Abstract
The chloroplast envelope plays critical roles in the synthesis and regulated transport of key metabolites, including intermediates in photosynthesis and lipid metabolism. Despite this importance, the biogenesis of the envelope membranes has not been investigated in detail. To identify the determinants of protein targeting to the inner envelope membrane (IM), we investigated the targeting of the nucleus-encoded integral IM protein, atTic40. We found that pre-atTic40 is imported into chloroplasts and processed to an intermediate size (int-atTic40) before insertion into the IM. Int-atTic40 is soluble and inserts into the IM from the internal stromal compartment. We also show that atTic40 and a second IM protein, atTic110, can target and insert into isolated IM vesicles in vitro. Collectively, our experiments are consistent with a “postimport” mechanism in which the IM proteins are first imported from the cytoplasm and subsequently inserted into the IM from the stroma.
Introduction
Chloroplasts are subdivided by three noncontiguous membrane systems into at least six suborganellar compartments that serve to segregate and organize several essential metabolic functions. The chloroplast outer and inner envelope membranes form the organelle boundary and effectively segregate chloroplast and cytoplasmic metabolism by controlling metabolite and ion transport (Joyard et al., 1998), whereas the internal thylakoid membrane performs the light harvesting and photophosphorylation reactions of photosynthesis. The vast majority of chloroplast membrane proteins are encoded in the nucleus (Bedard and Jarvis, 2005; Kessler and Schnell, 2006). As a result, the biogenesis of these membranes relies on the selective targeting and insertion of hundreds of proteins from their site of synthesis in the cytoplasm.
Except for the translocon component, Toc75, outer envelope membrane proteins are targeted directly from the cytoplasm to the membrane via targeting signals contained within and adjacent to their transmembrane helices (Kessler and Schnell, 2006). The majority of thylakoid membrane proteins contain cleavable N-terminal transit peptides that target them across the envelope. Import is mediated by the same translocon complexes within the outer (Toc) and inner (Tic) envelope membranes that mediate the import of soluble proteins (Schnell, 1998; Jarvis and Robinson, 2004). Upon import, thylakoid membrane proteins are released into the soluble stroma and processed to remove their transit peptides, and intrinsic secondary targeting signals direct them to the thylakoid (Jarvis and Robinson, 2004). The protein translocons at the thylakoid are homologous to protein export translocons found in prokaryotic cytoplasmic membranes (e.g., Sec, SRP, and TAT pathways), indicating conservation in these targeting systems from the original bacterial endosymbiont (Keegstra and Cline, 1999; Jarvis and Robinson, 2004).
The limited studies on targeting to the chloroplast inner envelope membrane (IM) leave open the question of whether nucleus-encoded proteins insert into the membrane during the import process via a stop-transfer mechanism or target to the membrane after the completion of import by inserting from the stromal side of the membrane. Two classes of nucleus-encoded integral IM proteins are known to exist. The first class contains proteins that lack cleavable transit peptides (Miras et al., 2002; Nada and Soll, 2004). Although the targeting determinants for these proteins have not been completely defined, they appear not to use the Toc–Tic translocons for import.
The second, larger class consists of IM proteins, is synthesized with cleavable transit peptides, and initially engages the Toc–Tic import machinery. The abundant inner membrane protein, Tic110, is a member of this class (Lubeck et al., 1996, 1997). The analysis of deletion mutants and fusion proteins indicates that the N-terminal region of Tic110, including its two transmembrane segments, is required for targeting to the IM (Lubeck et al., 1997). Interestingly, a fusion protein containing the N-terminal targeting determinants of Tic110 transiently accumulated as a soluble intermediate in the stroma before inserting into the IM (Lubeck et al., 1997). In a subsequent study, dominant-negative mutants of Tic110 that disrupt Tic complex formation resulted in the accumulation of normal Tic110 in the stroma in vivo (Inaba et al., 2005). Consistent with the fusion protein studies, chloroplasts isolated from these mutants transiently accumulated a soluble, mature form of Tic110 in in vitro import assays (Inaba et al., 2005). Collectively, the two studies suggested that Tic110 inserts into the membrane from the stroma after import.
In contrast to the studies with Tic110, in vitro import studies with several chloroplast polytopic membrane transporters led to the conclusion that IM proteins do not use stromal intermediates en route to the membranes (Li et al., 1992; Brink et al., 1995; Knight and Gray, 1995). In one case, soluble forms of fusion proteins to the envelope phosphate translocator were observed but were not shown to represent targeting intermediates (Knight and Gray, 1995). These results suggested that chloroplast IM proteins use a stop-transfer mechanism of targeting that results in direct insertion of the proteins into the inner membrane during import through the Toc–Tic system.
In this report, we wished to examine the process of targeting to the chloroplast IM by studying the import and insertion of a simple IM protein. To this end, we investigated the targeting of pre-atTic40, a nucleus-encoded chloroplast inner membrane protein with a single transmembrane helix (Stahl et al., 1999; Chou et al., 2003). We show that the import and membrane insertion of native pre-atTic40 involves a size intermediate that inserts into the inner membrane after import from the cytoplasm through the Toc–Tic machinery. Furthermore, we demonstrate that atTic40 and atTic110 can insert directly and selectively into isolated IM vesicles. These data are consistent with a pathway for the targeting of IM proteins that is independent of their import from the cytoplasm.
Results
Import of pre-atTic40 involves a processing intermediate
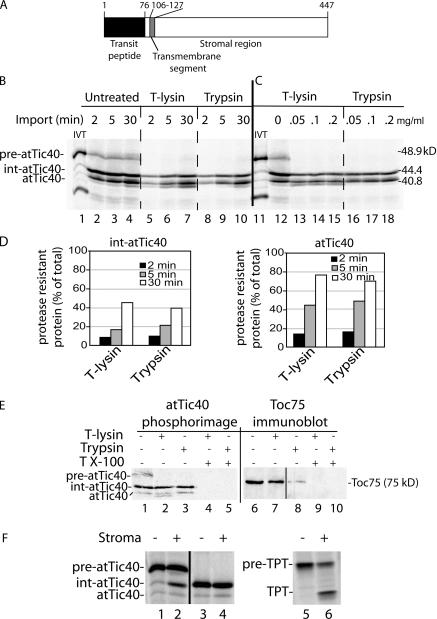
A schematic diagram of the structure of Arabidopsis thaliana pre-atTic40 is shown in Fig. 1 A. The protein contains a 76-amino-acid transit peptide that is removed upon import into chloroplasts. The single transmembrane helix is located within the N-terminal region between amino acids 106 and 127 of the preprotein with a C-terminal ∼35-kD soluble region extending into the stroma (Stahl et al., 1999; Chou et al., 2003). As a first step in examining the targeting of pre-atTic40, we performed a time course of import of in vitro–translated [35S]-labeled pre-atTic40 into isolated pea chloroplasts (Fig. 1 B, lanes 2–4). In addition to pre-atTic40, two other major forms of atTic40 are apparent in the import assays. The polypeptide with the highest mobility was confirmed to be mature atTic40 by comparing its mobility with that of endogenous atTic40 as detected by immunoblotting (unpublished data). In addition to mature atTic40, a major intermediate-sized form of the protein (int-atTic40) with a mobility between pre-atTic40 and atTic40 is observed in the import assays.
