Abstract
Nuclear translocation of Smad proteins is a critical step in signal transduction of transforming growth factor β (TGF-β) and bone morphogenetic proteins (BMPs). Using nuclear accumulation of the Drosophila Smad Mothers against Decapentaplegic (Mad) as the readout, we carried out a whole-genome RNAi screening in Drosophila cells. The screen identified moleskin (msk) as important for the nuclear import of phosphorylated Mad. Genetic evidence in the developing eye imaginal discs also demonstrates the critical functions of msk in regulating phospho-Mad. Moreover, knockdown of importin 7 and 8 (Imp7 and 8), the mammalian orthologues of Msk, markedly impaired nuclear accumulation of Smad1 in response to BMP2 and of Smad2/3 in response to TGF-β. Biochemical studies further suggest that Smads are novel nuclear import substrates of Imp7 and 8. We have thus identified new evolutionarily conserved proteins that are important in the signal transduction of TGF-β and BMP into the nucleus.
Introduction
Signal transduction of TGF-β cytokines is mediated by an evolutionarily conserved mechanism that depends on the Smad proteins to transduce extracellular stimulus into the nucleus (Raftery and Sutherland, 1999; Shi and Massagué, 2003). At unstimulated state, Smads spontaneously shuttle across the nuclear envelope and distribute throughout the cells (Inman et al., 2002; Xu et al., 2002; Reguly and Wrana, 2003; Nicolas et al., 2004). Upon TGF-β stimulation, the receptor-activated Smads (i.e., Smad2/3 downstream of TGF-β, and Smad1/5/8 downstream of bone morphogenetic proteins [BMPs]) are phosphorylated, assemble into complexes with Smad4, and become mostly localized in the nucleus. Such signal-induced nuclear translocation of activated Smads is essential for the TGF-β–dependent gene regulations that are critical for embryonic development and homeostasis. The molecular machinery responsible for this process, especially how the activated Smads are imported as complexes, is not entirely clear (Reguly and Wrana, 2003).
Previous studies on this subject used mostly in vitro methods, including reconstituted nuclear import assay which suggested either an importin-independent or importin β–mediated mechanism for nuclear import of Smads (Xiao et al., 2000; Xu et al., 2000, 2002; Kurisaki et al., 2001). The question is whether such conclusions apply to phosphorylated Smads in intact cells. Another broader issue is if additional factors, other than those mediating nuclear translocation by themselves, may be important for either activating Smads or targeting activated Smads into the nucleus. One example is recently demonstrated requirement of kinesin in guiding intra-cytoplasmic movement of Smads toward the cell surface receptor (Batut et al., 2007).
Forward genetic screens in Drosophila have been instrumental in identifying core components of the TGF-β pathway (Raftery et al., 1995). Recently, the RNAi technology offers a complementary cell-based approach to functionally identify molecules that mediate TGF-β signaling. Several critical elements of Decapentaplegic (Dpp; Drosophila BMP) signaling including phosphorylation of Mothers against decapentaplegic (Mad), nuclear accumulation of phospho-Mad and Medea, and transcriptional up-regulation of daughters against decapentaplegic (dad), have been characterized in Drosophila tissue culture cells (Das et al., 1998; Chen et al., 2006). This, together with a collection of dsRNAs targeting the entire annotated Drosophila genome, allowed us to genetically dissect the Dpp pathway and investigate molecular requirements for nuclear targeting of Smads upon stimulation (Armknecht et al., 2005).
In this study, we describe a genome-wide RNAi screening that uncovered moleskin (Msk) as a required component in nuclear import of Dpp-activated Mad. Both genetic and biochemical studies further validated this finding. Msk belongs to a family of proteins that were originally discovered for their ability to bind the small GTPase Ran, hence the name RanBP (Ran-binding protein) (Gorlich et al., 1997). Many RanBPs have been demonstrated to mediate nuclear import or export of various molecules and are since referred to as karyopherins (importins or exportins) (Mosammaparast and Pemberton, 2004; Stewart, 2007). We show that the mammalian Msk orthologues, Imp7 and Imp8 (also known as RanBP7 and 8), are responsible for nuclear import of both TGF-β and BMP-activated Smads in mammalian cells. Furthermore, we provide evidence that Smads are direct nuclear import cargoes of Msk/Imp7/8. Our data also revealed that in contrast to activated Smads, unphosphorylated Smads may enter the nucleus via Msk/Imp7/8-independent pathways, suggesting multiple routes for nucleocytoplasmic shuttling of Smads at basal state.
