoglycosides for a specific RNA target—the HIV-1 TAR (transactivating response element).
The aminoglycoside antibiotics were originally identified as low-molecular weight bactericidal compounds, which induce translational misreading (3). More recently, this misreading effect was correlated with the direct binding of aminoglycosides to a specific site within prokaryotic ribosomal RNA (4,5). Since then, numerous other RNA targets of potential therapeutic interest have been identified to which aminoglycosides bind, including tRNA (6), the hammerhead and Hepatitis Delta Virus ribozymes (7,8), the group I intron (9), and several Human Immunodeficiency Virus (HIV) RNAs (10–12). One of the HIV RNAs to which aminoglycosides bind, the TAR, stimulates the transcription of the full-length viral genome via its interactions with the viral protein Tat (13,14). In vitro aminoglycoside antibiotics bind to the TAR with moderate affinity (Kd ≈ 3 µM) and significantly inhibit the Tat–TAR interaction (11). Unfortunately, this interaction is not highly selective, as the binding affinity of aminoglycosides for the TAR is not sufficiently higher than for other RNA targets. In principle, if an aminoglycoside could be modified to selectively inhibit the Tat–TAR interaction in vivo, transcription of the viral genome, and thus viral replication, could potentially be blocked, as suggested (15,16).
To investigate the affinity and specificity of aminoglycoside binding to the TAR, an assay has been designed that utilizes TAR constructs strategically labeled with a pyrene fluorophore. Since the structure of the TAR changes significantly upon aminoglycoside binding (17,18), the fluorescence intensity of the pyrene also changes as a function of aminoglycoside concentration. Quantitation of the resultant binding isotherm yields a precise equilibrium dissociation constant and the stoichiometry for the binding of several different aminoglycosides to the TAR. By measuring these binding constants in the presence of other nucleic acid competitors, the binding specificity of the aminoglycosides for the TAR is also evaluated. The results show that electrostatic interactions provide a significant fraction of the binding energy for the TAR–aminoglycoside interaction, but these same electrostatic interactions decrease the binding specificity for the TAR, relative to other nucleic acids.
MATERIALS AND METHODS
Materials
All aminoglycosides were purchased as sulfate salts from Sigma (neomycin, paromomycin, ribostamycin and kanamycin B), or Calbiochem (kanamycin A), except for the free base form of tobramycin, which was a generous gift from Meiji Seika Kaishak, Ltd (Japan). Tris [tris (hydroxymethyl) aminomethane], HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] buffers and all inorganic salts were purchased from Fisher (enzyme grade). Nonidet P-40 was purchased from Fluka. Amberlite CG-50 type II resin was purchased from Sigma. DNA oligonucleotides were purchased from Integrated DNA Technologies (Iowa City, IA) and used without further purification. Yeast tRNA competitor was purchased from Sigma. Ribonuclease T1 was from Boeringher Mannheim (Germany) and ribonuclease V1 was from Ambion (Austin, TX).
Purification of aminoglycosides
All aminoglycosides were purified and isolated as the free base form using a 2 cm by 20 cm Amberlite CG-50 column (Type II, NH4+ form) as described (19). A general procedure involved loading 5 ml of aqueous aminoglycoside sulfate (∼50 mg/ml) onto a CG-50 column (NH4+ form) that had been equilibrated with 500 ml water. Following a wash of 500 ml 0.1 M NH4OH, each aminoglycoside was eluted with a 600 ml linear gradient of 0.1–1 M NH4OH. Aminoglycoside-containing fractions were identified by ninhydrin staining on a TLC plate and concentrated, followed by washing with 200 ml water, concentrating and lyophilization. 1H-NMR and mass spectral analysis confirmed the identity of each aminoglycoside.
Separation of neomycin B and C
Neomycin sulfate (B and C mixture) was protected as the t-butoxycarbonyl derivative as described (20), at which point the B and C isomers could be purified by silica gel flash column chromatography. Neomycin B eluted at 5.75% methanol in dichloromethane, while the C isomer eluted at 6.5% methanol in dichloromethane. Deprotection in trifluoroacetic acid as described (20) and subsequent purification via ion-exchange chromatography as with the other aminoglycosides yielded the free base form of each isomer.
Preparation of TAR oligonucleotides
All TAR oligonucleotides were purchased from Dharmacon RNA technologies (Lafayette, CO), deprotected as per manufacturer’s protocol, and lyophilized. The deprotected oligonucleotides were resuspended in water and purified using 20% denaturing PAGE. Excised gel slices were eluted overnight into a buffer containing 50 mM HEPES pH 7.5 and 1 M NaCl, followed by ethanol precipitation. After a 70% ethanol:water wash, the RNA pellets were dried and resuspended in water, quantitated by UV spectroscopy and their identities confirmed by MALDI-TOF mass spectrometry. For binding experiments, the TAR was refolded by heating a 40 µM solution of the pyrene-labeled TAR in the minimal Tris binding buffer (50 mM Tris pH 7.4, 100 mM NaCl, 1 mM MgCl2) to 95°C for 5 min, followed by cooling on ice for 20 min.
Fluorescence binding assay
For all fluorescence measurements, a Perkin-Elmer LS50B fluorimeter was used, with an excitation slit width of 12 nm and an emission slit width of 10 nm. Upon excitation at 345 nm, the spectrum between 360 and 450 nm (scan rate of 300 nm/min) was recorded four times and averaged for a composite spectrum.
For a typical binding experiment, the fluorescence spectrum of a 149 µl solution of the buffer in the absence of any RNA or aminoglycoside was recorded. This spectral blank, for which only Raman scatter was observed, was subtracted from all subsequent spectra within each binding experiment. This buffer blank included nucleic acid competitors, where applicable. Following determination of the buffer blank, 1 µl of a 40 µM solution of refolded, pyrene-labeled TAR was added (final concentration is 200 nM), the solution mixed, and the spectrum again recorded. Subsequent aliquots of 1 µl of an aqueous aminoglycoside solution (increasing concentrations from 2 µM to 44 mM) were added, and the fluorescence spectrum was recorded after each aliquot until the pyrene fluorescence reached saturation. Over the entire range of aminoglycoside concentrations, the emission maximum of 390 nm varied <1.5 nm. To correct for any possible contaminating fluorescence of each aminoglycoside, the full titration was repeated in the absence of pyrene-labeled TAR. Any observed ‘background’ emission at 390 nm was then subtracted from each corresponding point of the pyrene–TAR titration, and the resulting fluoresence intensity was corrected for dilution. This corrected fluorescence intensity value (I390) at each titration point was then normalized to the initial fluorescence of the pyrene-derivatized TAR in the absence of aminoglycoside (Iunbound) and plotted as a function of the dilution-corrected aminoglycoside concentration as shown in Figure 2.