Figure 1.
atTic40 targeting involves a size intermediate. (A) Schematic representation of the structure of pre-atTic40. The numbers indicate the positions of amino acid residues within the primary structure. (B) In vitro–translated [35S]pre-atTic40 was imported into isolated pea chloroplasts for the times indicated. Equivalent portions of each import reaction were treated in the absence (untreated) or presence of 100 μg/ml thermolysin (T-lysin) or 50 μg/ml trypsin to remove protein that was not fully imported. The total chloroplast fractions from each treatment and time point were analyzed by SDS-PAGE and phosphorimaging. (C) [35S]pre-atTic40 was imported into isolated pea chloroplasts for 5 min. Equivalent portions were treated with the indicated concentrations of thermolysin or trypsin. (D) Quantitative analysis of the levels of protease-resistant int-atTic40 and mature atTic40 from the import reactions in B. (E) Chloroplasts from a 5-min import reaction were treated with 200 μg/ml trypsin or thermolysin in the presence or absence of 1% Triton X-100 as indicated. The samples were analyzed by SDS-PAGE and phosphorimaging to detect imported [35S]pre-atTic40 (left) or immunoblotting to detect Toc75 (right). (F) Pre-atTic40 is processed to int-atTic40 by the SPP. In vitro–translated pre-atTic40 (lanes 1 and 2), int-atTic40 (lanes 3 and 4), or pretriose phosphate translocator (pre-TPT; lanes 5 and 6) was incubated in the presence (+) or absence (−) of a chloroplast stromal extract containing the SPP.
As a first step in defining the relationship between int-atTic40 and mature atTic40, we examined their suborganellar location. To this end, chloroplasts from each time point in import were treated with thermolysin or trypsin (Fig. 1 B). Thermolysin has been shown to digest proteins that are exposed at the chloroplast surface, but it does not penetrate the outer membrane (Cline et al., 1984). In contrast, trypsin can penetrate the outer membrane and access the intermembrane space, digesting envelope proteins that are not protected by the inner membrane (Cline et al., 1981, 1984; Jackson et al., 1998). Fig. 1 B demonstrates that mature atTic40 accumulates with time and becomes increasingly insensitive to either protease, consistent with targeting and insertion in the IM (Fig. 1 D). Similarly, the portion of int-atTic40 that is insensitive to protease digestion increases with time, rising from 8–10% at 2 min to 40–45% of the total int-atTic40 at 30 min (Fig. 1 D). Toc75, a protein deeply imbedded in the outer membrane, is not digested with thermolysin but is nearly completely degraded with trypsin (Fig. 1 E), confirming the differential accessibility of the two proteases to the envelope compartments. These data suggest that int-atTic40, like atTic40, accumulates inside the inner membrane, where neither thermolysin nor trypsin can access the polypeptide. Both pea and A. thaliana chloroplasts gave similar patterns of import, confirming that the intermediate-sized product was a true import product and not an artifact of the heterologous import system (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200605162/DC1). Pea chloroplasts were used for all subsequent experiments.
To ensure that the lack of atTic40 and int-atTic40 proteolysis was not a result of incomplete digestion, samples from the 5-min time point in import were treated with a range of thermolysin and trypsin concentrations (Fig. 1 C). Increasing the protease concentrations had no effect on the proportions of protease-insensitive atTic40 or int-atTic40. Furthermore, the addition of Triton X-100 to the samples before protease treatment resulted in complete degradation of int-atTic40 and atTic40 (Fig. 1 E), indicating that the proteins are not intrinsically protease resistant. These data confirm that both forms are localized inside the inner membrane.
Although atTic40 and int-atTic40 accumulate inside the chloroplast with time, a portion of each population is sensitive to both proteases. The highest levels of sensitive forms are observed at early time points in import (Fig. 1 D). Their sensitivity to both proteases indicates that they are exposed at the surface of the chloroplast and therefore appear to be in the process of import into the organelle. These forms are similar to import intermediates observed with stromal preproteins that are captured within the Toc–Tic machinery (Waegemann and Soll, 1991; Olsen and Keegstra, 1992; Schnell and Blobel, 1993).
To obtain additional evidence that int-atTic40 accessed the stroma, we tested whether the stromal processing peptidase (SPP) that cleaves the transit peptides of soluble proteins (Lamppa and Abad, 1987; Abad et al., 1989) was involved in converting pre-atTic40 to the intermediate form. Analysis of the pre-atTic40 sequence identifies a potential SPP processing site after amino acid 42 of the transit peptide (Emanuelsson et al., 1999). Incubation of in vitro–translated pre-atTic40 with a stromal extract results in the conversion of 35% of the precursor to an intermediate-size form coincident in size with int-atTic40 (Fig. 1 F). The processing by SPP is likely to occur at the predicted cleavage site (residue 42) because the mobility of a truncated form of pre-atTic40 lacking the first 42 residues is identical to int-atTic40 (Fig. S1 B). Furthermore, the truncated form is not cleaved by the stromal extract (Fig. 1 F), eliminating the possibility of processing at another site. As a control, we also tested cleavage of the precursor to the triose-phosphate translocator (pre-TPT; Knight and Gray, 1995). As expected, 50% of pre-TPT was converted to its mature form in the assay (Fig. 1 F). These data provide additional evidence that int-atTic40 is exposed to the stroma before IM insertion and processing to mature atTic40.
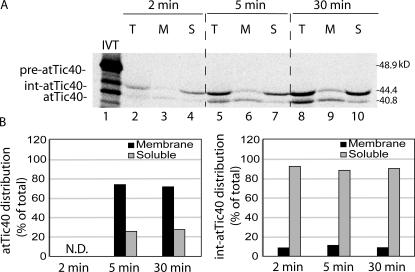
To determine if fully imported int-atTic40 and mature atTic40 had reached the IM, we fractionated chloroplasts from 2-, 5-, and 30-min import reactions. The samples were treated with trypsin to remove external or partially imported atTic40, osmotically lysed and separated into membrane and soluble fractions. As expected, the majority of mature atTic40 (∼75%) partitioned with the membrane fraction at the 5-min and 30-min time points (Fig. 2 B, left). In contrast, 88–94% of int-atTic40 was not associated with the membrane at the three time points (Fig. 2 B, right). Collectively, the data in Figs. 2 and 3 suggest that the protease-insensitive form of int-Tic40 is soluble and located in the chloroplast stroma.
Figure 2.
The int-atTic40 accumulates in a chloroplast soluble compartment. (A) [35S]pre-atTic40 was imported into isolated pea chloroplasts for the times indicated. After import, the chloroplasts were treated with 50 μg/ml trypsin and reisolated, and the total fraction (T) was separated into membrane (M) or soluble (S) fractions by osmotic lysis. The samples were analyzed by SDS-PAGE and phosphorimaging. Lane 1 contains 20% of [35S]-labeled pre-atTic40 (IVT) added to the original import reaction. (B) Quantitative analysis of the distribution of mature atTic40 (left) or int-atTic40 (right) between the soluble and membrane fractions of chloroplasts. N.D. indicates that the sample was below accurate detection levels.
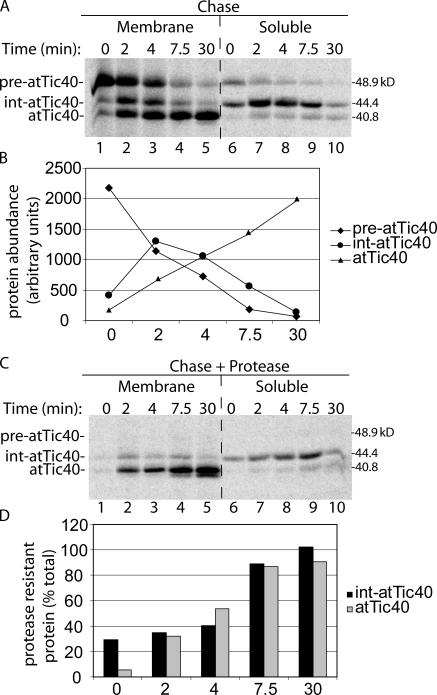
Figure 3.
