Abstract
Protein quality control in the endoplasmic reticulum (ER) involves recognition of misfolded proteins and dislocation from the ER lumen into the cytosol, followed by proteasomal degradation. Viruses have co-opted this pathway to destroy proteins that are crucial for host defense. Examination of dislocation of class I major histocompatibility complex (MHC) heavy chains (HCs) catalyzed by the human cytomegalovirus (HCMV) immunoevasin US11 uncovered a conserved complex of the mammalian dislocation machinery. We analyze the contributions of a novel complex member, SEL1L, mammalian homologue of yHrd3p, to the dislocation process. Perturbation of SEL1L function discriminates between the dislocation pathways used by US11 and US2, which is a second HCMV protein that catalyzes dislocation of class I MHC HCs. Furthermore, reduction of the level of SEL1L by small hairpin RNA (shRNA) inhibits the degradation of a misfolded ribophorin fragment (RI332) independently of the presence of viral accessories. These results allow us to place SEL1L in the broader context of glycoprotein degradation, and imply the existence of multiple independent modes of extraction of misfolded substrates from the mammalian ER.
Introduction
Quality control of newly synthesized glycoproteins involves recognition of misfolded proteins in the ER, where they are either returned to a productive folding pathway or are targeted for degradation (Ellgaard and Helenius, 2003). Terminally misfolded glycoproteins are transferred to the cytoplasm for proteasomal proteolysis, a process termed dislocation (Wiertz et al., 1996a,b).
How the cell distinguishes between newly synthesized proteins that have not yet acquired their correct folding state and proteins that are terminally misfolded remains a mystery. In yeast, genetic analysis has shown the involvement of a limited set of proteins that contribute to recognition of misfolded proteins and their subsequent degradation. The secretory protein carboxypeptidase Y (CPY), when engineered to yield a misfolded product, CPY*, has served as a substrate to identify the genetic factors that interfere with its disposal. Der1p was identified as a key player in clearing the yeast ER of misfolded CPY* (Knop et al., 1996; Hill and Cooper, 2000; Walter et al., 2001; Haynes et al., 2002). HMG-CoA reductase, which is a transmembrane protein, has similarly served as a reporter substrate, allowing Hampton et al. (1996) to define HRD1 and HRD3 as essential for its degradation (Gardner et al., 2000, 2001).
Hrd1p/Der3p has ubiquitin ligase activity (E3) and forms a complex predominantly with the ubiquitin-conjugating enzymes (E2s) Ubc7p and Ubc1p (Bays et al., 2001a), which are themselves recruited by the protein Cue1p (Biederer et al., 1997) to the site of degradation. Hrd3p is required for regulating the activity and stability of Hrd1p/Der3p (Plemper et al., 1999), but the function of Hrd3p in protein degradation remains obscure. Hrd3p has a large luminal domain that contains different sets of repeated regions that might be involved in substrate recognition or form complexes with chaperones. Apart from the Ring-H2 ligase Hrd1p/Der3p, there are additional ER membrane–resident E3s, such as Doa10p (Swanson et al., 2001).
Depending on the topology of the ER degradation substrates, different proteins are required for their clearance (Ahner and Brodsky, 2004). Substrates with defects in their cytosolic domain require Doa10p in yeast. Substrates with defects in their luminal portion require the ER lectin Htm1p/Mnl1p, the ubiquitin ligase Hrd1p/Der3p-Hrd3p, Der1p, and proteins involved in ER–Golgi trafficking (Vashist and Ng, 2004). The two pathways merge when leaving the ER; extraction of the ubiquitin-modified substrate occurs with the assistance of Cdc48p/p97 and its cofactors Ufd1p and Npl4p, culminating in delivery to the proteasome and proteolysis of the substrate (Meyer et al., 2000, 2002; Ye et al., 2001, 2003; Wang et al., 2004; Park et al., 2005). Recent studies analyzed the composition of the protein complexes involved. The Doa10p complex contains Ubc7p, Cue1p, Ubx2p, Cdc48p, and its cofactors Ufd1p and Npl4p. These proteins are mainly cytosolic, supporting Doa10p's role in clearing proteins with defects in their cytosolic domain. In addition to these proteins, the Hrd1p complex consists of Hrd3p, Der1p, the ER lectin Yos9p, and Usa1p (Carvalho et al., 2006; Denic et al., 2006). Yos9p has been shown to specifically bind misfolded glycoproteins (Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005). Ubx2p recruits Cdc48p to the membrane (Neuber et al., 2005). Usa1p is thought to link Der1p to the Hrd1p ligase and thereby assist in clearing luminally misfolded proteins from the ER (Ismail and Ng, 2006).
In mammalian cells, the dislocation pathway is more complex. Because of the lack of a genetic approach, the dissection of the degradation pathway in mammalian cells relies on the use of substrates, such as mutant versions of the cystic fibrosis chloride conductance channel (Ward et al., 1995; Bebok et al., 1998; Xiong et al., 1999; Kiser et al., 2001), of proteins considered terminally misfolded, such as the mutant null Hong Kong (NHK) version of the secretory glycoprotein α1 antitrypsin (Liu et al., 1999) or the truncated and misfolded version of the ER-resident glycoprotein ribophorin (RI), termed RI332 (Tsao et al., 1992; de Virgilio et al., 1998). Many membrane proteins fail to fold properly for lack of the correct partner subunits, such as the unpaired T cell receptor α chain (TCRα; Huppa and Ploegh, 1997a,b) or the free immunoglobulin μ chain (Fagioli et al., 2001).
Many parallels exist between the yeast and the mammalian glycoprotein quality control systems; however, the players in the mammalian system are more numerous. The mammalian version of HRD1 is also an ER membrane–resident ubiquitin ligase (Kaneko et al., 2002; Nadav et al., 2003; Kikkert et al., 2004), which forms a complex with SEL1L, a mammalian orthologue of yeast Hrd3p (Lilley and Ploegh, 2005). Additional ubiquitin ligases exist, such as gp78, which has similarity to HRD1 in its Ring finger and interacts with UBC7 via its CUE domain to ubiquitinate TCRα (Fang et al., 2001). There are at least three Der1p homologues in mammals, Derlin-1, -2, and -3, which play roles in the disposal of proteins from the ER (Lilley and Ploegh, 2004, 2005; Oda et al., 2006).