Results
Whole-genome RNAi screening identified factors involved in Dpp signaling
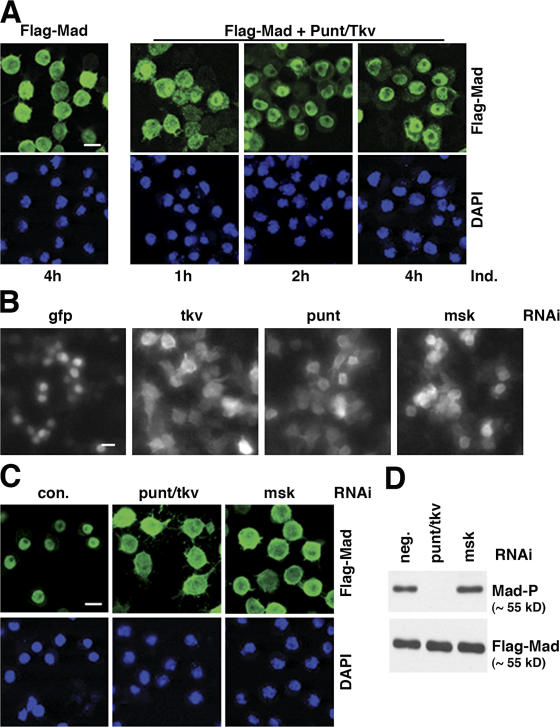
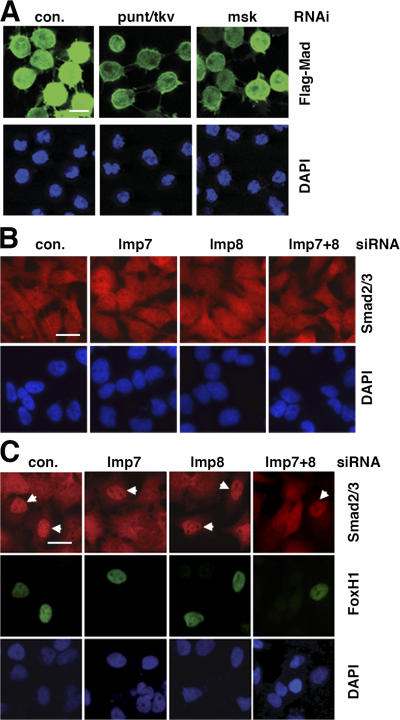
We used nuclear translocation of Mad as the readout in our RNAi screening because this is an early event in Dpp signaling. When Flag-Mad was conditionally expressed in S2R+ cells, it was detected diffusively throughout the cell (Fig. 1 A). In contrast, when the Dpp receptor kinases Punt and Thickvein (Tkv) were coexpressed, which caused Mad phosphorylation, the bulk of Flag-Mad gradually became predominantly localized to the nucleus (Fig. 1 A). With this cell line (Mad+R), we performed an RNAi screening in which the cells were treated with a library of ∼21,300 dsRNAs individually targeting over 95% of the annotated Drosophila genome (Armknecht et al., 2005). dsRNAs against the GFP and the punt/tkv combination were used as negative and positive controls, respectively. After 3 d of incubation with dsRNAs, the Mad+R cell line was induced to express Flag-Mad, Punt, and Tkv, and the subcellular location of Flag-Mad was visualized with anti-Flag immunofluorescence staining followed by high throughput automated microscopy. Upon visual inspection of images obtained from duplicate screenings, we identified 346 dsRNAs that caused diffused distribution of Flag-Mad throughout the cell compared with the negative control dsRNA. Many of the genes corresponding to the 346 dsRNAs contain domains suggestive of their functions, and can be broadly categorized as in Table I. The complete list of strong and weak hits in the primary screening can be accessed at the Drosophila RNAi Screening Center (DRSC) website (http://www.flyrnai.org).
Figure 1.
Whole-genome RNAi screening in S2R+ cells uncovered new components in the Dpp–Mad pathway. (A) Phosphorylation-dependent nuclear accumulation of Flag-Mad. S2R+ cells stably transfected with plasmids for Flag-Mad only, or Flag-Mad with punt and tkv were induced to express these proteins with CuSO4 for the indicated time. Flag-Mad was detected by immunofluorescence staining with anti-Flag, and the nuclei were marked with DAPI. Bar, 10 μm. (B) Flag-Mad distribution patterns in wells containing indicated dsRNA in genome-wide RNAi screening. S2R+ cells expressing Flag-Mad, Punt, and Tkv were stained with anti-Flag after indicated RNAi, and the images were obtained with Discovery1 automated microscopy (Molecular Devices). Bar, 10 μm. (C) In S2R+ cells induced to express Flag-Mad and Punt/Tkv, RNAi targeting either punt plus tkv or msk (with dsRNA different from that in B) blocked nuclear concentration of Flag-Mad. The cells and experimental procedure were as in A. Bar, 10 μm. (D) Same RNAi experiment as in C, and proteins were extracted from the cells and analyzed by immunoblotting with antibodies against phospho-Mad (Mad-P) or Flag.