Int-atTic40 is an intermediate in the import and membrane targeting of mature atTic40. (A) [35S]pre-atTic40 was incubated with isolated chloroplasts in the presence of 100 μM ATP to promote formation of an early import intermediate. The chloroplasts were isolated and incubated in the presence of 2 mM ATP to promote import of the bound pre-atTic40 for the times indicated (chase). The chloroplasts were separated into membrane and soluble fractions by osmotic lysis and analyzed by SDS-PAGE and phosphorimaging. (B) Quantitative analysis of the total amounts of [35S]-labeled pre-atTic40, int-atTic40, and mature atTic40 in the combined soluble and membrane fractions from A. (C) Samples from the same import experiment in A, but the intact chloroplasts were treated with 100 μg/ml thermolysin before separation into membrane and soluble fractions. (D) Quantitative analysis of the percentage of protease-resistant int-atTic40 or mature atTic40 (from C) relative to the total amount of each species (from A) at each time point in the import experiment. The samples in A and C were generated from the same import experiment and were analyzed on the same SDS-PAGE gel.
Int-atTic40 is the precursor to mature atTic40
To confirm that int-atTic40 is a true intermediate on the pathway of IM targeting, we tested whether int-atTic40 was the precursor to mature, membrane-integrated atTic40. For this experiment (Fig. 3 A), chloroplasts were incubated with pre- atTic40 in the presence of 0.1 mM ATP to form an early import intermediate that is inserted in the Toc–Tic machinery but has not fully crossed the envelope (pulse; Olsen et al., 1989; Olsen and Keegstra, 1992). The chloroplasts were isolated to remove any unbound translation product (Fig. 3 A, lane 1) and resuspended under import conditions to promote import of the early intermediate. Chloroplasts from each time point during the chase were lysed and separated into soluble and membrane fractions. As expected, bound pre-atTic40 was quantitatively converted to mature atTic40 after a 30-min chase (Fig. 3 A). Both forms were predominantly associated with the membrane fraction. The abundance of total int-atTic40 peaks at 2 min of chase and then largely disappears by 30 min (Fig. 3 B). The peak of int-atTic40 follows the decrease in pre-atTic40 and precedes the accumulation of mature atTic40 (Fig. 3 B), consistent with it representing an intermediate in the conversion of pre- atTic40 to atTic40. Furthermore, int-atTic40 accumulates in the soluble fraction of chloroplasts before conversion to atTic40 (Fig. 3 A). Treatment of the chloroplasts from each time point in the pulse-chase experiment with thermolysin demonstrates that protease-resistant forms of both int-atTic40 and mature atTic40 accumulate during the chase (Fig. 3 C). As expected, membrane-integrated atTic40 attains 90% protease resistance after the 30-min chase (Fig. 3 D). The soluble int-atTic40 is almost completely protease resistant at 7.5 min into the chase (Fig. 3 D), confirming that the intermediate is passing through the stroma en route to its insertion into the membrane.
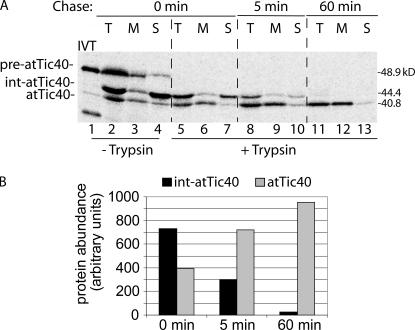
To test whether int-atTic40 was the direct precursor to atTic40, we performed a variation of the pulse-chase experiment. For this assay, isolated chloroplasts were incubated with pre-atTic40 for 5 min under import conditions to accumulate soluble int-atTic40 (Fig. 4 A). The chloroplasts were chilled to stop the import reaction and were treated with trypsin to remove pre- atTic40 and int-atTic40 that had not been completely imported into the organelle (Fig. 4 A). The treated chloroplasts were isolated to remove the protease and incubated again under import conditions for 5 or 60 min (Fig. 4 A). The soluble int-atTic40 that accumulated after 5 min of import could be quantitatively converted to membrane-integrated mature atTic40 by the additional incubation (Fig. 4 B). On the basis of these results, we conclude that the targeting of atTic40 involves a soluble intermediate that targets to the IM after import into the stroma.
Figure 4.
Soluble int-atTic40 is the immediate precursor to membrane-inserted atTic40. (A) [35S]pre-atTic40 was imported into isolated pea chloroplasts for 5 min. The chloroplasts were treated with trypsin, reisolated, and incubated under import conditions for the times indicated. After import, the total chloroplasts (T) were separated into soluble (S) and membrane (M) fractions. Lanes 2–4 contain total, membrane, and soluble fractions from the 5-min import reaction before protease treatment and subsequent incubation. (B) Quantitative analysis of the levels of total int-atTic40 and mature atTic40 at each time point in the postimport incubation of trypsin-treated chloroplasts.
The intermediate region of int-atTic40 is not essential for membrane targeting
A previous study on the import of pea Tic40 into isolated chloroplasts did not report a size intermediate in IM targeting (Stahl et al., 1999). Although we observed a soluble intermediate with characteristics similar to int-atTic40 when we repeated import with pea pre-Tic40 (unpublished data), the size of the intermediate was only slightly larger than the mature protein, indicating that the intermediate processing site is distinct from that in pre-atTic40. The difference in processing between pea and A. thaliana Tic40 raised questions about the role of the intermediate processing. Therefore, we tested whether the C-terminal region of the atTic40 transit peptide plays a role in IM targeting.
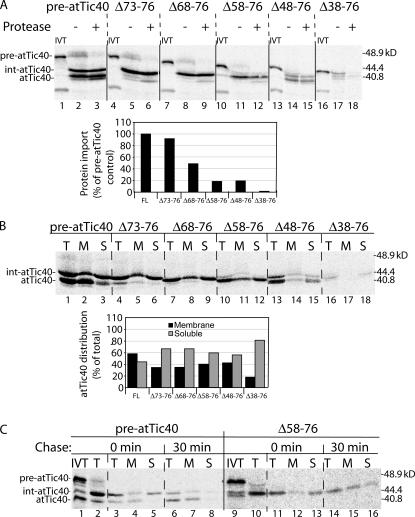
We generated a series of increasingly larger deletion mutants in the intermediate sequence (residues 43–76; Fig. 1 A) and examined their import and insertion properties. All deletions disrupt the putative processing site that gives rise to mature atTic40. Elimination of the four C-terminal residues of the transit peptide does not significantly affect import efficiency or processing to an intermediate size (Fig. 5 A). However, larger deletions dramatically reduce import efficiency in addition to inhibiting processing to the mature size (Fig. 5 A).
Figure 5.
The intermediate region of the int-atTic40 transit peptide is not required for membrane targeting. (A) [35S]-labeled pre-atTic40 (FL) and deletion mutants spanning the indicated amino acids of pre-atTic40 were imported into isolated chloroplasts for 30 min and subsequently treated in the presence (+) or absence (−) of trypsin. IVT, 20% of the in vitro–translated protein used in each reaction. The graph presents quantitative analysis of the import efficiency of each construct presented as a percentage of the amount of import observed with wild-type pre-atTic40. (B) Analysis of total (T), soluble (S), and membrane (M) fractions of chloroplasts from 30-min import reactions of [35S]-labeled pre-atTic40 and the transit peptide deletion mutants. The graph presents quantitative analysis of the distribution of the total atTic40 species between membrane and soluble fractions for each construct. (C) [35S]pre-atTic40 and [35S]pre-atTic40–Δ58-76 were imported into isolated pea chloroplasts for 5 min, treated with trypsin, reisolated, and incubated under import conditions for the times indicated. The total chloroplasts were separated into soluble and membrane fractions by alkaline extraction. Lanes 2 and 10 contain total (T) chloroplasts from the 5-min import reaction before protease treatment and subsequent incubation. Lanes 1 and 9 contain 20% of the translation product used for each of the import reactions.
Although import efficiency is reduced, deletions up to Δ58-76 appeared to target and insert into the IM, albeit with somewhat lower efficiencies than pre-atTic40 (Fig. 5 B). The effects of deletions larger than Δ58-76 on IM targeting could not be determined because of the combination of poor import efficiency and aberrant cleavage (Fig. 5 A), presumably because of disruption of the SPP processing site. To further ensure that the soluble forms of the deletions were productive intermediates in the targeting pathway, we performed a chase experiment similar to that shown in Fig. 4 using the Δ58-76 mutant (Fig. 5 C). The Δ58-76 soluble intermediate can be chased to the membrane with a 30-min incubation, similar to authentic int-atTic40. Collectively, these results indicate that the intermediate sequence of the transit peptide participates in import but is not essential for membrane insertion.