Among the better-studied routes of membrane glycoprotein degradation are the pathways used by human cytomegalovirus (HCMV) to destroy the class I major histocompatibility complex (MHC) heavy chains (HCs; Ahn et al., 1996). Class I MHC products serve as a warning system to alert cytotoxic T cells to the presence of virus-derived polypeptides inside the cell. The infected cell, thus, invites attack by the cytotoxic T cell as a means of eradicating the source of the virus (Tortorella et al., 2000). Large DNA viruses, such as HCMV, are under strong selective pressure to avoid recognition by the immune system. Although widespread amongst the [herpesviridae as a strategy to avoid detection, HCMV in particular has amassed a set of genes whose products evolved to interfere with assembly and intracellular transport of class I MHC products. Amid the HCMV-encoded immunoevasins, two, US2 and US11, which are small membrane glycoproteins that assist in the degradation of class I MHC HCs, stand out. US2 and US11 catalyze superficially similar reactions, characterized by complex formation of US2 or US11 with their target class I MHC HCs, and subsequent extraction of the class I MHC HCs from the ER, a process referred to as dislocation (Wiertz et al., 1996a,b). After dislocation, the class I HC is destroyed by the proteasome. In the presence of proteasome inhibitors, a diagnostic intermediate in this pathway is the result of attack by N-glycanase on the newly dislocated class I MHC HCs (Wiertz et al., 1996a,b; Hirsch et al., 2003; Blom et al., 2004). This intermediate consists of a fully cytoplasmic, yet intact, class I MHC HC, devoid of its N-linked glycan (Misaghi et al., 2004). It occurs not only in cells that express US2 or US11, but also in Daudi cells, which are unable to assemble class I MHC products for lack of the light chain β2-microglobulin (Hughes et al., 1997; Radcliffe et al., 2002). Similar deglycosylated intermediates have been reported for a misfolded fragment of RI (de Virgilio et al., 1998, 1999), but for most other glycoproteins examined, degradation occurs without the obvious production of deglycosylated intermediates.
These observations raise the question as to whether the pathways exemplified by US2- and US11-dependent degradation are indeed emblematic of glycoprotein turnover, as we have argued in the past. The furthest advanced is the characterization of the US11 pathway, for which we identified the Derlin-1 protein as an essential participant (Lilley and Ploegh, 2004; Ye et al., 2004). Derlin-1, in turn, forms a complex not only with itself and other members of the Derlin family, but also with the mammalian Hrd1p and Hrd3p (SEL1L) homologues, as well as with Cdc48p/p97 (Lilley and Ploegh, 2005; Ye et al., 2005). The US2 pathway is impervious to the action of a dominant-negative version of Derlin-1, whereas the US11 pathway is inhibited by it (Lilley and Ploegh, 2004). Direct involvement in mammalian glycoprotein degradation has now been suggested for components of this ER-resident complex, based on interference with degradation of α1 antitrypsin NHK by means of overexpression and knockdowns of Derlins (Oda et al., 2006). Still, in mammalian cells, much of the relevant data stem from the analysis of reactions that depend on the action of the viral accessories US11 and US2.
We analyze the contributions of a mammalian Hrd3p homologue, SEL1L. We show that reduction of the levels of SEL1L by RNA interference results in impairment of US11-mediated dislocation of class I MHC HC molecules. Expression of the same interfering small hairpin RNAs (shRNAs) against SEL1L, however, does not affect the US2 pathway. Expression of SEL1L shRNAs inhibits the degradation of RI332, which is a process that occurs independently of the presence of viral accessories. These results allow us to place SEL1L in a more general context of glycoprotein degradation, and suggest that SEL1L might be involved in substrate recognition of misfolded proteins in the ER.
Results
SEL1L is part of a mammalian ER multiprotein complex involved in dislocation (Lilley and Ploegh, 2005; Ye et al., 2005). SEL1L associates with Derlin-1, -2, p97, and HRD1, as well as with additional proteins that remain to be identified, some or all of which may also play a role in dislocation. Some of these proteins are presumably recruited to the site of dislocation through SEL1L. SEL1L is predicted to be a type I transmembrane protein (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200605196/DC1) with 5 N-linked glycans (Biunno et al., 1997). Because the bulk of the SEL1L protein is predicted to be in the ER lumen, it is possible that SEL1L first plays a role in substrate recognition and identification of misfolded proteins, and then recruits them to the site of dislocation. We performed coimmunoprecipitations from steady-state radioactive [35S]methionine/cysteine–labeled cells using mild lysis conditions (2% digitonin) to preserve ER membrane protein complexes, to analyze whether composition of protein complexes with SEL1L are conserved for the cell lines used in this study.
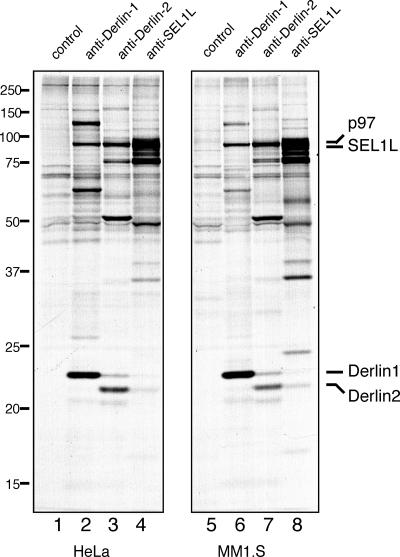
Our earlier observations concern the composition of protein complexes that include the Derlins in the astrocytoma cell line U373 (Lilley and Ploegh, 2005). We first verified that the types of complexes detected in U373 cells are not cell type–specific, but occur in other cell lines as well (Fig. 1). We were particularly interested in cell lines that would lend themselves to transient transfection experiments (Fig. 1, HeLa cells, lanes 1–4) and in multiple myeloma cells (Fig. 1, MM1.S cells, lanes 5–8), which represent cells with a high secretory capacity, and, presumably, a correspondingly pronounced requirement to clear misfolded proteins from the ER.
Figure 1.
SEL1L is part of a conserved complex in many cell types. Immunoprecipitation from digitonin lysates of HeLa (lanes 1–4) or MM1.S (lanes 5–8) cells pulse-labeled for 16 h (steady state) were performed with control rabbit IgG, anti–Derlin-1, anti–Derlin-2, and anti-SEL1L antibodies, respectively. The positions of the relevant proteins are indicated.
When we performed immunoprecipitations with antibodies against Derlin-1, -2, and SEL1L, the composition of the complexes obtained was very similar to that reported for U373 cells (Lilley and Ploegh, 2005). We conclude that the multiprotein complexes, which were initially identified for U373 cells, are representative of the complexes detected in other, unrelated cell types. The pattern of the protein complexes is equivalent in all tested cell lines, including U373 (astrocytoma), HeLa (epithelial), 293T, MM1.S (multiple myeloma), and C6 (Rat glioma) cells. SEL1L is likely to be a crucial component of a ubiquitous ER-dislocation complex.