Table I.
Preliminary hits and predicted functions
| Categories | Number |
|---|---|
| Total dsRNA screened | ∼21,300 |
| Scored positive in primary screening | 346 |
| Kinase | 7 |
| Cytoskeleton structure | 9 |
| Nuclear transport | 1 |
| Transcription factor | 13 |
| Enzymes | 13 |
| Other functions | 49 |
| Unknown functions | 67 |
| Annotation unconfirmed | 187 |
The candidate hits were selected in an anonymous manner (see Materials and methods). Indeed, among the hits that gave strong phenotype were punt and tkv, which confirmed that Mad phosphorylation is prerequisite for its nuclear accumulation and that the screening was robust (Fig. 1 B). Of particular note among the primary hits was msk, a karyopherin that was previously suggested to be required for nuclear import of activated Drosophila ERK (dERK) (Lorenzen et al., 2001). The RNAi library used here contains dsRNA targeting many molecules known to be involved in nuclear transport, including importins, exportins, and nucleoporins. But besides msk, none was among the 346 hits identified in the primary screening (Table I). In this study we focus on the analysis of msk; the validation and bioinformatics analyses of the other hits will be presented elsewhere.
Msk is required for nuclear accumulation of activated Mad and transcriptional activation of Dpp target genes
To verify the effect of msk RNAi in the primary screening, we designed and tested a second non-overlapping dsRNA against msk. Indeed, depletion of Msk by a different dsRNA also led to severely impaired nuclear concentration of Mad, and the effect was as potent as the positive control RNAi targeting Punt and Tkv (Fig. 1 C). This result strongly suggests that the block of Mad nuclear translocation we observed in the screening was not due to off-target effects of the dsRNA. In contrast to RNAi against Punt and Tkv, RNAi of msk did not affect C-terminal phosphorylation of Mad (Fig. 1 D), suggesting that Msk functions downstream of Mad phosphorylation, perhaps in transporting Mad into the nucleus.
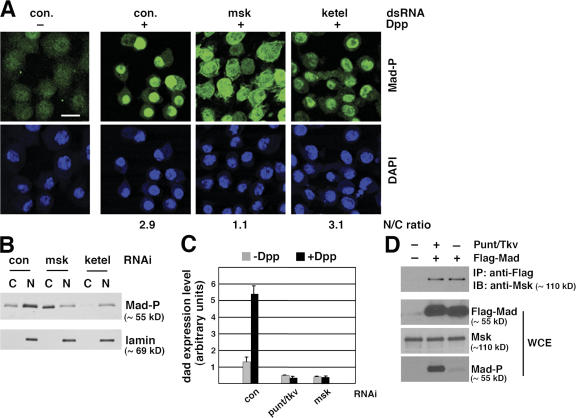
Because the screening was performed using a S2R+ cell line overexpressing exogenous Flag-Mad, we wanted to determine if endogenous Mad is under the same regulation by msk in a different Drosophila cell line. Dpp treatment of Drosophila S2 cells resulted in predominant nuclear distribution of phosphorylated Mad, as revealed by immunofluorescence staining using a phospho-Mad–specific antibody, PS1 (Fig. 2 A) (Tanimoto et al., 2000). Depletion of Msk by RNAi clearly resulted in more diffusive distribution of phospho-Mad (changing the nucleus/cytoplasm ratio from 2.9 to 1.1), while not affecting the level of Mad phosphorylation at its C terminus (Fig. 2 A and Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200703106/DC1).