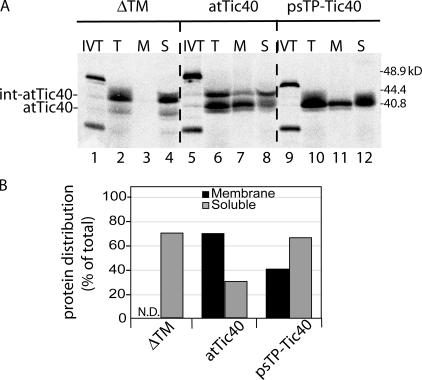
To further explore the determinants for pre-atTic40 targeting, we replaced its transit peptide with that of the small subunit of rubisco (psTP-Tic40). The import efficiency of this construct was comparable to pre-atTic40, but the protein was converted to mature atTic40 directly with no apparent intermediate processing (Fig. 6 A). Remarkably, the chimera targeted to the IM, albeit with approximately one half the efficiency of pre-atTic40 (Fig. 6 B). These results are consistent with the conclusion that the int-atTic40 intermediate sequence is not required for membrane integration but might facilitate the process. Finally, we tested whether the transmembrane helix of atTic40 was required for IM targeting. As expected, deletion of the transmembrane region (ΔTM-atTic40) had little effect on import but completely eliminated membrane integration (Fig. 6, A and B).
Figure 6.
The transmembrane helix of atTic40 is required for membrane targeting. (A) [35S]-labeled pre-atTic40 (atTic40), mature atTic40 fused to the transit peptide of SSU (psTP-Tic40), or a deletion mutant of pre-atTic40 lacking its transmembrane segment (ΔTM) were imported into chloroplasts for 30 min. After import, the chloroplasts were treated with thermolysin, reisolated, and total (T), membrane (M), or soluble (S) fractions were analyzed by SDS-PAGE and phosphorimaging. IVT, 20% of the in vitro–translated protein used in each reaction. (B) Quantitative analysis of distribution of each protein between the membrane and soluble fractions of chloroplasts after import. N.D. indicates that the sample was below accurate detection levels.
Int-atTic40 inserts into isolated IM vesicles
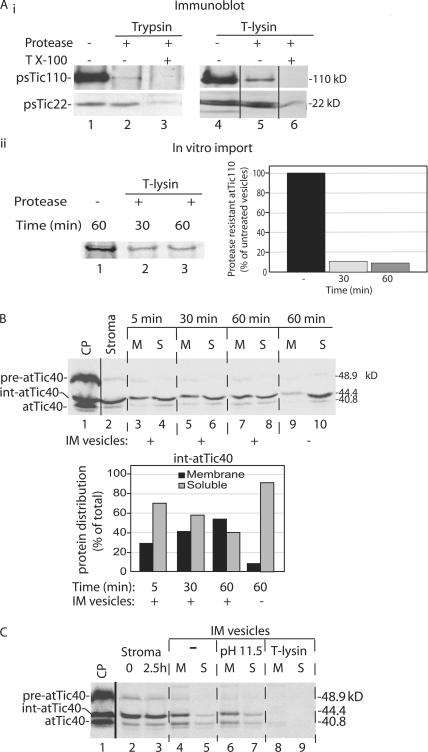
To test directly whether membrane insertion is independent of import, we studied the ability of int-atTic40 to integrate into isolated IM vesicles. Inside-out IM vesicles were purified from isolated chloroplasts using previously published procedures (Keegstra and Yousif, 1986; Young and McCarty, 1993). To ensure that the majority of the vesicles were inside out, thereby exposing the inner face of the IM to the external buffer, we tested the protease sensitivity of the IM marker proteins psTic110 and psTic22 (Cline et al., 1985; Shingles and McCarty, 1995; Fig. 7 A). The bulk of psTic110 (∼95 kD) extends into the stroma (Kessler and Blobel, 1996; Jackson et al., 1998), whereas psTi22 is bound to the outer face of the IM in the intermembrane space (Kouranov et al., 1999). The majority of psTic110 is degraded by either trypsin or thermolysin, whereas psTic22 is resistant to both proteases, indicating that the isolated vesicles are predominantly inside out (Fig. 7 A i). Disruption of the vesicles with Triton X-100 resulted in complete degradation of psTic110 and psTic22 (Fig. 7 A i). To quantify the proportion of inside- out vesicles in the population, we imported radiolabeled atTic110 into chloroplasts and treated isolated IM vesicles from these chloroplasts with thermolysin (Fig. 7 A ii). Greater than 90% of the radiolabeled atTic110 was degraded within 30 min, suggesting that at least 90% of the vesicle population was in the inside-out orientation.
Figure 7.
Int-atTic40 can insert into isolated inverted inner membrane vesicles. (A) Isolated IM vesicles are inside out. (i) IM vesicles (20 μg protein) were treated in the presence or absence of trypsin (1 μg trypsin/mg IM protein) or thermolysin (10 μg thermolysin/mg IM protein) on ice for 30 min. The samples were analyzed by SDS-PAGE and immunoblotting with psTic110 or psTic22 antiserum. (ii) IM vesicles were isolated from chloroplasts containing imported [35S]atTic110 and treated in the presence or absence of thermolysin as in panel i for the times indicated. The levels of [35S]atTic110 were detected and quantified using SDS-PAGE and phosphorimaging. (B) [35S]int-atTic40 from a stromal extract binds to IM vesicles. A stromal extract (50 μg protein) containing [35S]int-atTic40 was incubated with isolated IM vesicles (30 μg protein) for the times indicated. The quantitative analysis presents the distribution of int-atTic40 between soluble (S) and membrane (M) fractions at each time point in the insertion reaction. Lane 1 contains a sample of the total chloroplasts (CP) used as the source for the stromal extract. Lane 2 contains a sample of stromal extract equivalent to that added to the IM vesicle insertion reaction. Lanes 9 and 10 present the distribution of int-atTic40 from a reaction that lacked added IM vesicles. (C) IM vesicles from an int-atTic40 insertion reaction similar to that in B were incubated in the absence (−) or presence of alkaline buffer to extract peripheral proteins (pH 11.5) or with thermolysin to test int-atTic40 topology (T-lysin). The samples were separated into membrane and soluble fractions after the treatments.
As an initial substrate for the vesicle insertion reaction, a soluble stromal extract containing int-atTic40 (Fig. 7 B, lane 2) was isolated from chloroplasts after a 5-min pre-atTic40 import reaction (Fig. 7 B, lane 1). This extract was incubated with isolated IM vesicles, and the association of int-atTic40 and mature atTic40 with the vesicles was assayed by differential centrifugation. As evident in Fig. 7 B, int-atTic40 bound to IM vesicles. Binding was time dependent, with the majority of int-atTic40 (55%) associated with the vesicles during the incubation (Fig. 7 B, graph). A small but substantial fraction of the int-atTic40 is converted to a form with a mobility identical to mature atTic40, suggesting that a portion of int-atTic40 is processed to atTic40 upon association with vesicles. Extending the incubation to 2.5 h increased the proportion of int-atTic40 that was converted to atTic40 in the presence but not the absence of IM vesicles (Fig. 7 C). Extraction of the vesicles with alkaline buffer demonstrates that the membrane-associated forms of atTic40 are integrated into the membrane bilayer (Fig. 7 C). Furthermore, treatment of the vesicles with thermolysin after the insertion reaction results in complete degradation of the atTic40 proteins (Fig. 7 C), indicating that they correctly inserted with the bulk of their sequences exposed at the inner face of the IM.