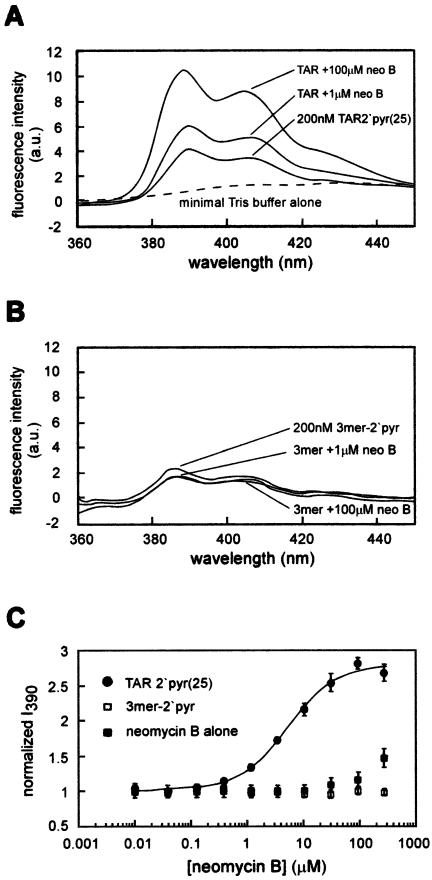
Figure 2.
(A) The fluorescence spectrum of TAR-2′pyr(25) in the absence and presence of purified neomycin B, in 50 mM Tris pH 7.4, 1 mM MgCl2 and 100 mM NaCl. The dotted line indicates the fluorescence spectrum of the buffer alone. This spectrum, with an excitation wavelength of 340 nm and slit widths of 12 nm for excitation and 10 nm for emission, has a λmax of 390 nm. (B) The fluorescence spectrum of a 5′-U-(2′-amino- butyryl-pyrene-uridine)-U 3mer in the presence and absence of purified neomycin B, in 50 mM Tris pH 7.4, 1 mM MgCl2 and 100 mM NaCl. The slight decrease in intensity reflects dilution. (C) An example of an isotherm for the binding of neomycin B to TAR-2′pyr(25) is shown in filled circles. The fluorescence intensity for this isotherm is normalized to the starting fluorescence as described (23), after correcting for dilution and for the residual fluorescence of neomycin B at higher concentrations (shown in filled squares). The open squares demonstrate that the fluorescence of a U-(2′-amino-butyryl-pyrene-uridine)-U 3mer does not increase over this range of neomycin concentrations. Error bars indicate the standard deviation of at least three independent measurements at each aminoglycoside concentration.
Since the binding isotherms indicate a monophasic binding interaction, the changes in fluorescence can be adequately described by a simple two-state equilibrium model:
and:
[TARbound] = Ka[TARunbound][ag]n
In the absence of aminoglycoside, the pyrene-derivatized TAR (TARunbound) has an initial fluorescence (Iunbound), which upon normalization is defined as 1. Upon titration, n molecules of aminoglycoside bind to the TAR forming a complex (TARbound • agn), with a relative fluorescence intensity of Ibound. Thus, the observed fluorescence is a sum of the fluorescence of the bound and unbound TAR as follows:
Note that in this system, [TAR] << Kd; consequently, the concentration of the TAR–aminoglycoside complex is much less than the total aminoglycoside concentration. Hence, for the purposes of equation 1, the assumption was made that the free aminoglycoside concentration, [ag], is equal to the total aminoglycoside concentration. Least-squares fitting to equation 1 using the Kaleidograph program determined Iunbound, Ibound, Kd and n as independent variables. For all binding isotherms measured herein, the Hill coefficient, n, was between 0.7 and 1.2, consistent with the 1:1 binding stoichiometry that was reported previously for this interaction (18). Therefore, since the TAR–aminoglycoside interaction is not expected to show significant cooperativity, n was fixed at 1 to determine the Kd values reported in the tables. The fit of the data to this equation was very good, with an R value exceeding 0.99. When n was fixed at other integer values, the quality of the fit was significantly lowered, further suggesting a 1:1 model for this interaction.
RNAse footprint of neomycin on the TAR
A trace concentration (<20 pM) of refolded, 5′-32P-labeled TAR was incubated with 0.004 U of ribonuclease V1 (Ambion) for 15 min at room temperature in a solution containing 50 mM Tris pH 7.4, 50 mM NaCl, 1 mM MgCl2, 0.04 µg/ml yeast torula RNA (Ambion), and the indicated concentrations of neomycin free base (Fig. 3). These reactions were quenched by dilution into an equal volume of loading dye (88% formamide with 0.02% bromophenol blue and xylene cyanol), followed by immediate separation with 20% denaturing PAGE.
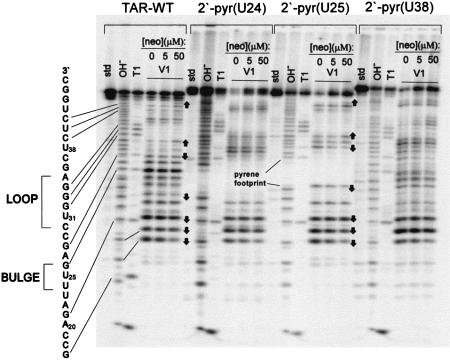
Figure 3.
Enzymatic footprint (RNAse V1) of neomycin binding to the four TAR constructs. ‘Std’ lanes are untreated RNA. ‘OH-’ lanes are an alkaline hydrolysis ladder of each RNA, and ‘T1’ lanes show cleavage by ribonuclease T1 under denaturing conditions, in the absence of neomycin. Similarities in the footprint between TAR WT and TAR-2′pyr(25) are noted as arrows pointing up or down, corresponding to either increases or decreases in cleavage, respectively, as the concentration of neomycin in increased.
Ribonuclease T1 reactions contained a trace concentration (<20 pM) of 5′-32P-labeled, refolded TAR and 1 U of ribonuclease T1 (Boerhinger Mannheim) in a solution of 20 mM sodium citrate pH 5.0, 1 mM EDTA, 3 M urea and 0.04 µg/µl yeast torula RNA (Ambion). These reactions were incubated at 55°C for 30 min, followed by dilution into an equal volume of loading dye and separation with 20% denaturing PAGE.
Alkaline hydrolysis reactions
In a buffer of 50 mM NaHCO3 pH 9.0, RNA solutions were incubated at 95°C for 7 min, followed by quenching on ice for 10 min. After dilution with an equal volume of loading dye, the reactions were separated on 20% PAGE. All polyacrylamide gels were run for ∼3 h at 15 W and analyzed using a Molecular Dynamics Storm Phosphorimager.
RESULTS
Assay design
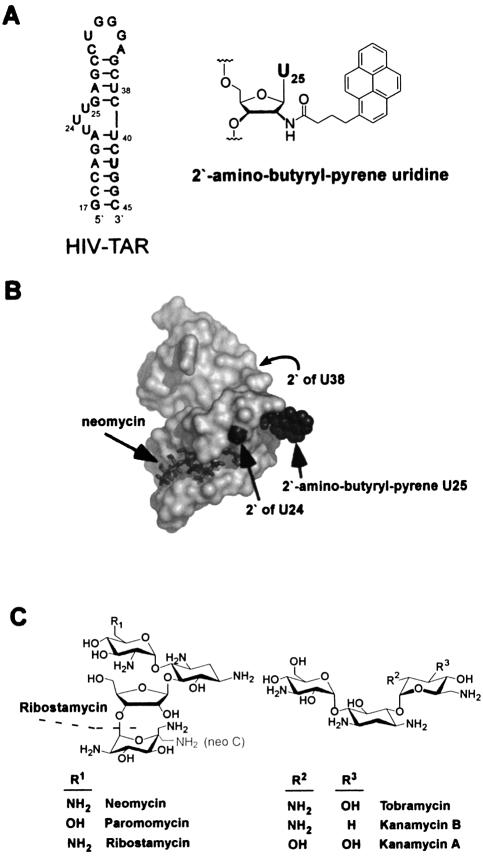
Circular dichroism, enzymatic footprinting and NMR experiments all indicate that the tertiary structure surrounding the bulged region of the TAR RNA (Fig. 1a) changes upon binding neomycin B (17,18). Since the pyrene fluorophore is exceptionally sensitive to its environment when incorporated into nucleic acids (21), we reasoned that a pyrene in this region would accurately report these structural changes by either an increase or a quenching of its fluorescence. Three versions of the HIV-1 TAR RNA were therefore designed. Each contains a single uridine to 2′-amino-butyryl-pyrene uridine (22) substitution at U24, U25 or U38 near the bulged region (Fig. 1a). At each of these three positions, examination of the NMR structure of a TAR–neomycin complex (18) suggests that the 2′-butyryl-pyrene modification can be accommodated without severe Van der Waals overlap (Fig. 1b), implying that these modifications should not drastically alter the tertiary fold. After purification and refolding, the fluorescence intensity of each of these modified TAR RNAs was measured as a function of aminoglycoside concentration.