Figure 2.
msk, but not the importin β homologue ketel, is required for nuclear accumulation of endogenous Mad in Dpp-treated Drosophila S2 cells. (A) S2 cells were treated with indicated dsRNA and then subject to Dpp stimulation (1 nM for 1 h). Distribution of phospho-Mad (Mad-P) was detected by immunofluorescence staining using the PS1 antibody. The phospho-Mad signal per unit area in the nucleus and cytoplasm was measured using NIH ImageJ, and the nucleus/cytoplasm (N/C) ratios are shown (>50 cells were counted per sample). Bar, 10 μm. (B) S2 cells treated with indicated dsRNA were stimulated with Dpp as in A. Subcellular fractions were prepared and examined for phospho-Mad (Mad-P) and lamin levels (C: cytoplasm; N: nucleus). (C) S2R+ cells were subject to indicated RNAi. The cells were then stimulated with Dpp (1 nM) for 2 h and the mRNA level of dad was measured by real-time RT-PCR. The expression level of Rp49 was used as the internal standard for quantitation. The plotted data are derived from multiple experiments. Error bars indicate SD. (D) Co-immunoprecipitation of endogenous Msk with Flag-Mad. Whole-cell extract (WCE) was prepared from S2 cells transfected with Flag-Mad and Punt/Tkv as indicated and subject to immunoprecipitation using anti-Flag antibody conjugated to agarose beads. The bound proteins as well as input extract were analyzed by immunoblotting with indicated antibodies.
Msk has been suggested to cooperate with the Drosophila importin β homologue Ketel in nuclear import of dERK, because mutations in either msk or ketel inhibited nuclear accumulation of dERK (Lorenzen et al., 2001). Moreover, the mammalian orthologues of Msk have been shown to function in conjunction with importin β (Gorlich et al., 1997; Jakel et al., 1999). Thus, we tested if ketel might also be involved in nuclear translocation of Mad. Knockdown of ketel by RNAi resulted in reduced phosphorylation of endogenous Mad (Fig. S1 A), but nevertheless phospho-Mad was still detected predominantly within the nucleus (Fig. 2 A). Quantitation of phospho-Mad staining intensity in the nucleus and cytoplasm confirmed that RNAi against ketel did not affect nuclear accumulation of phospho-Mad (Fig. 2 A; changing the nucleus/cytoplasm ratio from 2.9 to 3.1, n > 50). Similar observations were also made in S2R+ cells (unpublished data). Western blot analysis of cytoplasmic and nuclear fractions of S2 cells further validated that Msk, but not Ketel, is required for nuclear accumulation of phospho-Mad (Fig. 2 B). As shown in Fig. 2 B, although depletion of Ketel resulted in reduced amount of phospho-Mad through an unknown mechanism, the majority of phospho-Mad was still present in the nucleus (Fig. 2 B). Classic NLS–mediated nuclear import is dependent on importin β (Stewart, 2007). Indeed, RNAi against ketel clearly impaired nuclear accumulation of classic NLS–fused GFP, while depletion of Msk had no effect (Fig. S1 B). This result verified that RNAi against ketel was effective, and nuclear transport of Dpp-activated Mad is independent of the importin β homologue Ketel.
In both S2R+ and S2 cells, treatment with Dpp results in transcriptional activation of dad, a known Smad target gene in mammalian cells as well (Nakao et al., 1997; Tsuneizumi et al., 1997). When Msk was depleted by RNAi, the Dpp- induced increase in dad expression was completely abolished (Fig. 2 C). The blocking effect of msk RNAi on dad expression was as strong as that caused by punt/tkv RNAi (Fig. 2 C). Thus, consistent with being an essential factor for nuclear import of Mad, Msk is critical for the transcriptional output of Dpp.
Interaction between Msk and Mad
To address the question if Msk is directly involved in transporting phospho-Mad into the nucleus, we tested protein–protein interaction between endogenous Msk and Flag-tagged Mad. Indeed, endogenous Msk coimmunoprecipitated with Flag-Mad from S2 cell extract (Fig. 2 D). Under our experimental conditions, both basal state and phosphorylated Mad displayed comparable interaction with Msk (Fig. 2 D). This suggests that binding of Msk is not unique to phospho-Mad, and Msk alone may not account for why only phospho-Mad accumulates in the nucleus. Therefore, although Msk is crucial for phospho-Mad to enter the nucleus, additional factors are involved to retain only phospho-Mad in the nucleus.
The above observations in Drosophila cells identify msk as a new regulator in the Dpp pathway whose function is critical for nuclear accumulation of Dpp-activated Mad.