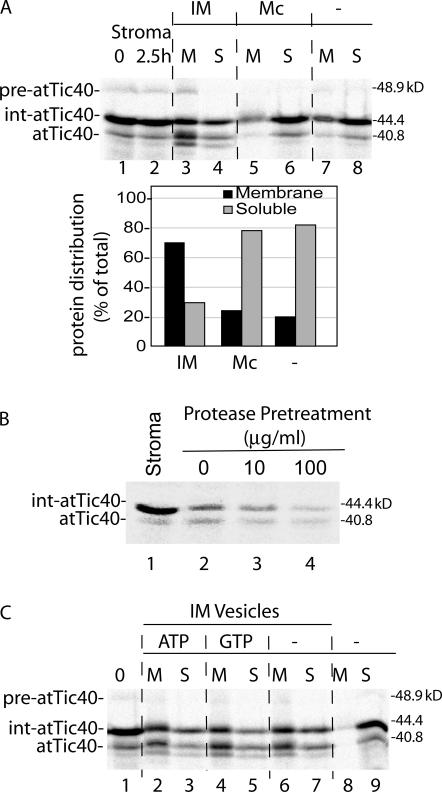
As a control for vesicle targeting specificity, we tested the binding and insertion of int-atTic40 to canine pancreatic microsomal membranes. Binding to the heterologous membranes was insignificant when compared with control reactions lacking membranes (Fig. 8 A). As an additional control, we tested the protein dependence of int-atTic40 insertion into IM vesicles by treating the IM vesicles with varying concentrations of thermolysin before the insertion reaction (Fig. 8 B). The protease treatments dramatically reduced the binding and processing of int-atTic40 in IM vesicles, demonstrating the requirement of proteinaceous components at the IM for insertion.
Figure 8.
The insertion of int-atTic40 into IM vesicles is selective and requires proteinaceous components at the membrane. (A) Int-atTic40 does not bind to canine pancreatic microsomes. A stromal extract containing [35S]int-atTic40 was incubated in the absence (−) or presence of isolated IM vesicles (30 μg protein) or canine pancreatic microsomes (Mc; 30 μg protein) for 2.5 h. After the reaction, the samples were separated into membrane (M) and soluble (S) fractions. The quantitative analysis presents the distribution of combined [35S]-labeled int-atTic40 and atTic40 between the membrane and soluble fractions from each reaction. (B) Int-atTic40 insertion into IM vesicles relies on protease-sensitive membrane components. Isolated IM vesicles were treated with the indicated concentrations of thermolysin before incubation with stromal extract containing [35S]int-atTic40. The fraction of [35S]int-atTic40 that associated with the membranes after alkaline extraction is shown in lanes 2–4. Lane 1 contains a sample of stromal extract equivalent to that added to each reaction. (C) Int-atTic40 targeting to IM vesicles does not require exogenous nucleoside triphosphates. A dialyzed stromal extract containing [35S]int-atTic40 was incubated with IM vesicles in the absence (−) or presence of 2 mM ATP or GTP as indicated. The reactions were separated into membrane and soluble fractions by alkaline extraction and analyzed by SDS-PAGE and phosphorimaging. Lane 1 contains a sample of stromal extract equivalent to that added to each reaction. Lanes 8 and 9 contains stromal extract that was incubated in the absence of IM vesicles and subsequently treated with alkaline buffer.
We also examined the energy dependence of the insertion reaction (Fig. 8 C). The stromal extract containing atTic40 was dialyzed to remove free nucleotides before the insertion reaction. As shown in Fig. 8 C, int-atTic40 bound and inserted into vesicles with similar efficiencies in the absence of added nucleoside triphosphates or in the presence of ATP or GTP. This observation suggests that direct insertion into the IM does not require nucleotide hydrolysis.
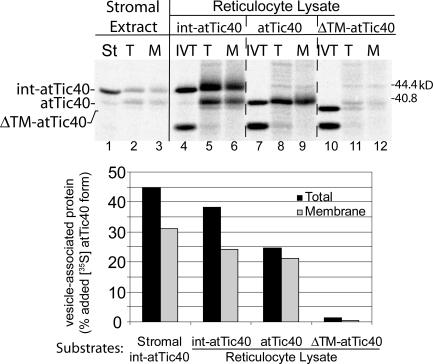
Finally, we examined the role of stromal factors in int-atTic40 targeting by testing whether in vitro–translated forms of atTic40 could target directly to vesicles. We compared the targeting of in vitro–translated int-atTic40, atTic40, or the ΔTM-atTic40 mutant to stromal int-atTic40 (Fig. 9). Remarkably, in vitro–translated int-atTic40 and atTic40 both associated with vesicles and inserted into the membranes with efficiencies only slightly lower than that observed for stromal int-atTic40 (Fig. 9, graph). Furthermore, in vitro–translated int-atTic40 was partially processed to mature atTic40. As expected, in vitro–translated mature atTic40 was not processed (Fig. 9). These data confirm that int-atTic40 processing to mature atTic40 occurs upon association with the IM. The ΔTM-atTic40 mutant showed no significant association or insertion into the membrane vesicles (Fig. 9), confirming the selectivity of the insertion assay. Collectively, these results indicate that in vitro–translated int-atTic40 is capable of inserting into isolated IM vesicles, suggesting that specific stromal factors are not required for the targeting reaction.
Figure 9.
atTic40 targeting to IM vesicles does not require stromal components. Stromal extract containing [35S]int-atTic40 or in vitro–translated [35S]int-atTic40, [35S]atTic40, or [35S]ΔTM-atTic40 (reticulocyte lysate) was incubated with isolated IM vesicles for 2.5 h as described in Fig. 7. The total (T) and alkaline-resistant fraction (M) that associates with IM vesicles is shown. IVT, 20% of the in vitro–translation products added to each targeting assay.
atTic110 also inserts into isolated IM vesicles
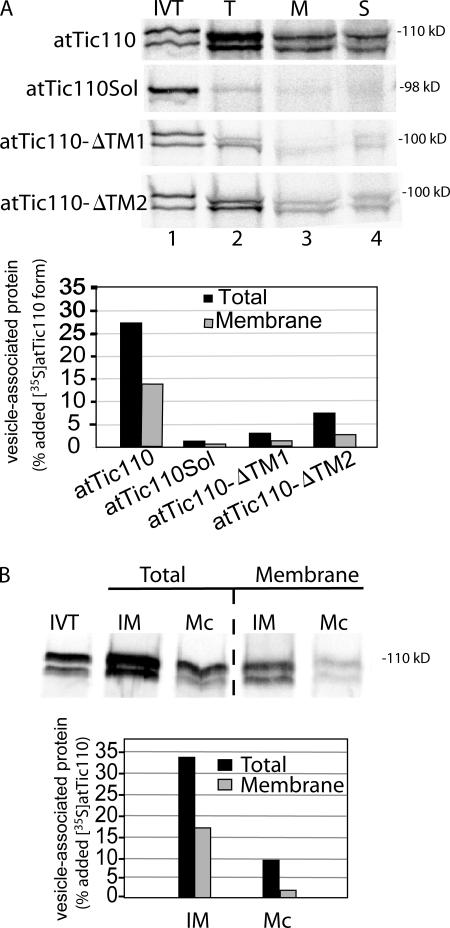
Previous studies on the import of Tic110 suggested that this IM protein also utilizes a soluble targeting intermediate (Lubeck et al., 1997; Inaba et al., 2005). Therefore, we examined the ability of in vitro–translated atTic110 to insert into IM vesicles. Fig. 10 A demonstrates that mature atTic110 binds to IM vesicles, and ∼50% of the bound protein is integrated into the membrane. As controls, we tested the binding and insertion of atTic110 lacking one (atTic110-ΔTM1 or atTic110-ΔTM2) or both of its transmembrane segments (atTic110Sol; Fig. 10 A). Consistent with previous in vitro import experiments, the binding and insertion of the deletion mutants was dramatically lower than for full-length atTic110. atTic110 showed considerably lower binding and no appreciable insertion into pancreatic microsomal membranes (Fig. 10 B), demonstrating the membrane specificity of the reaction. Overall, the data with atTic40 and atTic110 demonstrate that these proteins can insert directly into the IM from a soluble state independent of protein import from the cytoplasm.