When we analyzed the half-life of SEL1L, we found that it decays with a half-life of ∼180 min (Fig. 2, lanes 1, 3, and 5). SEL1L degradation appears to be proteasome-dependent because it is inhibited by inclusion of the proteasome inhibitor ZL3VS (Fig. 2, lanes 7, 9, and 11). SEL1L remains completely susceptible to endoglycosidase H (EndoH) treatment, suggesting that SEL1L is an ER protein and does not traffic through the secretory pathway (Fig. 2, lanes 2, 4, 6, 8, 10, and 12).
Figure 2.
SEL1L is an unstable ER membrane glycoprotein. Immunoprecipitation from NP-40 lysates of U373 cells pulse-labeled for 20 min and chased for the indicated time points in the absence (lanes 1–6) or presence (lanes 7–12) of ZL3VS were performed with anti-SEL1L antibodies. Half of the eluates were treated with EndoH (lanes 2, 4, 6, 8, 10, and 12).
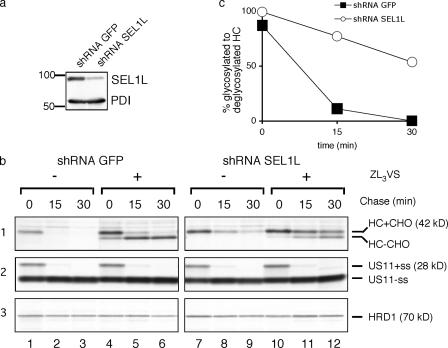
To address whether SEL1L plays a direct role in the process of dislocation from the ER, we generated small hairpin RNAs (shRNAs) that target SEL1L or a control unrelated protein (GFP) using the pRETRO-SUPER in vivo expression system (Brummelkamp et al., 2002). The shRNA plasmids reduced SEL1L protein levels to 30% compared with control cells (Fig. 3 a).
Figure 3.
shRNAs targeting SEL1L impair US11-mediated dislocation of class I MHC HC. (a) US11 cells were transduced with virus encoding shRNAs against GFP (control cells) or against SEL1L (knockdown cells) and analyzed for levels of SEL1L by immunoblotting with anti-SEL1L antibodies and anti–protein disulfide isomerase (PDI) antibodies as a loading control. (b) Control and knockdown cells were pulse-labeled for 10 min and chased for the indicated time points. Immunoprecipitation from SDS lysates for class I MHC HC (1), US11 (2), and HRD1 (3) were performed using respective antibodies. (c) Quantitation of the amount of glycosylated to deglycosylated HC from shRNA GFP (control) and shRNA SEL1L (knockdown) cells treated with ZL3VS.
We analyzed the stability of class I MHC HCs in cells expressing US11 and the SEL1L shRNAs. The fate of HC is best analyzed in a pulse-chase experiment in view of the rate of dislocation (t 1/2 = 2–5 min) in US11 cells (Wiertz et al., 1996a). In cells expressing US11 and shRNAs against GFP (control cells), HC disappears completely when proteasome inhibitors are not included (Fig. 3 b, 1, lanes 2 and 3). In cells expressing SEL1L shRNA, the rate of degradation of class I MHC HC is much reduced (Fig. 3 b, 1, lanes 8 and 9).
In the presence of the proteasome inhibitor ZL3VS, the deglycosylated class I MHC HC accumulates as the diagnostic intermediate that characterizes the dislocation reaction. In control shRNA US11 cells, complete conversion to the deglycosylated form of HC is seen after 30 min of chase (lane 6). In SEL1L knockdown cells, >50% of HC remains in its fully glycosylated form. This persistence of HC is attributable to compromised dislocation, as only ∼50% of HC accesses the cytosolically disposed N-glycanase (Fig. 3, b, 1 [lanes 11 and 12], and c).
This impairment in dislocation is not caused by reduced levels of US11 in SEL1L knockdown cells, as US11 is expressed at comparable levels, nor is it caused by aberrant ER insertion and processing of US11, as determined by normal cleavage of US11's signal sequence (Rehm et al., 2001) in SEL1L knockdown cells (Fig. 3 b, 2).
Because SEL1L is a mammalian homologue of yeast Hrd3p, we looked for possible parallels between the two systems that could account for the observed phenotype. Hrd1p, which is a yeast E3 ligase known to be necessary for degradation of some substrates (Bays et al., 2001b; Bordallo and Wolf, 1999; Hampton et al., 1996; Kikkert et al., 2004; Nadav et al., 2003), is regulated by Hrd3p and destabilized in Δhrd3 yeast (Gardner et al., 2000; Plemper et al., 1999). Therefore, we analyzed HRD1 levels in SEL1L knockdown cells to examine whether inhibition of dislocation is primarily caused by reduced levels of SEL1L or, alternatively, attributable to a reduction in HRD1 levels. We find that HRD1 is stable throughout the chase periods over which HC dislocation occurs (Fig. 3 b, 3).
A comparable reduction of SEL1L levels in US2 cells (Fig. 4 a) has no observable effect on HC dislocation. In US2 cells exposed to proteasome inhibitor, we see conversion of almost all class I MHC HCs to the deglycosylated species after 30 min (Fig. 4 b, 1, lane 6). In US2 cells, knockdown of SEL1L results in a rate of dislocation equal to that in control cells and yields a pattern noticeably distinct from that seen for the SEL1L knockdown in US11 cells (Fig. 4, b [1] and c). US2 levels are also comparable for control and SEL1L knockdown cells (Fig. 4 b, 2). Unimpaired degradation in US2 cells of class I MHC HCs is consistent with the idea that US11 co-opts a conserved complex to catalyze degradation of class I MHC HC, whereas US2 utilizes a distinct pathway (Lilley and Ploegh, 2004, 2005; Loureiro et al., 2006). This result is all the more striking because US2 and US11 target the same set of substrates, class I MHC products.
Figure 4.
shRNAs targeting SEL1L do not impair US2-mediated dislocation of class I MHC HC. (a) US2 cells were transduced with virus encoding shRNAs against GFP (control cells) or against SEL1L (knockdown cells) and analyzed for levels of SEL1L by immunoblotting with anti-SEL1L antibodies and anti-PDI antibodies as a loading control. (b) Control and knockdown cells were pulse-labeled for 10 min and chased for the indicated time points. Immunoprecipitation from SDS lysates for class I MHC HC (1) and US2 (2) were performed using respective antibodies. (c) Quantitation of the amount of glycosylated to deglycosylated HC from shRNA GFP (control) and shRNA SEL1L (knockdown) cells treated with ZL3VS.