Analysis of msk mutant cells in the eye imaginal disc
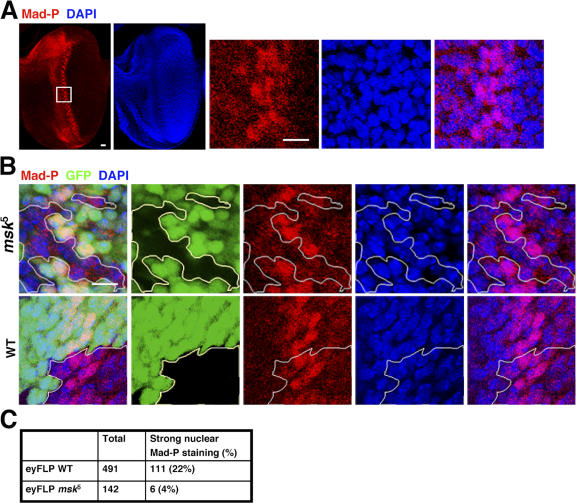
Mutations in the msk gene resulted in embryonic lethality (Baker et al., 2002; Vrailas et al., 2006). Thus, to evaluate the functions of Msk in vivo, we used ey:FLP to generate msk null (msk 5) clones in the developing eye imaginal disc (Baker et al., 2002; Vrailas et al., 2006). The clones were marked as GFP negative. In eye discs of third instar larvae, the PS1 antibody detected two stripes of phospho-Mad–containing cells around the morphogenetic furrow, consistent with the established role of Mad in eye development (Fig. 3 A) (Wiersdorff et al., 1996). The phospho-Mad signal in the anterior stripe is weaker and more diffused compared with that in the posterior stripe (Fig. 3 A). In the posterior stripe, phospho-Mad–positive cells span 5–6 cells wide, and quantitation of cell staining showed that 20.8% of the cells (n = 1,401) had phospho-Mad concentrated in the nucleus (Fig. 3 A). The rest of the cells within the posterior stripe have either undetectable or diffusive phospho-Mad staining (Fig. 3 A).
Figure 3.
msk null mutant cells in the developing eye imaginal disc did not have distinct nuclear staining of phospho-Mad. (A) Third instar eye imaginal discs (anterior to the left) were stained with PS1, which specifically recognizes phospho-Mad (Mad-P, red). The nuclei were marked with DAPI (blue). The boxed area was magnified and shown as the three panels on the right. Bars, 10 μm. (B) msk5 (null) or wild-type clones were generated using FLP recombinase driven by the eyeless promoter. The third instar eye imaginal discs were stained for phospho-Mad (red) and nuclei (blue). The clones were marked as negative for GFP signal (black) and are outlined. Bar, 10 μm. (C) msk5 or wild-type clonal cells falling within the posterior phospho-Mad–positive stripe (5–6 cells wide) were scored as having concentrated phospho-Mad staining in the nucleus or not. The numbers are obtained from more than eight discs in each case.
The msk 5 clones are small in size and number compared with the wild-type clones, consistent with previous reports that msk 5 mutation led to growth disadvantages (Baker et al., 2002; Vrailas et al., 2006). In msk 5 clones that straddle the posterior stripe, we detected a significantly smaller number of cells (4%, n = 142) with strong phospho-Mad staining concentrated in the nucleus (Fig. 3, B and C). In comparison, in wild-type clones generated by ey:FLP, the number of cells with high phospho-Mad signal distinctively in the nucleus is as high (22%, n = 491) as in genetically unmodified wild-type cells (20.8%, n = 1,401; Fig. 3, B and C).
The observed phenotype is consistent with our RNAi results in cell culture, which suggests a defect in nuclear import of phospho-Mad. Because we did not observe a considerable accumulation of cytoplasmic phospho-Mad, it is possible that in vivo the un-imported phospho-Mad is rapidly degraded or dephosphorylated. Although we cannot rule out other possibilities attributing to the observations in Fig. 3 B, it is clear that in vivo, cells with loss-of-function mutation in msk would have defects in phospho-Mad–mediated signaling.
The Msk orthologues in human are required for nuclear accumulation of Smad1 and transcriptional responses to BMP
Msk has two homologues in mammals, Imp7 and Imp8, each sharing over 50% identity in amino acid sequence with Msk. Imp7 and Imp8 themselves are ∼60% identical. Based on in vitro assays, Imp7 has been suggested to import ribosomal proteins, histone H1, HIV reverse transcription complexes, and glucocorticoid receptor into the nucleus (Jakel and Gorlich, 1998; Jakel et al., 1999; Fassati et al., 2003; Freedman and Yamamoto, 2004). Imp8 was recently shown to support nuclear import of the signal recognition particle 19 (SRP19) in vitro (Dean et al., 2001).