Figure 10.
The inner membrane protein, atTic110, also targets to isolated IM vesicles. (A) In vitro–translated [35S]-labeled atTic110 or atTic110 lacking one (atTic110-ΔTM1 or atTic110-ΔTM2) or both (atTic110Sol) transmembrane segments was incubated with isolated IM vesicles. The vesicles were recovered by differential centrifugation (T) and separated into membrane (M) and soluble (S) fractions by alkaline extraction. The quantitative analysis presents the amount of total or membrane-integrated [35S]-labeled protein that associated with IM vesicles. (B) In vitro–translated [35S]atTic110 was incubated with IM vesicles (30 μg protein) or canine pancreatic microsomal membranes (Mc; 30 μg protein). The vesicles were recovered by differential centrifugation (total) and extracted with alkaline buffer (membrane). The quantitative analysis presents the amount of total or membrane-integrated [35S]-labeled protein that associated with IM vesicles.
Discussion
We investigated the mechanism of import and insertion of atTic40 to better understand the process of protein targeting to the chloroplast IM. We demonstrate that the targeting and insertion of this simple single-pass transmembrane protein involves a soluble intermediate that inserts into the IM after it completes import from the cytoplasm. These observations were extended by our demonstration that atTic40 and a second IM protein, atTic110, can insert directly and selectively into isolated IM vesicles. These data are consistent with earlier observations with Tic110 (Lubeck et al., 1997; Inaba et al., 2005) and indicate that IM targeting of Tic110 and Tic40 involves soluble intermediates.
Both preTic40 and preTic110 appear to initially use the general Toc–Tic pathway for import into the organelle (Lubeck et al., 1996, 1997; Stahl et al., 1999) and are processed by SPP, generating soluble, stromal intermediates. The intermediates insert into the IM from the stroma side of the membrane. Insertion is dependent on information within the transmembrane helices of the proteins (Figs. 6, 9, and 10) and requires proteinaceous components of the inner membrane (Fig. 8). Our initial studies suggest that insertion occurs independent of nucleoside triphosphate hydrolysis (Fig. 8) or the need for stromal factors (Fig. 9).
The precise role of the two-step processing of the pre-atTic40 transit peptide in targeting, if any, remains to be determined. Our data indicate that the intermediate region, spanning amino acids 43–76 of the transit peptide, participates in import but is not required for IM insertion (Fig. 5). Furthermore, the intermediate processing site does not appear to be conserved because the pea orthologue of Tic40 appears to be initially processed at a site much closer to the C-terminal end of the transit peptide (Stahl et al., 1999; unpublished data). The initial cleavage to generate int-atTic40 during import is mediated by SPP (Fig. 1). The data in Figs. 7, 8, and 9 indicate that the final processing step requires association with the inner membrane. Therefore, the processing could be performed by a specific envelope protease. Although the protease responsible for processing int-atTic40 to mature atTic40 remains to be identified, two potential processing peptidases have been identified at the envelope (Inoue et al., 2005; Bolter et al., 2006).
Although our data demonstrate that import and IM targeting are independent processes, one observation suggests that the two reactions can be coupled under some circumstances. The protease protection studies of pre-atTic40 import demonstrate that both int-atTic40 and mature atTic40 are largely exposed to the chloroplast surface at the earliest time points in import and, therefore, have not completed the import process (Fig. 1). Protease-resistant forms corresponding to fully imported int-atTic40 and mature atTic40 accumulate with time in the standard import assay (Fig. 1) and in our import pulse-chase experiment (Fig. 3), reaching their maximum levels at later time points. Collectively, these observations suggest that IM targeting can initiate while int-atTic40 is in the process of import. The fact that the intermediate accumulates in the stroma with time suggests that the rate of import of pre-atTic40 from the cytoplasm exceeds the rate of insertion into the inner membrane, resulting in the inability of the insertion reaction to keep pace with import.
Our studies provide compelling evidence that the import and membrane insertion of atTic40 and atTic110 are independent processes. However, we cannot rule out the possibility that other IM proteins (e.g., polytopic transporters) use a stop-transfer mechanism. In mitochondria, both processes appear to operate for targeting of proteins to the IM (Herrmann, 2003; Herrmann and Neupert, 2003; Herrmann and Bonnefoy, 2004; Preuss et al., 2005). Nucleus-encoded mitochondrial IM proteins appear to initially engage the general import machinery of the outer mitochondrial membrane (TOM complex; Rehling et al., 2004). A small number of nucleus-encoded mitochondrial IM proteins are inserted into the membrane from the matrix after they complete import (Herrmann, 2003; Herrmann and Neupert, 2003; Herrmann and Bonnefoy, 2004). The Oxa1 pathway mediates insertion of several of these proteins (Herrmann et al., 1997; Baumann et al., 2002; Herrmann and Bonnefoy, 2004). This pathway is a member of the Oxa1/Alb3/YidC family of protein export pathways that are conserved from prokaryotes. Oxa1 and a second pathway also mediate the insertion of mitochondria-encoded IM proteins (Herrmann, 2003; Frazier et al., 2006). However, for the majority of mitochondrial IM proteins, complete translocation into the matrix is interrupted by stop-transfer sequences, resulting in the lateral insertion of the proteins into the lipid bilayer via the Tim22 or Tim23 complexes (Herrmann and Neupert, 2003; Rehling et al., 2004). Additional studies with a wider array of chloroplast IM proteins should demonstrate whether stop-transfer mechanisms also exist in chloroplasts.
Although the pathway of Tic40 and Tic110 targeting in chloroplasts and the Oxa1 pathway in mitochondria are analogous at first examination, it is unclear whether the chloroplast proteins use a conservative targeting pathway. Genomic and proteomic studies fail to identify proteins homologous to bacterial export pathways in the chloroplast IM (Ferro et al., 2002, 2003; Froehlich et al., 2003; Jarvis, 2004; Gerdes et al., 2006). Furthermore, the conserved protein export pathways, including the Oxa1/YidC homologue, Alb3, are all present at the thylakoid membrane (Jarvis and Robinson, 2004). Therefore, the relationship of the chloroplast IM targeting translocon to the conserved translocons at the molecular level remains to be determined. Identification of the factors involved in IM targeting in the chloroplast will provide more definitive evidence for the molecular mechanism of this novel membrane protein targeting system.
Materials and methods
cDNAs and construction of atTic40 mutants
Complete pre-atTic40 (available from GenBank/EMBL/DDBJ under accession no. BT006595) and pre-psTic40 (available from GenBank/DMBL/DDBJ under accession no. AY157668) coding regions were amplified from total seedling cDNA by RT-PCR. The atTic40 cDNA was modified by overlap extension PCR to introduce a silent mutation that eliminated the internal Nco1 restriction site. Both cDNAs were cloned into the Nco1 and BamH1 sites of pET21d.
The atTic40 deletion mutants were generated by overlap extension PCR of the atTic40 cDNA to eliminate the specific residues indicated in the figures. The psTP-Tic40 construct (pET21d-pssuTP-matTic40) corresponds to the 57 amino acids of the complete transit peptide of the pea small subunit of rubisco (Pain and Blobel, 1987), fused to mature atTic40. The recombinant int-atTic40 (pET21d-int-atTic40) and mature atTic40 (pET21d-atTic40) constructs for in vitro translation lack the N-terminal 42 and 76 amino acids of pre-atTic40, respectively. The int-atTic40 construct contains an additional Gly codon following the initiation codon to introduce a 5′ Nco1 restriction site. The atTic110ΔTM1 (pET21d-mature atTic110-ΔTM1-His) and atTic110ΔTM2 (pET21d-mature atTic110-ΔTM2-His) constructs contained deletions of amino acids 43–62 and 70–89 of mature atTic110, respectively. The atTic110 and atTic110Sol constructs were previously described (Inaba et al., 2005).