Given the homology of SEL1L to Hrd3p, it is likely that SEL1L plays a general role in glycoprotein turnover, similar to what was shown for the Der1p homologues of the Derlin proteins (Lilley and Ploegh, 2004; Oda et al., 2006). Thus far, the only substrates that for their dislocation depend on Derlin-1 are class I MHC HCs in US11-expressing cells and US2 itself (Lilley and Ploegh, 2004). Overexpression and knockdowns of Derlin-2 and -3 have been reported to affect the half-life of α1 antitrypsin NHK (Oda et al., 2006).
Therefore, we examined other model substrates of ER dislocation for their susceptibility to inhibition by a knockdown of SEL1L. We analyzed the effect of SEL1L shRNAs on the degradation of a mutant version of RI, RI332. The RI332 mutant lacks the C terminus of RI; unlike the very stable RI protein, the RI332 fragment has a half-life of ∼90 min (Tsao et al., 1992; de Virgilio et al., 1998, 1999). We chose RI332 as substrate because its mode of disposal includes an intermediate that is the product of N-glycanase activity (Kitzmuller et al., 2003). We transiently transfected RI332 into HeLa cells, which had been stably transduced with shRNAs against either GFP or SEL1L (Fig. 5 a). SEL1L knockdown cells showed a reduction of SEL1L levels to 40% of controls. We transfected a GFP expression plasmid into both GFP shRNA and SEL1L shRNA HeLa cells as a transfection control, as well as a control for shRNA-mediated knockdown in the HeLa shRNA GFP control cells. The percentage of cells expressing GFP was equivalent (∼40%), but, as expected, the intensity of the GFP signal was much lower in GFP shRNA cells (unpublished data).
Figure 5.
Dislocation of truncated RI (RI332) is impaired by shRNAs targeting SEL1L. (a) HeLa cells were stably transduced with either shRNAS against GFP (control cells) or SEL1L (knockdown cells) and analyzed for levels of SEL1L by immunoblotting with anti-SEL1L antibodies and anti-PDI antibodies as a loading control. (b) Control and knockdown cells were transiently transfected with RI332. 36 h after transfection, these cells were pulse-labeled for 20 min and chased for the indicated time points. Immunoprecipitation from SDS lysates were performed using anti-RI I antibodies. (c) The bar diagram shows quantitation of the experiment. Error bars represent the SD of three independent experiments.
Two days after transfection, cells were harvested, pulsed for 20 min with [35S]methionine, and chased for 0, 90, and 180 min. Immunoprecipitation with anti-RI antibody retrieves both the truncated and wild-type RI. In control shRNA GFP HeLa cells, ∼90% of truncated RI332 is degraded after 180 min, whereas in SEL1L knockdown cells, we see a considerable stabilization of RI332; ∼40% of RI332 remains (Fig. 5, b [lanes 3 and 6] and c). The observed stabilization of RI332 is all the more noteworthy as the achieved stable knockdown of SEL1L in HeLa cells is ≥40% (Fig. 5 a). Wild-type RI (a stable protein in the ER) remains unaffected, and expression of SEL1L shRNA does not affect overall glycosylation as assessed by class I MHC HC and US11 maturation and glycosylation earlier.
Does SEL1L specifically interact with misfolded proteins in the ER?
Because SEL1L has a long luminal domain with tetratricopeptide repeats (TPRs; Sel1 repeats of the TPR family; Fig. S1), a domain structure suggested to mediate protein–peptide interactions, we determined whether SEL1L interacts with misfolded proteins, and whether SEL1L can discriminate between properly folded and misfolded proteins. We examined association of RI332 with SEL1L by immunoprecipitating for SEL1L from extracts of cells transiently transfected with RI332 and lysed under mild conditions. We found that RI332 interacts with SEL1L (Fig. 6 b, lane 1), and we confirmed this interaction in re-immunoprecipitation experiments (Fig. 6 b, lane 2). We were unable to detect the full length, and presumably properly folded, form of RI in the re-immunoprecipitation, although the presence of trace amounts of full-length RI remains a possibility, as misfolded RI332 is in excess over endogenous RI (Fig. 6 a). As a second specificity control, we confirmed that the deglycosylated dislocation intermediate of RI332, which accumulates in the cytosol when cells are treated with proteasome inhibitor, does not interact with SEL1L (Fig. 6 and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200605196/DC1).
Figure 6.
SEL1L interacts with RI332 293T cells transfected with RI332 that were pulse-labeled to steady-state for 5 h in the presence of 10 μM of the proteasome inhibitor ZL3VS. Immunoprecipitations from digitonin lysates were performed with anti-SEL1L antibodies. The anti-SEL1L immunoprecipitate was either analyzed directly (b, lane 1) or reimmunoprecipitated sequentially using anti-RI I antibodies (b, lane 2) or anti-SEL1L antibodies (b, lane 3). (a) Input levels of endogenous RI and overexpressed RI332. Immunoprecipitates were resolved on a 12% SDS polyacrylamide gel. Asterisk represents a presumably processed version of RI332 in the ER.
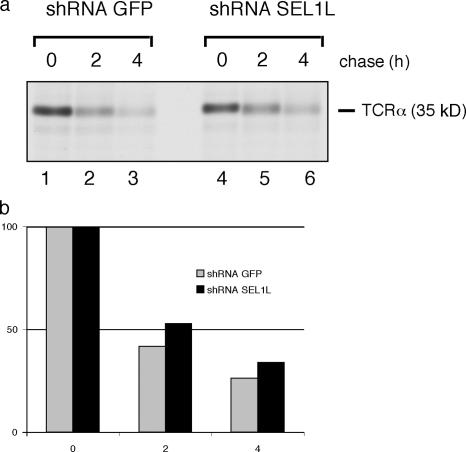
We transiently transfected the same HeLa cell lines that had been stably transduced with shRNAs targeting either SEL1L or GFP with TCRα. TCRα is likely degraded because of the unpaired charged residues in its transmembrane domain when its proper partner subunits (TCRβ and CD3 complex) are not coexpressed (Cosson et al., 1991). Cells were labeled for 20 min with [35S]methionine and chased for 0, 2, and 4 h (Fig. 7 a), and the amount of TCRα remaining at each time point was quantitated (Fig. 7 b). We see a moderate effect of a SEL1L knockdown on TCRα degradation; after 4 h of chase, 25% of TCRα remains in the shRNA GFP control cells, whereas in SEL1L knockdown cells, 35% remains (Fig. 7 b). The effect is likely to be less pronounced for TCRα than for RI332 because TCRα would be classified as misfolded because of recognition of unpaired charges in its transmembrane domain, whereas RI332 is an entirely luminal protein. From yeast, we know that the site of misfolding is crucial for the recruitment of specific substrate recognition proteins (Ahner and Brodsky, 2004; Carvalho et al., 2006). Another explanation for the poor interference with TCRα degradation could be the level of SEL1L knockdown to no more than 40%. Interference with the degradation of TCRα might require a greater reduction in SEL1L levels than attained with the present knockdown strategies.