Figure 1.
(A) The sequence and secondary structure of the HIV-1 TAR RNA. ‘TAR WT’ is the all 2′-hydroxy version of this sequence. TAR-2′pyr(24), TAR-2′pyr(25) and TAR-2′pyr(38) each have a single uridine to 2′-amino-butyryl-pyrene uridine substitution at residue 24, 25 or 38, respectively. The chemical structure of 2′-amino-butyryl-pyrene is shown. (B) The NMR structure of the HIV-1 TAR RNA when bound to neomycin as determined by Faber et al. (18), showing the context of the 2′-positions modified. The 2′-amino-butyryl-pyrene modification has been modeled onto this structure at U25 in an orientation that permits it to protrude into solution. Although this orientation is arbitrary, it demonstrates that conformations are available at this position, which do not incur significant Van der Waals overlap with other parts of the structure. The 2′ position of U38 is on the back of the structure, as indicated by the arrow. (C) Chemical structures of the aminoglycosides tested. Ribostamycin does not contain the fourth ring. Neomycin B and C differ only in the chirality at the 5′′′-carbon.
Neomycin binding increases the fluorescence intensity of TAR-2′pyr(25)
Upon excitation at 345 nm, a 200 nM solution of refolded TAR 2′pyr(25) has a fluorescence spectrum typical of pyrene-labeled nucleic acids (Fig. 2a) (21,23). Upon addition of increasing concentrations of purified neomycin B, the intensity of both emission maxima (390 and 407 nm) increases by ∼3-fold, saturating near 50–100 µM, while the emission maximum at 390 nm varies <1.5 nm. In contrast, the emission spectrum of an RNA 3mer containing a single 2′-amino-butyryl-pyrene uridine shows little change upon addition of neomycin (Fig. 2b), confirming that the observed fluorescence changes reflect binding to the TAR RNA, rather than an unrelated interaction with the pyrene label.
Faber et al. (18) reported modest aggregation of the HIV-1 TAR, particularly at high neomycin concentrations. Since this could potentially affect the fluorescence, it was conceivable that a fraction of the fluorescence change was due to aggregation. To address this, the fluorescence intensity of increasing concentrations of TAR-2′pyr(25) was measured in the absence of neomycin under a range of solution conditions. It is likely that any aggregation would be evident from a non-linear relationship between the observed fluorescence and the TAR concentration. Indeed, at NaCl concentrations below 40 mM, a non-linear relationship was observed, with significant quenching of the fluorescence as the RNA concentration was increased (not shown). This non-linearity was observed even when additional non-fluorescent TAR (TAR WT) was added. Similarly, below 40 mM NaCl some quenching was also observed at neomycin concentrations above 200 µM. This suggests that under low-salt conditions the TAR either aggregates or experiences an alternate folding transition. At NaCl concentrations above 40 mM, however, in the presence of 50 mM Tris pH 7.4 and 1 mM MgCl2, the emission at 390 nm did increase linearly with the concentration of TAR-2′pyr(25) between 100 and 800 nM (not shown), and the addition of higher concentrations of TAR WT did not affect the fluorescence. In addition to higher NaCl concentrations, MgCl2 (2.5 mM) was the most effective at relieving the non-linear behavior among several salts and detergents tested. For the initial studies, therefore, a minimal buffer in which the TAR is ‘well-behaved’ was chosen, which contains 50 mM Tris pH 7.4, 1 mM MgCl2 and 100 mM NaCl.
An equilibrium titration curve for the binding of neomycin B to TAR 2′-pyr(25), spanning over four orders of magnitude of neomycin B concentration (Fig. 2c), in this minimal buffer was constructed. To minimize dilution effects, small aliquots of concentrated aqueous neomycin B were added to the binding reaction and manually mixed for 1 min, such that for the entire concentration range the total volume increase was <6%. One potential concern of this manual mixing method is whether the binding interaction has completely reached equilibrium when the fluorescence spectrum is measured. The rate at which equilibrium is reached, kequil, is a sum of the association and dissociation rate constants (kequil = kon [RNA] + koff) (24,25). If kon is near the diffusion limit, as suggested (26,27), koff should be very fast for a binding interaction with an affinity constant in the micromolar range (Kd = koff/kon), and the rate of equilibration (kequil) should also be very fast. Nevertheless, to ensure that this is the case, the fluorescence at each concentration point was measured three times, with 1 min of manual mixing between measurements. In all cases, the variance in emission was <1% at each neomycin concentration, confirming that equilibration is fast.
Another potential concern was that the addition of neomycin B increases the observed fluorescence intensity in a way unrelated to the pyrene fluorescence. This led to the observation that in the absence of the pyrene-labeled TAR, high concentrations of neomycin B do, in fact, show a small increase in fluorescence intensity (Fig. 2c), despite rigorous purification. It is probable that this increase in fluorescence represents either a fluorescent contamination or, more likely, a change in the light scattering properties of the solution. At any rate, these reproducible changes are only observed at high neomycin concentrations, rather than in the transitory portion of the binding isotherm. Nevertheless, to account for this, the fluorescence of a neomycin B solution alone was quantitated and subtracted from that of the pyrene-labeled TAR at each point of the titration.
Another possible concern of this equilibrium titration method is photobleaching. Specifically, the repeated mixing during the titration could potentially resaturate the cuvette with dissolved oxygen before each emission scan, thereby accelerating excited-state fluorophore breakdown. To assess this possibility, repeated aliquots of water were added and manually mixed as above, and the fluorescence measured. In each case, repeated dilution and mixing did not cause any observable changes in the fluorescence intensity, precluding any serious photobleaching effects within the time-frame of a typical binding experiment.
After considering each of these important corrections and normalizing the fluorescence intensity to that of the unbound TAR, the resultant binding isotherm can be accurately fit to a previously described equation for the binding of Mg2+ to the Tetrahymena group I intron (23), yielding an equilibrium dissociation constant (Kd) of 5.8 ± 1 µM and a Hill coefficient of 0.99 for the TAR–neomycin B interaction (Fig. 2c, Table 1). This binding affinity agrees favorably with the previously reported values of Kd ranging from 1 to 5.9 µM for this interaction (17,18), and the Hill coefficient is consistent with the reported 1:1 stoichiometry (18). Finally, the TAR–neomycin B Kd was compared over a range of TAR-2′pyr(25) concentrations. Between 100 and 500 nM TAR-2′pyr(25), the measured affinity was invariant, further confirming that under these conditions, aggregative effects are minimal.