Chloroplast isolation and in vitro import assays
Import substrates were generated using a coupled in vitro transcription–translation system derived from reticulocyte lysate (Promega). Chloroplast isolation from pea and A. thaliana and the in vitro import experiments were performed as previously described (Smith et al., 2002). For the time course of in vitro import, reactions were stopped at the indicated time points by rapid dilution with three volumes of ice-cold 50 mM Hepes-KOH, pH 7.5, and 330 mM Sorbitol (HS buffer). Protease treatments were performed by dividing the import reactions into three equal parts and diluting each with threefold excess ice-cold HS buffer alone or buffer containing trypsin (50 μg/ml final concentration) or thermolysin (100 μg/ml final concentration). The reactions were incubated on ice for 30 min, and proteolysis was stopped with 1 mM PMSF, 0.05 mg/ml TLCK, 0.1 mg/ml soybean trypsin inhibitor, and 2 μg/ml aprotinin (trypsin inhibitor; Jackson et al., 1998) or 10 mM EDTA (thermolysin inhibitor). The chloroplasts were isolated through 40% Percoll silica gel containing the corresponding protease inhibitor, washed once with HS buffer, and processed for SDS-PAGE analysis. All quantitative analysis of radiolabeled samples was performed with a FLA-5000 phosphorimager and Multi Gauge v. 3.0 software (Fujifilm).
Chloroplast lysis and fractionation
Chloroplasts were lysed by suspension in HS buffer to a concentration of 0.5–1 mg chlorophyll/ml and diluted with five volumes of 2 mM EDTA. The lysate was mixed vigorously and incubated on ice for 10 min. The samples were adjusted to 0.2 M NaCl, and the membrane fraction was collected by centrifugation at 18,000 g for 30 min at 4°C (Smith et al., 2002). For alkaline extraction, the membrane pellet was resuspended with a small volume of HS buffer and diluted with 20 volumes of 0.2 M Na2CO3, pH 12. The samples were homogenized with a Teflon homogenizer (Kontes Glass Co.) and incubated at room temperature for 10 min, and the membrane fraction was collected by centrifugation at 100,000 g for 15 min. The soluble fractions were removed and concentrated by precipitation in 20% trichloroacetic acid.
SPP assay
The stromal extract was prepared as previously described (Abad et al., 1989; Tranel and Keegstra, 1996). For the SPP processing assays, 1 μl of in vitro–translation product was incubated with 20 μl stromal extract at 26°C for 90 min. As a control, 1 μl of in vitro–translation product was mock treated with 20 μl of 5 mM Hepes-KOH, pH 8.0.
Pulse-chase experiments
For the pre-atTic40 binding and chase experiments in Fig. 3, isolated chloroplasts were depleted of internal ATP by incubation in the dark at room temperature for 15 min. In vitro–translated pre-atTic40 was depleted of nucleotides by gel filtration. The early import intermediate was generated by incubating chloroplasts with pre-atTic40 in the presence of 100 μM ATP under import conditions (Olsen et al., 1989; Schnell and Blobel, 1993). The chloroplasts were reisolated through 40% Percoll silica gel, washed once with ice-cold HS buffer, and resuspended in ice-cold HS buffer containing 50 mM KOAc and 4 mM MgOAc (import buffer) without ATP. A fraction was removed as the 0-min sample, and the remaining chloroplasts were diluted into 10 volumes of import buffer containing 2 mM ATP at 26°C. Equal fractions of the import reaction were removed at 2, 4, 7.5, and 30 min, reisolated, and separated into membrane and soluble fractions.
For the experiment in Fig. 4 to test int-atTic40 conversion to atTic40, chloroplasts (400 μg chlorophyll) were incubated with pre-atTic40 for 5 min under standard import conditions. The reaction was stopped, and chloroplasts were treated in the absence or presence of trypsin as described in Chloroplast isolation and in vitro import assays. After recovery through Percoll silica gel, the chloroplasts were suspended to 2 ml in prewarmed (26°C) import buffer containing 2 mM ATP. Equal fractions of the reaction mixture were collected at 5 and 60 min, reisolated, and separated into membrane and soluble fractions.
In vitro insertion of int40 into isolated IM vesicles
The chloroplast IM vesicles were isolated as described previously (Keegstra and Yousif, 1986). The membranes were recovered by differential centrifugation and stored in HS buffer containing 1 mM dithiothreitol at −80°C.
The stromal extract containing int-atTic40 was isolated using chloroplasts from a 5-min pre-atTic40 import reaction as described in SPP assay. The chloroplasts (1–2 mg chlorophyll/ml final concentration) were lysed hypertonically with 10 mM Hepes-KOH, pH 8.0, and 10 mM MgCl2 (Yuan et al., 1991) on ice for 10 min. The stromal fraction was separated from the membrane fraction by centrifugation at 40,000 g for 30 min, adjusted to 50 mM Hepes-KOH, pH 7.5, 50 mM KOAc, and 4 mM MgCl2, and stored at −80°C.
For a typical in vitro insertion assay, 15 μl of isolated stromal extract (50 μg protein) or 1 μl in vitro–translated atTic40 or atTic110 protein was incubated with 30 μg inner membrane vesicles under conditions identical to those used for standard chloroplast import assays. The vesicle insertion was performed at 26°C for 1 or 2.5 h. Then, three volumes of ice-cold HS buffer were added and the vesicles were reisolated by spinning at 100,000 g for 20 min. The vesicles were washed once with HS buffer and resuspended in 30 μl HS buffer before further treatments, such as carbonate extraction.
Online supplemental material
Fig. S1 shows that the int-atTic40 targeting intermediate is observed during import into both pea and A. thaliana chloroplasts. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200605126/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Takehito Inaba, Dr. Laura Green, Fei Wang, and Caleb Rounds for their expert technical assistance and helpful discussions.
This work was supported by National Science Foundation grant MCB-0090727 and National Institutes of Health grant GM-61893 to D.J. Schnell.
Abbreviations used in this paper: IM, inner envelope membrane; SPP, stromal processing peptidase.