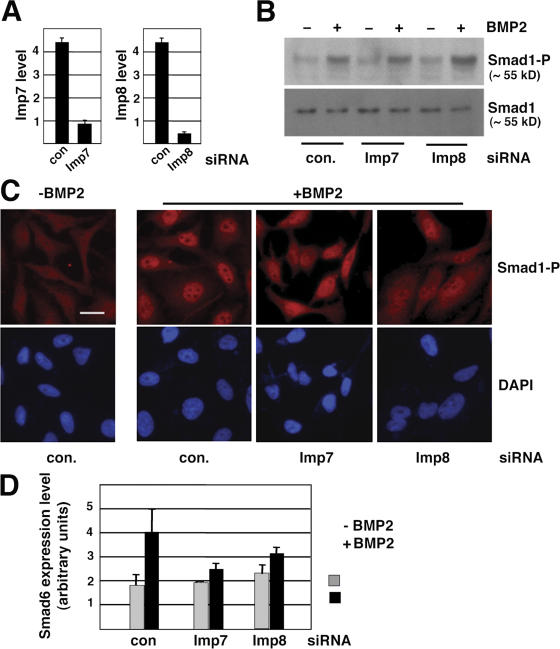
To investigate the roles of Imp7 and 8 in nuclear transport of Smads in mammalian cells, we designed siRNA duplexes that are effective in knocking down Imp7 or 8 individually (Fig. 4 A). Although Imp7 and 8 siRNA duplexes had no effects on BMP2-induced phosphorylation of Smad1 (Fig. 4 B), immunofluorescent staining with phospho-Smad1–specific antibody showed that knockdown of either Imp7 or 8 resulted in a more diffusive distribution of phospho-Smad1 after BMP2 stimulation, while in control siRNA transfected cells phospho-Smad1 was mostly present in the nucleus (Fig. 4 C). Corresponding to this defect in nuclear accumulation of Smad1, the BMP2-induced transcriptional activation of Smad6 was also suppressed in cells transfected with siRNA against either Imp7 or 8 (Fig. 4 D). Therefore, similar to their Drosophila counterpart, Imp7 and 8 are critical for nuclear translocation of BMP-activated Smad1 in mammalian cells.
Figure 4.
Imp7 and 8 are required for nuclear accumulation of BMP-activated Smad1 in HeLa cells. (A) HeLa cells transfected with indicated siRNA duplexes (40 nM) were analyzed for mRNA levels of Imp7 or Imp8 by real-time RT-PCR. GAPDH was used as the internal standard for quantitation. Error bars indicate SD. (B) HeLa cells transfected with siRNAs were treated with BMP2 (100 ng/ml, 1 h) as indicated and analyzed by immunoblotting using antibodies specific for phospho-Smad1 or total Smad1. (C) HeLa cells transfected and treated as in B were immunostained with the PS1 antibody recognizing phospho-Smad1 and analyzed by fluorescence microscopy. Bar, 10 μm. (D) BMP2-induced transcriptional up-regulation of Smad6 was inhibited by siRNA targeting Imp7 or Imp8. HeLa cells transfected and treated as in B were analyzed for Smad6 mRNA level by real-time RT-PCR, with GAPDH serving as the standard. The plotted data are derived from three experiments and the error bars indicate SD.
Imp7 and 8 are required for nuclear import of TGF-β–activated Smad2/3 and transcriptional activation of their target genes
TGF-β and BMP pathways are similar in the general signaling mechanism, but differ in the receptor kinases and Smads that are used for signaling (Shi and Massagué, 2003). We thus investigated if Imp7 and 8 are shared by TGF-β and BMP pathways in transporting different receptor-activated Smads into the nucleus. Again, knockdown of either Imp7 or 8 severely inhibited nuclear accumulation of Smad2 and 3 in response to TGF-β stimulation (Fig. 5 A). Such observation was made in both HeLa and HaCaT cells, and Smad2/3 phosphorylation in response to TGF-β was not affected by the same siRNA against Imp7 or 8 (Fig. 5 B). The block of Smad2/3 nuclear accumulation was also manifested in substantially reduced transcriptional activation of the TGF-β target gene Smad7 (Fig. 5 C). Therefore, Smads downstream of TGF-β also depend on Imp7 or 8 for nuclear translocation.
Figure 5.