Table 1. The binding of neomycin B to each TAR construct.
| Mean Kd (μM)a | nb | Fold maximal increase of Iobs | |
|---|---|---|---|
| TAR 2′pyr(24) | 3.2 ± 0.5 | 1.1 | 2.2 |
| TAR 2′pyr(25) | 5.8 ± 1 | 0.99 | 3.0 |
| TAR 2′pyr(38) | 9.0 ± 0.9 | 1.0 | 3.8 |
aTolerances indicate the standard deviation of at least three independent measurments.
bHill coefficient from curve fit (see Materials and Methods).
The binding interaction of neomycin B with TAR-2′pyr(24) and TAR-2′pyr(38)
An equilibrium titration of neomycin B with TAR-2′pyr(24) or TAR-2′pyr(38) reveals that all three TAR constructs bind neomycin B with similar affinities (Table 1). Also, the Hill coefficients for the binding of neomycin B to all three constructs are consistent with a 1:1 binding interaction. It is interesting to note that for each of the three TAR constructs, the total increase in fluorescence intensity upon saturation varies from 2.2 for TAR-2′pyr(24) up to 3.8 for TAR-2′pyr(38). This may reflect the different solvation environments the fluorophore experiences in each construct. More likely, these variations may indicate the different degrees to which the RNA conformation near each fluorophore changes upon neomycin binding. This explanation is consistent with the observation that the changes that neomycin induces in the electron paramagnetic resonance spectral width of a nitroxide label are different when the probe is at U23, U25 or U38 (28). Specifically, that study indicated that U38 undergoes the most structural and dynamical changes upon neomycin binding, while U23 undergoes the least. This hierarchy agrees well with our trend in the maximal fluorescence changes that neomycin B induces (U38 > U25 > U24). Again, this implies the link between a change in the fluorescence intensity and binding-induced changes in the structure of the TAR.
Enzymatic footprint of neomcyin–TAR binding
An important question to consider is whether the large 2′-amino-butyryl-pyrene modification changes how neomycin binds to the TAR; that is, does the pyrene alter either the binding site or the structural changes that accompany neomycin binding? To address this, the enzymatic footprint of neomycin binding to each of the TAR constructs was characterized, using Ribonuclease V1, which cleaves primarily double-stranded or strongly stacked regions (29). To varying degrees, each of the TAR constructs shows similar protection patterns as the unmodified TAR (Fig. 3). Notable among these similarities are those at C19, A20 and G21, adjacent to the bulge as observed previously (17). Interestingly, some additional neomycin-dependent changes in the cleavage pattern are observed distal to the binding site that were not described previously. For instance, neomycin B binding increases the degree to which RNase V1 cleaves at U31, which probably reflects the binding-induced global changes of the TAR fold. Accordingly, a comparison of the structures of the unbound TAR and the neomycin-bound TAR (30) reveals that neomycin binding does in fact cause this stacked region of the loop to become more solvent exposed. Of the three constructs, TAR-2′pyr(25) shows the most similar protection pattern to the all-2′-hydroxyl TAR (similarities noted as arrows in Fig. 3). Furthermore, for TAR-2′pyr(25), these cleavage-protection patterns change with neomycin concentration the same as for the all-2′-hydroxyl TAR, confirming that the attached pyrene does not significantly alter the binding site, RNA morphology or the affinity of neomycin for the TAR. For these reasons, TAR-2′pyr(25) was used for the further characterization of aminoglycoside–TAR binding.
Binding affinities of different aminoglycosides to TAR-2′pyr(25)
To further characterize the thermodynamic basis of the aminoglycoside–TAR interaction, the binding of a series of different natural aminoglycosides to TAR-2′pyr(25) was measured at pH 7.4 in the presence of 1 mM MgCl2 and 100 mM NaCl (Fig. 1c and Table 2). As with neomycin B, at high concentrations in the absence of RNA, each of the aminoglycosides displays a modest increase in fluorescence for which the data must be corrected. The TAR binding affinities among this series range over nearly two orders of magnitude and indicate several important points. As observed for other aminoglycoside–RNA interactions, the binding affinities correlate with the number of amines (31–33), with roughly a 1.25 kcal mol–1 favorable contribution to the binding free energy (ΔG298K) per amine. This implies that much of the energetic impetus for binding is electrostatic, as typically observed for aminoglycoside–RNA interactions (2). To a lesser extent, the structural context of the amines within the aminoglycoside also affects binding. On average, among aminoglycosides with the same number of amines, the 4,6-linked derivatives (tobramycin and kanamycin A) bind with higher affinities than the respective 4,5-linked derivatives (paromomycin and ribostamycin).
Table 2. The binding of each aminoglycoside to TAR-2′pyr(25).
| Number of amines | Mean Kd (μM)a | Fold maximal increase of Iobs | |
|---|---|---|---|
| Neomycin B | 6 | 5.8 ± 1 | 3.0 |
| Neomycin C | 6 | 8.2 ± 4 | 3.0 |
| Paromomycin | 5 | 72 ± 20 | 5.2 |
| Kanamycin B | 5 | 89 ± 10 | 4.0 |
| Tobramycin | 5 | 43 ± 4 | 3.8 |
| Kanamycin A | 4 | 107 ± 13 | 3.6 |
| Ribostamycin | 4 | 355 ± 33 | 1.7 |
aTolerances indicate the standard deviation of at least three independent measurements.
It is also interesting to note that each of the aminoglycosides causes a different maximal increase in fluorescence intensity upon binding TAR-2′pyr(25), ranging from 1.7- to 5.2-fold increase by ribostamycin or paromomycin, respectively. One explanation for these differences is that each aminoglycoside has a different effect on the local solvation environment near the pyrene fluorophore, with resultant differences in fluorescence intensity. Since there is no identifiable correlation between the number of amines and the maximal fluorescence change, however, this seems unlikely. Rather, the maximal fluorescence change likely reflects the degree to which the RNA conformation changes upon binding.
Ionic strength dependence of binding
The correlation between binding affinity and the number of amines of an aminoglycoside prompted a closer examination of the effect that ionic strength has on binding. The binding affinity for neomycin B was measured over a range of NaCl concentrations from 20 to 250 mM in the presence of 50 mM Tris 7.4 and 2 mM MgCl2. Above 250 mM NaCl, the binding affinity is sufficiently weak that experimental error prevents an accurate determination of Kd. As with other RNA–polyamine interactions, the binding affinity decreases with increasing ionic strength over this range (Fig. 4). This result underscores the thermodynamic importance of electrostatic interactions to the interaction of aminoglycosides with RNA.
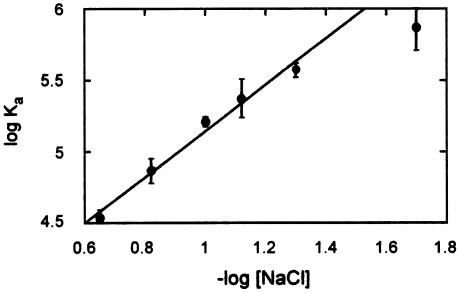
Figure 4.