References
- Abad, M.S., S.E. Clark, and G.K. Lamppa. 1989. Properties of a chloroplast enzyme that cleaves the chlorophyll a/b binding protein precursor. Plant Physiol. 90:117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, F., W. Neupert, and J.M. Herrmann. 2002. Insertion of bitopic membrane proteins into the inner membrane of mitochondria involves an export step from the matrix. J. Biol. Chem. 277:21405–21413. [DOI] [PubMed] [Google Scholar]
- Bedard, J., and P. Jarvis. 2005. Recognition and envelope translocation of chloroplast preproteins. J. Exp. Bot. 56:2287–2320. [DOI] [PubMed] [Google Scholar]
- Bolter, B., A. Nada, H. Fulgosi, and J. Soll. 2006. A chloroplastic inner envelope membrane protease is essential for plant development. FEBS Lett. 580:789–794. [DOI] [PubMed] [Google Scholar]
- Brink, S., K. Fischer, R.-B. Klosgen, and U.-I. Flugge. 1995. Sorting of nuclear-encoded chloroplast membrane proteins to the envelope and the thylakoid membrane. J. Biol. Chem. 270:20808–20815. [DOI] [PubMed] [Google Scholar]
- Chou, M.L., L.M. Fitzpatrick, S.L. Tu, G. Budziszewski, S. Potter-Lewis, M. Akita, J.Z. Levin, K. Keegstra, and H.M. Li. 2003. Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22:2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., J. Andrews, B. Mersey, E.H. Newcomb, and K. Keegstra. 1981. Separation and characterization of inner and outer envelope membranes of pea chloroplasts. Proc. Natl. Acad. Sci. USA. 78:3595–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., M. Werner-Washburne, J. Andrews, and K. Keegstra. 1984. Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 75:675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., K. Keegstra, and L.A. Staehelin. 1985. Freeze-fracture electron microscopic analysis of ultrarapidly frozen envelope membranes on intact chloroplasts and after purification. Protoplasma. 125:111–123. [Google Scholar]
- Emanuelsson, O., H. Nielsen, and G. von Heijne. 1999. ChloroP, a neural network- based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8:978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro, M., D. Salvi, H. Riviere-Rolland, T. Vermat, D. Seigneurin-Berny, D. Grunwald, J. Garin, J. Joyard, and N. Rolland. 2002. Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc. Natl. Acad. Sci. USA. 99:11487–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro, M., D. Salvi, S. Brugiere, S. Miras, S. Kowalski, M. Louwagie, J. Garin, J. Joyard, and N. Rolland. 2003. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics. 2:325–345. [DOI] [PubMed] [Google Scholar]
- Frazier, A.E., R.D. Taylor, D.U. Mick, B. Warscheid, N. Stoepel, H.E. Meyer, M.T. Ryan, B. Guiard, and P. Rehling. 2006. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 172:553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich, J.E., C.G. Wilkerson, W.K. Ray, R.S. McAndrew, K.W. Osteryoung, D.A. Gage, and B.S. Phinney. 2003. Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Proteome Res. 2:413–425. [DOI] [PubMed] [Google Scholar]
- Gerdes, L., T. Bals, E. Klostermann, M. Karl, K. Philippar, M. Hunken, J. Soll, and D. Schunemann. 2006. A second thylakoid membrane-localized Alb3/Oxa1/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. J. Biol. Chem. 281:16632–16642. [DOI] [PubMed] [Google Scholar]
- Herrmann, J.M. 2003. Converting bacteria to organelles: evolution of mitochondrial protein sorting. Trends Microbiol. 11:74–79. [DOI] [PubMed] [Google Scholar]
- Herrmann, J.M., and W. Neupert. 2003. Protein insertion into the inner membrane of mitochondria. IUBMB Life. 55:219–225. [DOI] [PubMed] [Google Scholar]
- Herrmann, J.M., and N. Bonnefoy. 2004. Protein export across the inner membrane of mitochondria: the nature of translocated domains determines the dependence on the Oxa1 translocase. J. Biol. Chem. 279:2507–2512. [DOI] [PubMed] [Google Scholar]
- Herrmann, J.M., W. Neupert, and R.A. Stuart. 1997. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 16:2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, T., M. Alvarez-Huerta, M. Li, J. Bauer, C. Ewers, F. Kessler, and D.J. Schnell. 2005. Arabidopsis tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell. 17:1482–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., A.J. Baldwin, R.L. Shipman, K. Matsui, S.M. Theg, and M. Ohme-Takagi. 2005. Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 171:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.T., J.E. Froehlich, and K. Keegstra. 1998. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273:16583–16588. [DOI] [PubMed] [Google Scholar]
- Jarvis, P. 2004. Organellar proteomics: chloroplasts in the spotlight. Curr. Biol. 14:R317–R319. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., and C. Robinson. 2004. Mechanisms of protein import and routing in chloroplasts. Curr. Biol. 14:R1064–R1077. [DOI] [PubMed] [Google Scholar]
- Joyard, J., E. Teyssier, C. Miege, D. Berny-Seigneurin, E. Marechal, M.A. Block, A.J. Dorne, N. Rolland, G. Ajlani, and R. Douce. 1998. The biochemical machinery of plastid envelope membranes. Plant Physiol. 118:715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and A.E. Yousif. 1986. Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 118:316–325. [Google Scholar]
- Keegstra, K., and K. Cline. 1999. Protein import and routing systems of chloroplasts. Plant Cell. 11:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, F., and G. Blobel. 1996. Interaction of the protein import and folding machineries in the chloroplast. Proc. Natl. Acad. Sci. USA. 93:7684–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, F., and D.J. Schnell. 2006. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic. 7:248–257. [DOI] [PubMed] [Google Scholar]
- Knight, J.S., and J.C. Gray. 1995. The N-terminal hydrophobic region of the mature phosphate translocator is sufficient for targeting to the chloroplast inner envelope membrane. Plant Cell. 7:1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov, A., H. Wang, and D.J. Schnell. 1999. Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J. Biol. Chem. 274:25181–25186. [DOI] [PubMed] [Google Scholar]
- Lamppa, G.K., and M.S. Abad. 1987. Processing of a wheat light-harvesting chlorophyll a/b protein precursor by a soluble enzyme from higher plant chloroplasts. J. Cell Biol. 105:2641–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.-M., T.D. Sullivan, and K. Keegstra. 1992. Information for targeting to the chloroplastic inner envelope membrane is contained in the mature region of the maize Bt1-encoded protein. J. Biol. Chem. 267:18999–19004. [PubMed] [Google Scholar]
- Lubeck, J., J. Soll, M. Akita, E. Nielsen, and K. Keegstra. 1996. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Lubeck, J., L. Heins, and J. Soll. 1997. A nuclear-encoded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J. Cell Biol. 137:1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras, S., D. Salvi, M. Ferro, D. Grunwald, J. Garin, J. Joyard, and N. Rolland. 2002. Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem. 277:47770–47778. [DOI] [PubMed] [Google Scholar]
- Nada, A., and J. Soll. 2004. Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J. Cell Sci. 117:3975–3982. [DOI] [PubMed] [Google Scholar]
- Olsen, L.J., and K. Keegstra. 1992. The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267:433–439. [PubMed] [Google Scholar]
- Olsen, L.J., S.M. Theg, B.R. Selman, and K. Keegstra. 1989. ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 264:6724–6729. [PubMed] [Google Scholar]
- Pain, D., and G. Blobel. 1987. Protein import in chloroplasts requires a chloroplast ATPase. Proc. Natl. Acad. Sci. USA. 84:3288–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, M., M. Ott, S. Funes, J. Luirink, and J.M. Herrmann. 2005. Evolution of mitochondrial oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J. Biol. Chem. 280:13004–13011. [DOI] [PubMed] [Google Scholar]
- Rehling, P., K. Brandner, and N. Pfanner. 2004. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 5:519–530. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J. 1998. Protein targeting to the thylakoid membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:97–126. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J., and G. Blobel. 1993. Identification of intermediates in the pathway of protein import into chloroplasts and their localization to envelope contact sites. J. Cell Biol. 120:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingles, R., and R.E. McCarty. 1995. Production of membrane vesicles by extrusion: size distribution, enzyme activity, and orientation of plasma membrane and chloroplast inner-envelope membrane vesicles. Anal. Biochem. 229:92–98. [DOI] [PubMed] [Google Scholar]
- Smith, M.D., L.M. Fitzpatrick, K. Keegstra, and D.J. Schnell. 2002. In vitro analysis of chloroplast protein import. In Current Protocols in Cell Biology. M. Dasso, J.S. Bonifacino, J. Lippincott-Schwartz, J.B. Harford, and K.M. Yamada, editors. John Wiley and Sons, Inc., New York. 11.16.1–11.16.21. [DOI] [PubMed]
- Stahl, T., C. Glockmann, J. Soll, and L. Heins. 1999. Tic40, a new “old” subunit of the chloroplast protein import translocon. J. Biol. Chem. 274:37467–37472. [DOI] [PubMed] [Google Scholar]
- Tranel, P.J., and K. Keegstra. 1996. A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell. 8:2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann, K., and J. Soll. 1991. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1:149–158. [Google Scholar]
- Young, X.K., and R.E. McCarty. 1993. Assay of proton-coupled glycolate and D-glycerate transport into chloroplast inner envelope membrane vesicles by stopped-flow fluorescence. Plant Physiol. 101:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J., K. Cline, and S.M. Theg. 1991. Cryopreservation of chloroplasts and thylakoids for studies of protein import and integration. Plant Physiol. 95:1259–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.