Imp7 and Imp8 are required for TGF-β–activated Smad2/3 to translocate into the nucleus. (A) HeLa or HaCaT cells transfected with indicated siRNAs (40 nM) were analyzed by immunostaining using anti-Smad2/3 antibody, with or without prior TGF-β stimulation as indicated (100 pM, 30 min). The nuclei were marked by DAPI. Bars, 10 μm. (B) HaCaT cells transfected and treated as in A were examined by immunoblotting using antibodies recognizing phospho-Smad2 or total Smad2 and 3. (C) Total RNA was isolated from the same HaCaT cells as in B and the mRNA level of Smad7 was measured by quantitative real-time PCR. The plotted data are derived from three experiments and the error bars indicate SD. (D) HeLa cells were transfected with siRNA against Imp7 or Imp8 individually or in combination (20 nM each) and were stimulated with TGF-β before immunostaining as in A. Non-targeting control siRNA was used to balance the final concentration of total siRNA in each transfection (40 nM final). Bar, 10 μm. (E) Cells in D were categorized as nuclear (Smad2/3 predominantly in the nucleus) or cytoplasmic (Smad2/3 evenly distributed in the cytoplasm and nucleus) based on anti-Smad2/3 immunostaining pattern. Over 400 cells were counted in each case.
Because both Imp7 and Imp8 are required for nuclear transport of Smads, we next examined the relative contributions from these two. The efficiency of Smads nuclear translocation could be quantitated by counting the number of cells exhibiting “nucleus only” versus “cytoplasmic” distribution of Smads. We found that indeed when transfected at the same final concentration, combining siRNAs targeting Imp7 and 8 was more effective in inhibiting nuclear accumulation of Smad2/3 than individual siRNA against either Imp7 or 8 alone (Fig. 5, D and E). This suggests that Imp7 and 8 are likely to act in parallel in mediating nuclear translocation of TGF-β–activated Smad2/3.
Overexpression of Imp7 and 8 rescued siRNA effects
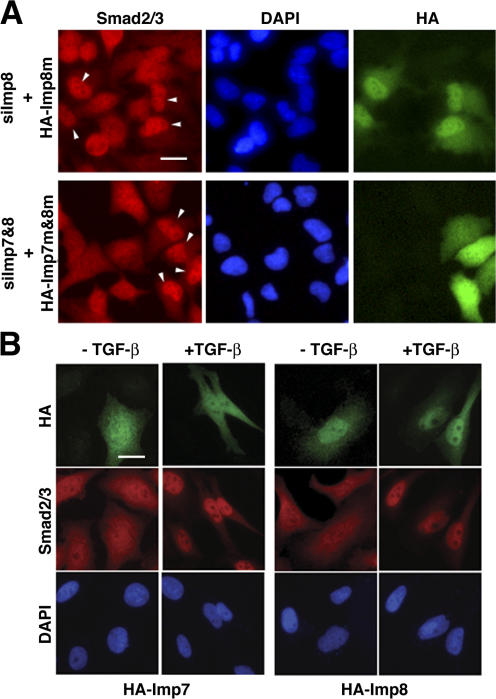
Because siRNAs against Imp7 and 8 are highly effective in blocking nuclear accumulation of Smad2/3, we examined if reintroducing Imp7 and 8 cDNAs would rescue the RNAi phenotype. To this end, we generated silent mutations in Imp7 and 8 sequences and verified that the mutants are no longer targeted by the Imp7 or Imp8 siRNA, respectively (not depicted; see Materials and methods). Such mutant cDNAs were transfected into HeLa cells 2 d after the siRNA transfection. Indeed, upon TGF-β stimulation, Smad2/3 accumulation in the nucleus was restored only in cells that expressed the rescuing HA-tagged Imp7 or 8 plasmids (Fig. 6 A). This result validated that the defects in Smads nuclear translocation observed in Figs. 4 and 5 were specifically due to depletion of endogenous Imp7 and 8.
Figure 6.
Effects of Imp7 and 8 overexpression on Smad localization. (A) Overexpression of siRNA-insensitive Imp7 and 8 rescued the defect in Smad2/3 nuclear accumulation in response to TGF-β. HeLa cells were transfected with siRNAs targeting endogenous Imp8 (top) or Imp7 and 8 combined (bottom) first, followed with expression vectors encoding HA-tagged mutant Imp7 and 8 that are no longer recognized by the siRNAs. After TGF-β stimulation (100 pM, 30 min), cells were co-stained with anti-Smad2/3 (red) and anti-HA (green) antibodies, and DAPI for the nuclei (blue). Cells expressing HA-tagged mutant Imp7 and 8 are marked with arrows. Bar, 10 μm. (B) HA-tagged wild-type Imp7 or 8 were transfected into Hela cells. After indicated treatments, cells were stained with antibodies and DAPI as in A. Bar, 10 μm.