The ionic strength dependence of neomycin B binding to TAR-2′pyr(25). The linear fit shown, with a slope of 1.61, does not include the point at 20 mM NaCl. As mentioned in the text, significant aggregation is sometimes observed at this low salt concentration, affecting the measured Ka. The line indicates a least-squares linear fit. Error bars indicate the standard deviation of at least three independent measurements at each NaCl concentration.
Binding specificity of the aminoglycoside–TAR interaction
To estimate the RNA-to-DNA binding specificity of the TAR–aminoglycoside interaction, binding affinities were measured in the presence of a 100-fold nucleotide excess of various DNA competitors. Initial attempts to use calf thymus DNA as a competitor were unsuccessful, due to large increases in the measured fluorescence intensity as the aminoglycoside concentration was increased. Since these large increases persisted in the absence of pyrene-labeled TAR, it was reasoned that they might represent changes in the light scattering properties of the solution as a result of aminoglycoside-induced DNA aggregation, akin to the aggregative effects of other polyamines on DNA (34). By using a shorter 17 bp DNA duplex in competition studies, these aggregative light- scattering effects were alleviated, and an accurate assessment of TAR:DNA specificity was possible (Table 3). In the minimal binding buffer (50 mM Tris pH 7.4, 1 mM MgCl2, 100 mM NaCl) a 100-fold nucleotide excess of duplex DNA (17.1 µM duplex; 200 nM TAR) decreased the affinity of neomycin B for the TAR by 2.4-fold. This high RNA:DNA specificity, typical for aminoglycosides, was also observed for short DNA duplexes with different sequences (not shown) and agrees quite favorably with the RNA:DNA specificity of the aminoglycoside for the rev response element (RRE) (35). Interestingly, as the number of amines in the aminoglycoside decreases (tobramycin and kanamycin A), the competitive effect of DNA also decreases. This likely reflects the decreased ability that an aminoglycoside with fewer amines would have for forming non-specific electrostatic interactions.
Table 3. Equilibrium dissociation constants (Kd, µM) for aminoglycosides to TAR-2′pyr(25) in the presence of nucleic acid competitors.
| Minimal conditionsa | ‘Physiological’ conditionsb | |||||
|---|---|---|---|---|---|---|
| No competitor | +dsDNAc | +tRNAmixd | No competitor | +dsDNAc | +tRNAmixd | |
| Neomycin B | 5.8 ± 1 | 14 ± 3 | 94 ± 12 | 9.0 ± 1 | 15 ± 5 | 84 ± 18 |
| Tobramycin | 43.4 ± 4 | 44 ± 6 | 115 ± 7 | 22 ± 4 | 44 ± 2 | 137 ± 10 |
| Kanamycin A | 107 ± 13 | 142 ± 2 | 141 ± 2 | 84 ± 15 | 122 ± 13 | 157 ± 25 |
a50 mM Tris pH 7.4, 100 mM NaCl, 1 mM MgCl2.
b30 mM HEPES pH 7.4, 100 mM KCl, 10 mM Na2HPO4, 20 mM guanidinium HCl, 2 mM MgCl2, 20 mM NaCl, 0.5 mM EDTA, 0.001% non-ionic detergent P-40.
cdsDNA competitor sequence: 5′-GCTGAATACATAAGACG-3′, and its complement.
dIndicate apparent Kd (see text).
The specificity with which aminoglycosides bind the TAR in the presence of other structured RNAs was assessed by using 100-fold nucleotide excess of a mixture of natural tRNA (tRNAmix). Although some apparently aggregative effects were observed with the tRNA, they were modest, and could be corrected for without significant error. Unlike the DNA competitors, the 100-fold excess of tRNAmix not only significantly decreased the affinity of the aminoglycosides for the TAR, but also induced a much steeper binding isotherm (not shown). For this reason, the quality of the fit to our single binding site model dropped slightly (R ≈ 0.97). Thus, the binding constants in the presence of tRNAmix reported in Table 3 reflect an apparent Kd. A 100-fold excess of tRNAmix decreased the apparent affinity of neomycin B for the TAR by 16-fold, a significantly larger decrease than reported for neomycin B binding to the RRE (35). This may reflect the slightly higher affinity of neomycin B for the RRE than for the TAR (10,32,35–37). As with DNA competitors, the competitive effect of tRNAmix decreases as the number of amines decreases (tobramycin to kanamycin A), reflecting the decrease in non-specific electrostatic binding energy. This trend is also consistent with the inherently decreased affinity of these aminoglycosides for tRNA (6). A similar correlation between binding specificity and the number of amines was also observed for the prokaryotic ribosomal A-site RNA (33).
To better understand the physiological relevance of these binding specificities, the competition studies were repeated in a complex binding mixture that more closely approximates ionic conditions inside the cell (38). It is conceivable that the TAR–aminoglycoside interaction could behave very differently in a cellular environment, and that the specificities measured in the minimal buffer do not accurately represent the case in vivo. Interestingly, the binding affinities and specificities of the aminoglycosides for the TAR were, however, largely unchanged in this ‘physiological’ binding buffer (Table 3), which contains not only a higher ionic strength but also a more diverse collection of ions. The only apparent differences in affinities were observed for tobramycin and kanamycin A, which bind with slightly higher affinities under these more physiological conditions. These results imply that the affinities and particularly the specificities measured with this assay are reasonably representative of the TAR–aminoglycoside interaction in vivo.
DISCUSSION
For RNA-binding small-molecule therapeutics to be useful, it is imperative that they possess high target-specificity. To rapidly understand and improve the elements of a small molecule that can confer this target specificity, it is important to have an assay that accurately measures the binding affinities of the interaction and can be easily generalized to different RNA targets and small molecules. To that end, a pyrene-based fluorescent assay has been developed to report the binding affinities and specificities of small-molecular weight ligands for the HIV-1 TAR.
Previously, a pyrene fluorophore was post-synthetically attached to a single 2′-amino-modified nucleotide to monitor the tertiary folding of the Tetrahymena group I intron P4–P6 domain (23). Here, three versions of the HIV-1 TAR were designed, each containing a single uridine to 2′-amino-butyryl-pyrene uridine modification. After deprotection, purification and refolding of the RNAs, the fluorescence intensity of the pyrene fluorophore was monitored as a function of aminoglycoside concentration. As the concentration of aminoglycoside was increased, the fluorescence intensity of the pyrene fluorophore also increased, yielding a monophasic and saturable binding isotherm. Analysis of the isotherms thus obtained gave an accurate and reproducible measure of the binding affinity for each aminoglycoside–TAR interaction.
For the data to be useful in the development of novel TAR-binding molecules, it is important that the pyrene-label does not dramatically perturb the TAR structure or the aminoglycoside–TAR interaction. Examination of the structure of the TAR–neomycin complex (30) reveals that in each of the three different TAR constructs, the pyrene is in a very different structural context relative to the neomycin-binding site. It is reasonable to predict, therefore, that if the pyrene does perturb the binding interaction, it would do so very differently among the three constructs, with resultant differences among the affinities. Yet, the affinities of neomycin for all three TAR constructs are very close, suggesting against any such perturbation. Furthermore, the range of neomycin affinities measured (Kd ranging from 3.2 to 9.0 µM) is very similar to the range reported in the literature (Kd ranging from 1 to 5.9 µM) (17,18), again arguing against any deformative effects of the pyrene fluorophore. Most importantly, the enzymatic footprint of neomycin binding to each construct compares well with that of the unmodified all 2′-hydroxyl-TAR. This confirms that neomycin not only binds the pyrene-labeled TARs in the same region as the unmodified TAR, but also induces similar structural changes upon binding.