Figure 7.
TCRα degradation is moderately slowed by shRNAs targeting SEL1L. The same control and knockdown cells used in Fig. 5 were transiently transfected with TCRα. 36 h after transfection these cells were pulse-labeled for 20 min and chased for the indicated time points. Immunoprecipitation from SDS lysates were performed using anti–TCRα-antibodies. (c) The histogram shows mean quantitation of two independent experiments.
Discussion
SEL1L is essential for dislocation of class I MHC molecules from the ER in cells that express US11. Upon reduction of the levels of the SEL1L protein, fully glycosylated, EndoH-sensitive class I MHC HCs accumulate in the ER. In contrast, the US2 pathway is not perturbed by reduction of SEL1L protein levels. These results underscore the relevance of other observations that place US11 and US2 in distinct pathways, e.g., based on their susceptibility to interference with Derlin-1 function; a Derlin-1GFP fusion protein impairs degradation of class I MHC molecules via the US11-dependent, but not via the US2-dependent, pathway (Lilley and Ploegh, 2004).
There are several lines of evidence to suggest that US2 and US11 use different principles to accelerate degradation of class I MHC molecules, in addition to the aforementioned difference in Derlin-1 dependency. US2 must exploit features in its relatively short cytoplasmic tail, whereas tailless US11 remains dislocation-competent (Furman et al., 2002; Loureiro et al., 2006). The preference of the US2 and US11 pathways for folded and unfolded class I MHC molecules appears to be different (Blom et al., 2004), as is the requirement for elements in the tail of the class I MHC molecules themselves (Wiertz et al., 1996b). Our data now establish that the US11, but not the US2, pathway is sensitive to a SEL1L knockdown. These distinctions are all the more remarkable given the similarities of the substrates targeted for degradation, the identical class I MHC HCs in the same parent cell line.
We also report that down-regulation of SEL1L affects the degradation of a RI fragment whose route of degradation is similar to what we have described for class I MHC products targeted by the HCMV immunoevasins (Hughes et al., 1997; Kitzmuller et al., 2003). In the presence of proteasome inhibitors, a deglycosylated RI332 species, the product of N-glycanase digestion, is observed (Kitzmuller et al., 2003). Our data, thus, support the notion that human SEL1L is the orthologue of yeast Hrd3p because reduction of SEL1L levels perturbs the degradation of a misfolded substrate, RI332.
We conclude that SEL1L and its associated partners operate in a pathway that is neither restricted to class I MHC products, nor strictly dependent on the involvement of virus-encoded proteins. The concept of physically extracting proteins from the ER and their delivery to the cytosolic proteasome achieves the compartmentalization required to spare nascent and properly folded ER proteins from premature degradation. The advantages of studying the pathways for the HCMV US2 and US11 proteins are the speed with which degradation occurs and the involvement of well-characterized substrates, the class I MHC products. The available antibodies allow an easy distinction between various folding intermediates, and the occurrence of the tell-tale deglycosylated degradation intermediate is an accurate reporter for its localization and for the dislocation reaction per se (Wiertz et al., 1996a,b). We have taken advantage of the fact that US2 and US11 attract very similar substrates in one and the same cell, yet apparently do so by mechanistically different pathways (Furman et al., 2002; Lilley and Ploegh, 2004; Loureiro et al., 2006).
This does raise the question, however, of whether the results obtained for these HCMV immunoevasins and class I MHC substrates can be extrapolated to other substrates, and to pathways that operate independently of viral accessories.
To our knowledge, the observation that SEL1L is involved in the degradation of RI332, a known dislocation substrate, is the first example for mammalian cells that directly places SEL1L in a pathway of protein degradation. The dominant-negative version of Derlin-1, Derlin-1GFP, had no effect on RI332 degradation (unpublished data), making the mammalian degradation pathways even more complex. The different degradation pathways that, in yeast, process distinct types of substrates (luminally vs. cytoplasmically exposed proteins) likely operate in mammals, too, but involve more factors that comprise the different complexes. In addition to the multiple Derlin proteins, ubiquitin ligases, and lectins involved in mammalian ER degradation, there is an additional SEL1L-like protein in mammals that bears homology to Hrd3p (Fig. S1). Whether this protein binds to HRD1 and participates in the degradation of misfolded proteins remains to be determined.
In yeast, substrates that require Der1p for degradation require Hrd1p/Hrd3p and usually belong to the set of completely luminal substrates. In mammals, depending on the substrate, Derlin-1 and SEL1L can act in concert with each other, as we show for class I MHC HC, but SEL1L can also assist in the degradation of substrates independently of Derlin-1, as is the case for RI332. A recent study suggests that, in yeast, Hrd3p and Der1p can recruit substrates independently of each other (Gauss et al., 2006b).
We also examined a substrate that is recognized as misfolded by means of charged residues in its transmembrane domain, TCRα. We observed modest stabilization when SEL1L expression is compromised. The lesser effect on this substrate compared with RI332, which is an entirely luminal substrate, further suggests that the nature of the substrate determines recruitment of an otherwise conserved complex. We cannot exclude that different degradation substrates require different levels of SEL1L. Perhaps TCRα degradation usually proceeds in the presence of very few SEL1L molecules.
The ER machinery involved in recognition of substrates is likely to be specific for certain substrates, but the details of recognition of misfolded substrates in yeast are no better resolved than in mammalian cells. In yeast, Yos9p, a putative lectin protein, is involved in the degradation of a membrane-bound version of CPY* (Buschhorn et al., 2004) and might be involved in identifying and targeting substrates for degradation (Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005). A recent study suggests that proteins that are misfolded in yeast bind to Hrd3p, which itself binds to Yos9p. Yos9p then ensures that only terminally misfolded proteins are being degraded (Gauss et al., 2006a). Another avenue to identification of misfolded glycoproteins might be through their high-mannose–containing, N-linked glycan modifications (Helenius and Aebi, 2004). Mannose residues are trimmed in the ER by the enzyme α-mannosidase I (Hosokawa et al., 2003; Wu et al., 2003), generating a Man8GlcNac2 tag implicated in targeting glycoproteins for degradation (Ellgaard and Helenius, 2003). Proteins with this tag bind to calnexin and other lectins, the ER degradation-enhancing mannosidase-like (EDEM) proteins, EDEM1 and EDEM2 (Hosokawa et al., 2001, 2003; Mast et al., 2005; Olivari et al., 2005), which then target some glycoproteins for degradation (Molinari et al., 2003; Ahner and Brodsky, 2004; Oda et al., 2006) There is a mammalian orthologue of Yos9p, OS-9, whose function is unknown but might be critical in degradation of certain substrates.
Similar to EDEM, SEL1L might be involved in substrate recognition and bind to a subset of misfolded proteins. SEL1L is remarkably conserved, and contains several repetitive structural and functional domains (sel1-like repeats of the TPR family) and a type II fibronectin domain in its large luminal part that could bind to chaperones or misfolded proteins. Alternatively, SEL1L might recruit substrate recognition proteins such as EDEM or OS-9 through its type II fibronectin domain at its N terminus. In yeast, Hrd3p's N-terminal domain is essential for its function in ER degradation, and its central region required for interaction with Hrd1p (Gardner et al., 2001).
SEL1L forms a 1:1 stoichiometric complex with HRD1 in mammalian cells (Lilley and Ploegh, 2005), but until now, SEL1L's direct involvement in protein degradation has not been shown. Thus, we conclude that SEL1L, based on its associated partners, its direct involvement in glycoprotein degradation, and its structural relationship to yeast Hrd3p, helps select proteins for entry into a degradative pathway. Our data provide the first functional evidence to support the notion that SEL1L is indeed the orthologue of yeast Hrd3p.
Materials and methods
Antibodies, DNA constructs, and cell lines
Anti-HC, anti-US2, anti-US11, anti–Derlin-1, anti–Derlin-2, and anti-SEL1L antibodies have been described previously (Tortorella et al., 1998; Gewurz et al., 2002; Lilley et al., 2003; Lilley and Ploegh, 2004, 2005).
Anti-RI332 antibodies and the RI332 DNA construct were a gift from N.E. Ivessa (University of Vienna, Vienna, Austria) and G. Kreibich (New York University Medical Center, New York, NY), anti-TCRα antibodies were purchased from Sigma-Aldrich, and anti-HRD1 antibodies were a gift from E. Wiertz (Leiden University, Leiden, Netherlands; Kikkert et al., 2004).
The TCRα expression construct was described previously (Huppa and Ploegh, 1997a).
For stable shRNA expression, the pRETRO-SUPER vector was used (Brummelkamp et al., 2002).
U373-MG astrocytoma/glioblastoma cells, US2, US11, 293T, and HeLa cells were cultured as previously described (Rehm et al., 2001; Lilley et al., 2003).
MM1.S cells were cultured in RPMI containing 10% FCS.
Metabolic labeling, immunoprecipitation, endoglycosidase H digestion, gel electrophoresis, immunoblotting, and transient transfections
These procedures were similar to those described in Lilley and Ploegh (2005). Treatment of cells with the proteasome inhibitor ZL3VS has been previously described (Shamu et al., 1999). Analysis of class I MHC HC stability in US11 and US2 cells, pulse-labeling, cell lysis (SDS and digitonin), and re-immunoprecipitations have been previously described (Lilley et al., 2003). NEM (N-ethylmaleimide) was included in digitonin and SDS lysis at a concentration of 2.5 mM. SDS PAGE has been previously described (Ploegh, 1995).
293T cells were transiently transfected with Trans-IT (Takara Mirus Bio) according to the manufacturer, using 2–5 μg of DNA for transient transfections and 10 μg of total DNA for virus production in 10-cm dishes. HeLa cells were transiently transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer, using 2–5 μg for transient transfections in 10-cm dishes.
Immunoprecipitates were treated with EndoH according to the manufacturer (New England Biolabs).
The program ImageJ (National Institutes of Health) was used for quantitating bands on film.
SEL1L knockdown
19-nt target sequences in SEL1L were chosen according to the online design software available from the Whitehead Institute webpage (http://jura.wi.mit.edu/siRNAext/). Sense and antisense strands were annealed to form the shRNA template insert and ligated into the retroviral vector pRETRO SUPER (Brummelkamp et al., 2002) to generate shRNA construct s2 for stable shRNA expression and for transient transfection (19mer sequences; GGCTATACTGTGGCTAGAA and TTCTAGCCACAGTATAGCC, respectively). As a control, the unrelated construct (GFP shRNA) targeting the enhanced GFP reporter was used (Lilley and Ploegh, 2005). All constructs were sequence-confirmed and used for virus production in HEK 293T cells (Soneoka et al., 1995). pRETRO-transduced U373 cells and HeLa cells were selected with 0.375 μg/ml and 1 μg/ml puromycin, respectively, as previously described (Lilley and Ploegh, 2005).
Online supplemental material
Fig. S1 shows a schematic of the Hrd3p homologues in mammals. Fig. S2 shows SEL1L interacting with RI332 293T cells transfected with RI332. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200605196/DC1.
Supplementary Material
Acknowledgments
We thank N. E. Ivessa and G. Kreibich for providing the RI332 expression construct and for the anti-RI antibodies, and members of the Ploegh laboratory for critical reading of the manuscript and for technical advice. B. Mueller is a fellow of the Boehringer Ingelheim Fonds. B.N. Lilley was a Howard Hughes Medical Institute Predoctoral Fellow.
This work was funded by National Institutes of Health grant AI033456 to H.L. Ploegh.
B.N. Lilley's present address is Dept. of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138.
Abbreviations used in this paper: CPY, carboxypeptidase Y; EDEM, ER degradation-enhancing mannosidase-like; HC, heavy chain; MHC, major histocompatibility complex; NHK, null Hong Kong; PDI, protein disulfide isomerase; shRNA, small hairpin RNA; HCMV, human cytomegalovirus; RI, ribophorin; TPR, tetratricopeptide repeat.