It is interesting to consider the structural basis for the increase in pyrene fluorescence upon aminoglycoside binding. One possible explanation is that upon binding neomycin, the TAR becomes more structured, permitting a closer association of the pyrene with the RNA bases than possible in the less structured unbound form. This would result in less solvent quenching of pyrene fluorescence than the more solvated environment of a disordered TAR structure. This explanation is in line with the increased structural organization observed for a nitroxide-labeled TAR upon binding (28). The positional hierarchy of fluorescence increases observed here (U38 > U25 > U24) agrees with the proposed trend in structural ordering (28). A converse explanation is that in the unbound TAR structure, pyrene may be near a pyrimidine base that quenches its fluorescence by a known electron-transfer mechanism (39,40). In this case, the TAR structural changes upon neomycin binding would move the pyrene away from the quenching pyrimidine, increasing the fluorescence. Thus, the degree to which binding increases fluorescence at each position would reflect proximity to other pyrimidines in the unbound TAR structure. It is likely that a combination of these effects may explain the increases in pyrene fluorescence.
The binding of aminoglycosides to RNA has been described by an electrostatic compensation model (2,41), in which positively charged ammonium ions form favorable interactions with the negatively charged field surrounding the RNA fold (41). In accordance, we observe that the affinities of a series of aminoglycosides to the TAR correlate well with the number of amines in the antibiotic, with roughly a 10-fold reduction in the binding affinity (or ∼1.3 kcal mol–1 of binding energy) per amine. The ionic strength dependence of binding for the TAR–neomycin B complex further underscores the importance of electrostatic interactions (Fig. 4).
One effect that can potentially explain the ionic strength dependence is the necessary displacement of inorganic salt ions from both the aminoglycoside and the RNA to form an intermolecular ‘ion pair’. This ion displacement can be considered from the standpoint of classical polyelectrolyte theory developed to describe DNA–polylysine interactions (42). This theory assumes that the energetic effect of ion displacement from polylysine (or in the present case an aminoglycoside) is negligible, and that ion displacement from the nucleic acid energetically predominates. Consequently, the slope of a plot of log Ka versus log [M+] is equal to –mΨ, where m is the number of ions displaced from the nucleic acid (essentially the number of intermolecular ion pairs) and Ψ is the fractional probability of a counterion being thermodynamically associated with each phosphate of the RNA (Fig. 4). For the TAR–neomycin B interaction this slope is 1.61. Using estimates for Ψ ranging from 0.68 to 0.89 for single- or double-stranded nucleic acids, respectively (43), m ranges from 1.9 to 2.1. This prediction of two ionic contacts compares favorably with a similar determination of two to three contacts for an RNA aptamer selected to bind neomycin B (44), but is significantly less than the four ionic contacts predicted for the ribosomal A-site RNA (45). This prediction also agrees well with the NMR structure of the TAR–neomycin complex, which would predict one to three amine–phosphate interactions.
It is clear from this ionic strength dependence that electrostatic interactions are thermodynamically very important to the aminoglycoside–RNA interaction. Extrapolation to 1 M salt, where electrostatic interactions should be negligible, gives a Kd ≌ 0.3 mM. This non-electrostatic binding energy of –4.8 kcal mol–1 is only 68% of the total binding energy observed in the two buffer conditions reported here (ΔG ≌ –7.0 kcal mol–1). This is significantly less than the values typically observed for the non-electrostatic binding energy of protein–RNA interactions (∼80%) (46). Clearly, electrostatic interactions are integral to the binding of aminoglycosides to RNA.
Strikingly, the detailed structure of the aminoglycoside appears less significant than the overall charge. For instance, the epimeric neomycin B and C bind to the TAR equally well, despite the different absolute configuration at C5′′′ (Fig. 1). Inspection of the structure of the TAR–neomycin B complex shows that the 6′′′ amine protrudes into the solvent, whereas for neomycin C, this amine would face more directly toward the RNA phosphate backbone near G44 and could be predicted to bind more tightly. The observation that the two aminoglycosides bind to the TAR with the same affinity diminishes the importance of local structure, and is consistent with a structural electrostatic complementarity model (47).
Even more striking is the comparison of kanamycin B and paromomycin. Both have five amines, but the relative placement of these amines is very different. Despite these differences, both bind the TAR with similar affinity. Thus, in this case the number of amines supercedes their relative position within the structure. This may be explained by the inherent conformational flexibility of the aminoglycosides (48–50), which could permit these two very different molecules to adapt to the same RNA fold equally well. Furthermore, given the highly electrostatic basis of the binding interaction, it is conceivable that multiple conformational states of the aminoglycoside could bind to a given RNA equally well (48), or at a slightly different binding site. It is clear, therefore, how the conformational adaptability of the aminoglycosides would imply that the overall charge should generally be more important for binding than the local structure.
This study also highlights the challenges of designing a highly target-specific RNA-directed small molecule. Consider the case of a specific mRNA target. On average, <50 copies of a given mRNA are present in the cytoplasm at any given time (38). In addition to an estimated 360 000 other mRNAs, there are likely to be >500 000 tRNA or rRNA molecules in the cytoplasm (38). The hopeful therapeutic molecule must, therefore, face the daunting task of finding and binding to its target to the exclusion of more than a 10 000-fold excess of other potential targets. The data presented here suggest that, in their current forms, aminoglycosides are not yet up to this task. When challenged with a mere 100-fold nucleotide excess of tRNA competitors, the binding affinity of neomycin B (the tightest binder measured herein) for the TAR is decreased by at least 9-fold (‘physiological’ conditions). This means first that the concentration of neomycin would need to be dramatically higher to elicit a cellular TAR response. More importantly, at such concentrations, the significant binding to other RNAs could cause significant negative effects on normal cellular function. Clearly, we must learn how to dramatically enhance the target specificity of aminoglycoside-based ligands.
The trends observed here suggest that one adversary to gaining high target specificity with aminoglycosides is their electrostatic interactions. As the overall positive charge of the aminoglycosides is decreased, the competitive effect of an excess of tRNA also decreases. Intuitively this trend makes sense, given that much of the solvent exposed surface of RNA is negatively charged. The well-documented conformational flexibility of both the RNA and the aminoglycosides (49–51) further permits the molecules to adapt to one another, thereby increasing the frequency with which these two oppositely-charged molecules are conformationally compatible for binding. It follows that decreasing the overall charge– charge interactions could increase the target specificity of small molecules. However, it is also evident that the binding energy provided by these interactions must in some way be compensated. Although tobramycin shows significantly higher TAR:tRNA specificity than neomycin, its binding affinity is also substantially lower, which would limit effectiveness. The obvious conclusion is that it is desirable to develop aminoglycoside derivatives in which the electrostatic binding elements have been partially replaced by other high-affinity binding elements that are capable of higher specificity.