References
- Ahn, K., A. Angulo, P. Ghazal, P.A. Peterson, Y. Yang, and K. Fruh. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. USA. 93:10990–10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahner, A., and J.L. Brodsky. 2004. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 14:474–478. [DOI] [PubMed] [Google Scholar]
- Bays, N.W., R.G. Gardner, L.P. Seelig, C.A. Joazeiro, and R.Y. Hampton. 2001. a. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3:24–29. [DOI] [PubMed] [Google Scholar]
- Bays, N.W., S.K. Wilhovsky, A. Goradia, K. Hodgkiss-Harlow, and R.Y. Hampton. 2001. b. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell. 12:4114–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebok, Z., C. Mazzochi, S.A. King, J.S. Hong, and E.J. Sorscher. 1998. The mechanism underlying cystic fibrosis transmembrane conductance regulator transport from the endoplasmic reticulum to the proteasome includes Sec61beta and a cytosolic, deglycosylated intermediary. J. Biol. Chem. 273:29873–29878. [DOI] [PubMed] [Google Scholar]
- Bhamidipati, A., V. Denic, E.M. Quan, and J.S. Weissman. 2005. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell. 19:741–751. [DOI] [PubMed] [Google Scholar]
- Biederer, T., C. Volkwein, and T. Sommer. 1997. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 278:1806–1809. [DOI] [PubMed] [Google Scholar]
- Biunno, I., V. Appierto, M. Cattaneo, B.E. Leone, G. Balzano, C. Socci, S. Saccone, A. Letizia, G. Della Valle, and V. Sgaramella. 1997. Isolation of a pancreas-specific gene located on human chromosome 14q31: expression analysis in human pancreatic ductal carcinomas. Genomics. 46:284–286. [DOI] [PubMed] [Google Scholar]
- Blom, D., C. Hirsch, P. Stern, D. Tortorella, and H.L. Ploegh. 2004. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. EMBO J. 23:650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo, J., and D.H. Wolf. 1999. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae. FEBS Lett. 448:244–248. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. [DOI] [PubMed] [Google Scholar]
- Buschhorn, B.A., Z. Kostova, B. Medicherla, and D.H. Wolf. 2004. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 577:422–426. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., V. Goder, and T.A. Rapoport. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373. [DOI] [PubMed] [Google Scholar]
- Cosson, P., S.P. Lankford, J.S. Bonifacino, and R.D. Klausner. 1991. Membrane protein association by potential intramembrane charge pairs. Nature. 351:414–416. [DOI] [PubMed] [Google Scholar]
- de Virgilio, M., H. Weninger, and N.E. Ivessa. 1998. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J. Biol. Chem. 273:9734–9743. [DOI] [PubMed] [Google Scholar]
- de Virgilio, M., C. Kitzmuller, E. Schwaiger, M. Klein, G. Kreibich, and N.E. Ivessa. 1999. Degradation of a short-lived glycoprotein from the lumen of the endoplasmic reticulum: the role of N-linked glycans and the unfolded protein response. Mol. Biol. Cell. 10:4059–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic, V., E.M. Quan, and J.S. Weissman. 2006. A luminal Surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 126:349–359. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191. [DOI] [PubMed] [Google Scholar]
- Fagioli, C., A. Mezghrani, and R. Sitia. 2001. Reduction of interchain disulfide bonds precedes the dislocation of Ig-mu chains from the endoplasmic reticulum to the cytosol for proteasomal degradation. J. Biol. Chem. 276:40962–40967. [DOI] [PubMed] [Google Scholar]
- Fang, S., M. Ferrone, C. Yang, J.P. Jensen, S. Tiwari, and A.M. Weissman. 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 98:14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman, M.H., H.L. Ploegh, and D. Tortorella. 2002. Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules. J. Biol. Chem. 277:3258–3267. [DOI] [PubMed] [Google Scholar]
- Gardner, R.G., A.G. Shearer, and R.Y. Hampton. 2001. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol. Cell. Biol. 21:4276–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R.G., G.M. Swarbrick, N.W. Bays, S.R. Cronin, S. Wilhovsky, L. Seelig, C. Kim, and R.Y. Hampton. 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss, R., E. Jarosch, T. Sommer, and C. Hirsch. 2006. a. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat. Cell Biol. 8:849–854. [DOI] [PubMed] [Google Scholar]
- Gauss, R., T. Sommer, and E. Jarosch. 2006. b. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 25:1827–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewurz, B.E., H.L. Ploegh, and D. Tortorella. 2002. US2, a human cytomegalovirus-encoded type I membrane protein, contains a non-cleavable amino-terminal signal peptide. J. Biol. Chem. 277:11306–11313. [DOI] [PubMed] [Google Scholar]
- Hampton, R.Y., R.G. Gardner, and J. Rine. 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 7:2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, C.M., S. Caldwell, and A.A. Cooper. 2002. An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 158:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019–1049. [DOI] [PubMed] [Google Scholar]
- Hill, K., and A.A. Cooper. 2000. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 19:550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, C., D. Blom, and H.L. Ploegh. 2003. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22:1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, N., I. Wada, K. Hasegawa, T. Yorihuzi, L.O. Tremblay, A. Herscovics, and K. Nagata. 2001. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, N., L.O. Tremblay, Z. You, A. Herscovics, I. Wada, and K. Nagata. 2003. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 278:26287–26294. [DOI] [PubMed] [Google Scholar]
- Hughes, E.A., C. Hammond, and P. Cresswell. 1997. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc. Natl. Acad. Sci. USA. 94:1896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa, J.B., and H.L. Ploegh. 1997. a. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 7:113–122. [DOI] [PubMed] [Google Scholar]
- Huppa, J.B., and H.L. Ploegh. 1997. b. In vitro translation and assembly of a complete T cell receptor-CD3 complex. J. Exp. Med. 186:393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, N., and D.T. Ng. 2006. Have you HRD? Understanding ERAD is DOAble! Cell. 126:237–239. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., M. Ishiguro, Y. Niinuma, M. Uesugi, and Y. Nomura. 2002. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532:147–152. [DOI] [PubMed] [Google Scholar]
- Kikkert, M., R. Doolman, M. Dai, R. Avner, G. Hassink, S. van Voorden, S. Thanedar, J. Roitelman, V. Chau, and E. Wiertz. 2004. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279:3525–3534. [DOI] [PubMed] [Google Scholar]
- Kim, W., E.D. Spear, and D.T. Ng. 2005. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell. 19:753–764. [DOI] [PubMed] [Google Scholar]
- Kiser, G.L., M. Gentzsch, A.K. Kloser, E. Balzi, D.H. Wolf, A. Goffeau, and J.R. Riordan. 2001. Expression and degradation of the cystic fibrosis transmembrane conductance regulator in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 390:195–205. [DOI] [PubMed] [Google Scholar]