In summary, the study presented here validates the use of a pyrene-labeled RNA to thermodynamically characterize the binding interactions between RNA and low-molecular weight ligands. This method presents several advantages over previously described small-molecule RNA binding assays. Protocols which measure the fluorescence anisotropy changes of a fluorescently tagged aminoglycoside risk the serious possibility of the fluorophore perturbing the binding interaction. Further, to measure a wide variety of small molecules one must either undertake impractical derivatization of each molecule to be tested or measure binding indirectly through a competition assay. Another method, measuring peptide displacement caused by a small molecule, does not permit measurement of binding stoichiometry and cannot be rapidly generalized to different RNA targets. In contrast, using only relatively basic laboratory instrumentation—a fluorimeter—and commercially available pyrene-derivatized oligonucleotides, the assay described herein can be rapidly generalized to measure the binding of a wide variety of small-molecule ligands to a diverse set of RNA targets. Further, the high sensitivity of the pyrene fluorophore to structural changes strengthens the case for general applicability to other small-molecule RNA interactions (52,53). Additionally, preliminary experiments indicate that this assay will also be useful for larger RNA ligands (Tat peptide; unpublished data). By adapting this assay to measure the binding of potential therapeutics to several different RNA targets, the characteristics that allow them to select among the targets could be revealed. It is our hope, therefore, that this type of assay will provide the means to more accurately assess small-molecule RNA binding affinities and specificities.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Nathan Luedtke and Susan Seaman for critical reading of the manuscript. Supported in part by NIH grant AI47673 (Y.T.) and Ruth Kirschtein National Research Service Award 1 F32 GM065652-01A1 (K.B.).
REFERENCES
- 1.Hermann T. and Westhof,E. (1998) RNA as a drug target: chemical, modeling and evolutionary tools. Curr. Opin. Biotechnol., 9, 66–73. [DOI] [PubMed] [Google Scholar]
- 2.Hermann T. (2000) Strategies for the design of drugs targeting RNA and RNA–protein complexes. Int. Ed. Engl., 39, 1891–1905. [DOI] [PubMed] [Google Scholar]
- 3.Davies J., Gorini,L. and Davis,B.D. (1965) Misreading of RNA code words induced by aminoglycoside antibiotics. Mol. Pharmacol., 1, 93–106. [PubMed] [Google Scholar]
- 4.Moazed D. and Noller,H.F. (1987) Interactions of antibiotics with functional sites in 16S ribosomal RNA. Nature, 327, 389–394. [DOI] [PubMed] [Google Scholar]
- 5.Woodcock J., Moazed,D., Cannon,M., Davies,J. and Noller,H.F. (1991) Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J., 10, 3099–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirk S.R. and Tor,Y. (1999) tRNA(Phe) binds aminoglycoside antibiotics. Bioorg. Med. Chem., 7, 1979–1991. [DOI] [PubMed] [Google Scholar]
- 7.Stage T.K., Hertel,K.J. and Uhlenbeck,O.C. (1995) Inhibition of the hammerhead ribozyme by neomycin. RNA, 1, 95–101. [PMC free article] [PubMed] [Google Scholar]
- 8.Earnshaw D.J. and Gait,M.J. (1998) Hairpin ribozyme cleavage catalyzed by aminoglycoside antibiotics and the polyamine spermine in the absence of metal ions. Nucleic Acids Res., 26, 5551–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Ahsen U., Davies,J. and Schroeder,R. (1991) Antibiotic inhibition of group I ribozyme function. Nature, 353, 368–370. [DOI] [PubMed] [Google Scholar]
- 10.Zapp M.L., Stern,S. and Green,M.R. (1993) Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell, 74, 969–978. [DOI] [PubMed] [Google Scholar]
- 11.Mei H.Y., Mack,D.P., Galan,A.A., Halim,N.S., Heldsinger,A., Loo,J.A., Moreland,D.W., Sannes-Lowery,K.A., Sharmeen,L., Truong,H.N. et al. (1997) Discovery of selective, small-molecule inhibitors of RNA complexes—I. The Tat protein/TAR RNA complexes required for HIV-1 transcription. Bioorg. Med. Chem., 5, 1173–1184. [DOI] [PubMed] [Google Scholar]
- 12.Ennifar E., Paillart,J.C., Marquet,R., Ehresmann,B., Ehresmann,C., Dumas,P. and Walter,P. (2003) HIV-1 RNA dimerization initiation site is structurally similar to the ribosomal A-site and binds aminoglycoside antibiotics. J. Biol. Chem., 278, 2723–2730. [DOI] [PubMed] [Google Scholar]
- 13.Frankel A.D. (1992) Activation of HIV transcription by Tat. Curr. Opin. Gen. Devo., 2, 293–298. [DOI] [PubMed] [Google Scholar]
- 14.Jones K.A. and Peterlin,B.M. (1994) Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem., 63, 717–743. [DOI] [PubMed] [Google Scholar]
- 15.Karn J., Keen,N.J., Churcher,M.J., Aboul-ela,F., Varani,G., Hamy,F., Felder,E.R., Heizmann,G. and Kimkait,T. (1998) The HIV Tat-TAR interaction, a novel target for drug discovery. In Van der Goot,H. (ed.), Pharmacochemistry Library. Elsevier Health Sciences, Netherlands, Vol. 29, pp. 121–132. [Google Scholar]
- 16.Froeyen M. and Herdewijn,P. (2002) RNA as a target for drug design, the example of Tat-TAR interaction. Curr. Top. Med. Chem., 10, 1957–1861. [DOI] [PubMed] [Google Scholar]
- 17.Wang S., Huber,P.W., Cui,M., Czarnik,A.W. and Mei,H.Y. (1998) Binding of neomycin to the TAR element of HIV-1 RNA induces dissociation of Tat protein by an allosteric mechanism. Biochemistry, 37, 5549–5557. [DOI] [PubMed] [Google Scholar]
- 18.Faber C., Sticht,H., Schweimer,K. and Rosch,P. (2000) Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin B. J. Biol. Chem., 275, 20660–20666. [DOI] [PubMed] [Google Scholar]
- 19.Umezawa H. and Kondo,S. (1975) Ion-exchange chromatography of aminoglycoside antibiotics. Methods Enzymol., 43, 263–278. [DOI] [PubMed] [Google Scholar]
- 20.Michael K., Wang,H. and Tor,Y. (1999) Enhanced RNA binding of dimerized aminoglycosides. Bioorg. Med. Chem., 7, 1361–1371. [DOI] [PubMed] [Google Scholar]
- 21.Kierzek R., Li,Y., Turner,D.H. and Bevilacqua,P.C. (1993) 5′-Amino pyrene provides a sensitive, nonperturbing fluorescent probe of RNA secondary and tertiary structure formation. J. Am. Chem. Soc., 115, 4985–4992. [Google Scholar]
- 22.Silverman S.K., Deras,M.L., Woodson,S.A., Scaringe,S.A. and Cech,T.R. (2000) Multiple folding pathways for the P4-P6 RNA domain. Biochemistry, 39, 12103–12512.11015187 [Google Scholar]