- Kitzmuller, C., A. Caprini, S.E. Moore, J.P. Frenoy, E. Schwaiger, O. Kellermann, N.E. Ivessa, and M. Ermonval. 2003. Processing of N-linked glycans during endoplasmic-reticulum-associated degradation of a short-lived variant of ribophorin I. Biochem. J. 376:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., A. Finger, T. Braun, K. Hellmuth, and D.H. Wolf. 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Lilley, B.N., and H.L. Ploegh. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 429:834–840. [DOI] [PubMed] [Google Scholar]
- Lilley, B.N., and H.L. Ploegh. 2005. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 102:14296–14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, B.N., D. Tortorella, and H.L. Ploegh. 2003. Dislocation of a type I membrane protein requires interactions between membrane-spanning segments within the lipid bilayer. Mol. Biol. Cell. 14:3690–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., P. Choudhury, C.M. Cabral, and R.N. Sifers. 1999. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J. Biol. Chem. 274:5861–5867. [DOI] [PubMed] [Google Scholar]
- Loureiro, J., B.N. Lilley, E. Spooner, V. Noriega, D. Tortorella, and H.L. Ploegh. 2006. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature. 441:894–897. [DOI] [PubMed] [Google Scholar]
- Mast, S.W., K. Diekman, K. Karaveg, A. Davis, R.N. Sifers, and K.W. Moremen. 2005. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 15:421–436. [DOI] [PubMed] [Google Scholar]
- Meyer, H.H., J.G. Shorter, J. Seemann, D. Pappin, and G. Warren. 2000. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19:2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, H.H., Y. Wang, and G. Warren. 2002. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21:5645–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi, S., M.E. Pacold, D. Blom, H.L. Ploegh, and G.A. Korbel. 2004. Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turnover. Chem. Biol. 11:1677–1687. [DOI] [PubMed] [Google Scholar]
- Molinari, M., V. Calanca, C. Galli, P. Lucca, and P. Paganetti. 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 299:1397–1400. [DOI] [PubMed] [Google Scholar]
- Nadav, E., A. Shmueli, H. Barr, H. Gonen, A. Ciechanover, and Y. Reiss. 2003. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun. 303:91–97. [DOI] [PubMed] [Google Scholar]
- Neuber, O., E. Jarosch, C. Volkwein, J. Walter, and T. Sommer. 2005. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 7:993–998. [DOI] [PubMed] [Google Scholar]
- Oda, Y., T. Okada, H. Yoshida, R.J. Kaufman, K. Nagata, and K. Mori. 2006. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 172:383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari, S., C. Galli, H. Alanen, L. Ruddock, and M. Molinari. 2005. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 280:2424–2428. [DOI] [PubMed] [Google Scholar]
- Park, S., R. Isaacson, H.T. Kim, P.A. Silver, and G. Wagner. 2005. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 13:995–1005. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., J. Bordallo, P.M. Deak, C. Taxis, R. Hitt, and D.H. Wolf. 1999. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell Sci. 112:4123–4134. [DOI] [PubMed] [Google Scholar]
- Ploegh, H.L. 1995. Trafficking and assembly of MHC molecules: how viruses elude the immune system. Cold Spring Harb. Symp. Quant. Biol. 60:263–266. [DOI] [PubMed] [Google Scholar]
- Radcliffe, C.M., G. Diedrich, D.J. Harvey, R.A. Dwek, P. Cresswell, and P.M. Rudd. 2002. Identification of specific glycoforms of major histocompatibility complex class I heavy chains suggests that class I peptide loading is an adaptation of the quality control pathway involving calreticulin and ERp57. J. Biol. Chem. 277:46415–46423. [DOI] [PubMed] [Google Scholar]
- Rehm, A., P. Stern, H.L. Ploegh, and D. Tortorella. 2001. Signal peptide cleavage of a type I membrane protein, HCMV US11, is dependent on its membrane anchor. EMBO J. 20:1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu, C.E., C.M. Story, T.A. Rapoport, and H.L. Ploegh. 1999. The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J. Cell Biol. 147:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneoka, Y., P.M. Cannon, E.E. Ramsdale, J.C. Griffiths, G. Romano, S.M. Kingsman, and A.J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary, R., R. Bielmann, M. Nita-Lazar, P. Burda, and C.A. Jakob. 2005. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell. 19:765–775. [DOI] [PubMed] [Google Scholar]
- Tortorella, D., C.M. Story, J.B. Huppa, E.J. Wiertz, T.R. Jones, I. Bacik, J.R. Bennink, J.W. Yewdell, and H.L. Ploegh. 1998. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 142:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella, D., B. Gewurz, D. Schust, M. Furman, and H. Ploegh. 2000. Down-regulation of MHC class I antigen presentation by HCMV; lessons for tumor immunology. Immunol. Invest. 29:97–100. [DOI] [PubMed] [Google Scholar]
- Tsao, Y.S., N.E. Ivessa, M. Adesnik, D.D. Sabatini, and G. Kreibich. 1992. Carboxy terminally truncated forms of ribophorin I are degraded in pre-Golgi compartments by a calcium-dependent process. J. Cell Biol. 116:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist, S., and D.T. Ng. 2004. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, J., J. Urban, C. Volkwein, and T. Sommer. 2001. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 20:3124–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., A. Satoh, G. Warren, and H.H. Meyer. 2004. VCIP135 acts as a deubiquitinating enzyme during p97–p47-mediated reassembly of mitotic Golgi fragments. J. Cell Biol. 164:973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, C.L., S. Omura, and R.R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 83:121–127. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., T.R. Jones, L. Sun, M. Bogyo, H.J. Geuze, and H.L. Ploegh. 1996. a. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 84:769–779. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T.R. Jones, T.A. Rapoport, and H.L. Ploegh. 1996. b. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 384:432–438. [DOI] [PubMed] [Google Scholar]
- Wu, Y., M.T. Swulius, K.W. Moremen, and R.N. Sifers. 2003. Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc. Natl. Acad. Sci. USA. 100:8229–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, X., E. Chong, and W.R. Skach. 1999. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J. Biol. Chem. 274:2616–2624. [DOI] [PubMed] [Google Scholar]
- Ye, Y., H.H. Meyer, and T.A. Rapoport. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656. [DOI] [PubMed] [Google Scholar]
- Ye, Y., H.H. Meyer, and T.A. Rapoport. 2003. Function of the p97–Ufd1–Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y., Y. Shibata, C. Yun, D. Ron, and T.A. Rapoport. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 429:841–847. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Y. Shibata, M. Kikkert, S. van Voorden, E. Wiertz, and T.A. Rapoport. 2005. Inaugural article: recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 102:14132–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.