- 23.Silverman S.K. and Cech,T.R. (1999) RNA tertiary folding monitored by fluorescence of covalently attached pyrene. Biochemistry, 38, 14224–14237. [DOI] [PubMed] [Google Scholar]
- 24.Conlan L.H. and Dupureur,C.M. (2002) Multiple metal ions drive DNA association by PvuII endonuclease. Biochemistry, 41, 14848–14855. [DOI] [PubMed] [Google Scholar]
- 25.Beebe J.A. and Fierke,C.A. (1994) Relaxation Kinetics. Academic Press, New York, NY. [Google Scholar]
- 26.Baliga R. and Crothers,D.M. (2000) On the kinetics of distamycin binding to its target sites on duplex DNA. Proc. Natl Acad. Sci. USA, 97, 7814–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alberty R.A. and Hammes,G.G. (1958) Application of the theory of diffusion-controlled reactions to enzyme kinetics. J. Phys. Chem., 62, 154–159. [Google Scholar]
- 28.Edwards T.E. and Sigurdsson,S.T. (2002) Electron paramagnetic resonance dynamic signatures of TAR RNA-small molecule complexes provide insight into RNA structure and recognition. Biochemistry, 41, 14843–14847. [DOI] [PubMed] [Google Scholar]
- 29.White J.S. and White,D.C. (1997) Source Book of Enzymes. CRC Press, Boca Raton, FL. [Google Scholar]
- 30.Aboul-ela F., Karn,J. and Varani,G. (1996) Structure of the HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res., 24, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen N.E., Brannvall,M., Virtanen,A. and Kirsebom,L.A. (1999) Inhibition of RNase P RNA cleavage by aminoglycosides. Proc. Natl Acad. Sci. USA, 96, 6155–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrix M., Priestley,E.S., Joyce,G.F. and Wong,C.-H. (1997) Direct observation of aminoglycoside-RNA interactions by surface plasmon resonance. J. Am. Chem. Soc., 119, 3641–3648. [DOI] [PubMed] [Google Scholar]
- 33.Wong C.H., Hendrix,M., Priestley,E.S. and Greenberg,W.A. (1998) Specificity of aminoglycoside antibiotics for the A-site of the decoding region of ribosomal RNA. Chem. Biol., 5, 397–406. [DOI] [PubMed] [Google Scholar]
- 34.Pelta J., Livolant,F. and Sikorav,J.-L. (1996) DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem., 271, 5656–5662. [DOI] [PubMed] [Google Scholar]
- 35.Luedtke N.W. and Tor,Y. (2000) A novel solid-phase assembly for identifying potent and selective RNA ligands. Int. Ed. Engl., 39, 1788–1790. [DOI] [PubMed] [Google Scholar]
- 36.Kirk S.R., Luedtke,N.W. and Tor,Y. (2000) Neomycin-acridine conjugate: a potent inhibitor of Rev-RRE binding. J. Am. Chem. Soc., 122, 980–981. [Google Scholar]
- 37.Leclerc F. and Cedergren,R. (1998) Modeling RNA-ligand interactions: the Rev-binding element RNA-aminoglycoside complex. J. Med. Chem., 41, 175–182. [DOI] [PubMed] [Google Scholar]
- 38.Alberts B. (1994) Molecular Biology of the Cell, 3rd Edn. Garland Publishing, New York, NY. [Google Scholar]
- 39.Netzel T.L., Zhao,M., Nafisi,K., Headrick,J., Sigman,M.S. and Eaton,B.E. (1995) Photophysics of 2′-deoxyuridine (dU) nucleosides covalently substituted with either 1-pyrenyl or 1-pyrenoyl: observation of pyrene-to-nucleoside charge-transfer emission in 5-(1-Pyrenyl)-dU. J. Am. Chem. Soc., 117, 9119–9128. [Google Scholar]
- 40.Manoharan M., Tivel,K.L., Zhao,M., Nafisi,K. and Netzel,T.L. (1995) Base-sequence dependence of emission lifetimes for D141018–30–6NA oligomers and duplexes covalently labeled with pyrene: relative electron-transfer quenching efficiencies of A, G, C and T nucleosides toward pyrene. J. Phys. Chem., 99, 17461–17472. [Google Scholar]
- 41.Hermann T. and Westhof,E. (1999) Docking of cationic antibiotics to negatively charged pockets in RNA folds. J. Med. Chem., 42, 1250–1261. [DOI] [PubMed] [Google Scholar]
- 42.Lohman T.M., deHaseth,P.L. and Record,M.T.,Jr (1980) Pentalysine-deoxyribonucleic acid interaction: a model for the general effects of ion concentrations on the interactions of proteins with nucleic acids. Biochemistry, 19, 3522–3530. [DOI] [PubMed] [Google Scholar]
- 43.Record M.T. Jr, Anderson,C.F. and Lohman,T.M. (1978) Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening and ion effects on water activity. Q. Rev. Biophys., 11, 103–178. [DOI] [PubMed] [Google Scholar]
- 44.Jiang L., Majumdar,W.H., Jaishree,T.J., Weijun,X. and Patel,D.J. (1999) Saccharide-RNA recognition in a complex formed between neomycin B and and RNA aptamer. Structure, 7, 817–827. [DOI] [PubMed] [Google Scholar]
- 45.Kaul M. and Pilch,D. (2002) Thermodynamics of aminoglycoside-rRNA recognition: the binding of neomycin-class aminoglycosides to the A site of 16S rRNA. Biochemistry, 41, 7695–7706. [DOI] [PubMed] [Google Scholar]
- 46.Carey J. and Uhlenbeck,O.C. (1983) Kinetic and thermodynamic characterization of the R17 coat protein-ribonucleic acid interaction. Biochemistry, 22, 2610–2615. [DOI] [PubMed] [Google Scholar]
- 47.Tor Y., Hermann,T. and Westhof,E. (1998) Deciphering RNA recognition: aminoglycoside binding to the hammerhead ribozyme. Chem. Biol., 5, R277–283. [DOI] [PubMed] [Google Scholar]
- 48.Michael K. and Tor,Y. (1998) Designing novel RNA binders. Chem. Eur. J., 4, 2091–2098. [Google Scholar]
- 49.Hermann T. (2002) Rational ligand design for RNA: the role of static structure and conformational flexibility in target recognition. Biochimie, 84, 869–875. [DOI] [PubMed] [Google Scholar]
- 50.Daniels P.J.L., Mallams,A.K., McCombie,S.W., Morton,J.B., Nagabhushan,T.L., Rane,D.F., Reichert,P. and Wright,J.J. (1981) Semisynthetic aminoglycoside antibacterials. Part 11. Solution conformations of semisynthetic and naturally occurring aminoglycoside antibiotics. J. Chem. Soc. Perkin Trans., 1, 2209–2227. [Google Scholar]
- 51.Imberty A. and Perez,S. (2000) Structure, conformation and dynamics of bioactive oligosaccharides: theoretical approaches and experimental validations. Chem. Rev., 100, 4567–4588. [DOI] [PubMed] [Google Scholar]
- 52.Yamana K., Iwase,R., Furutani,S., Tsuchida,H., Zako,H., Yamaoka,T. and Murakami,A. (1999) 2′-Pyrene modified oligonucleotide provides a highly sensitive fluorescent probe of RNA. Nucleic Acids Res., 27, 2387–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kostenko E., Dobrikov,M., Pyshnyi,D., Petyuk,V., Komarova,N., Vlassov,V. and Zenkova,M. (2001) 5′-Bis-pyrenylated oligonucleotides displaying excimer fluorescence provide sensitive probes of RNA sequence and structure. Nucleic Acids Res., 29, 3611–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]