When overexpressed in HeLa cells, Imp7 was diffusively distributed throughout the cell while more Imp8 was detected in the nucleus than in the cytoplasm (Fig. 6 B). Such patterns of Imp7 and 8 localization remained the same upon TGF-β stimulation (Fig. 6 B). Overexpression of Imp7 or 8 had no detectable effects on the distribution of endogenous Smad2/3 at both basal and TGF-β–stimulated states (Fig. 6 B). This suggests that in HeLa cells, endogenous Imp7 and 8 are not limited in quantity to support nuclear translocation of Smad2/3.
Smads are direct nuclear transport substrates of Imp7 and 8
We next investigated Smads interactions with Imp7 and 8 by coimmunoprecipitation experiments. Flag-tagged Smad1 or Smad2 were overexpressed in 293T cells and immunoprecipitated with anti-Flag antibody. In both cases, HA-tagged Imp7 or Imp8 coimmunoprecipitated with either Smad1 or Smad2 (Fig. 7, A and B). Constitutively active BMP receptor (ALK3-QD) or TGF-β receptor kinase (ALK5-TD) was cotransfected to induce C-terminal phosphorylation of Smad1 and Smad2, respectively. Such phosphorylation of Smads did not affect their interaction with Imp7 or 8 (Fig. 7, A and B). Because phosphorylated Smad1, 2, and 3 readily assemble into complexes, our results suggest that monomeric and multimeric forms of Smads have similar interactions with Imp7 or 8 (Wu et al., 2001; Chacko et al., 2004). These observations are consistent with our finding in Drosophila cells (Fig. 2 D).
Figure 7.
Interaction of Smads with Imp7 and Imp8, and the regulation by Ran-GTP. (A) Co-immunoprecipitation of Smad1 with Imp7 and Imp8. 293T cells were transfected with indicated expression plasmids and the whole-cell extract (WCE) was immunoprecipitated with anti-Flag antibody. Protein A/G bead was used as the control (c). The bound proteins and the input extract (WCE) were analyzed by immunoblotting as indicated. (B) Co-immunoprecipitation of Smad2 with Imp7 and Imp8. Same experimental design as in A, but with different expression plasmids transfected as indicated. (C) Mapping of Smad3 domains involved in interaction with Imp7/8. Recombinant GST fusions of indicated Smad3 or Smad2 fragments were used to pull down endogenous Imp7/8 in HeLa cells. The bound proteins were analyzed by an antibody that recognized both Imp7 and 8. Comparable amount of GST proteins was used in the pull down as judged by the Coomassie stain intensity. The arrowheads mark GST fusion proteins on the SDS-PAGE gel. S3MH1: aa 1–155; S2MH1: aa 1–185; S3MH2: aa 231–425; S3(L+MH2): aa 145–425; S3FL: full-length. Schematic drawing of Smad3 is also shown. (D) Purified GST-Imp8 on glutathione beads was used to pull down purified recombinant Smad1 and Smad3. The bound proteins were examined by immunoblotting using indicated antibodies. GST was used as the control. (E) Ran-GTP interrupts association between Smad3 and Imp8. GST-fusion of full-length Smad3 (GST-S3FL) was used in a pull-down experiment as in C. The bound proteins were further incubated with RanQ69L-GTP or BSA, and proteins released into the supernatant were collected and analyzed at indicated time points (15 min and 45 min elution). At the 45-min time point, the beads were washed again and proteins remaining bound to GST-S3FL (bound) were also examined by anti-HA immunoblotting.
For detailed analysis of Smad-Imp7/8 interaction, we focused on Smad3. We produced GST-fusions of the MH1, MH2, and linker plus MH2 domains of Smad3 in Escherichia coli and tested their ability to pull down endogenous Imp7 and 8 in Hela cells. When comparable amount of GST fusion proteins were used, both the MH1 (aa 1–155) and the linker plus MH2 (aa 146–425) domains were able to bind endogenous Imp7/8, with the MH1 domain exhibiting stronger interaction (Fig. 7 C). The same assay barely detected any interaction between the Smad3 MH2 (aa 231–425) domain and Imp7/8 (Fig. 7 C). Therefore, interaction with Imp7/8 appears to involve multiple interfaces in the MH1 and linker regions of Smad3. The MH1 domains of Smad2 (aa 1–185) and Smad3 are highly similar except for two insertions in Smad2 that prevent Smad2 from binding to DNA (Zawel et al., 1998). But apparently such differences did not affect Smad2 binding to Imp7/8 through the MH1 domain (Fig. 7 C). Bacterially produced GST-Imp8 was able to pull down purified recombinant Smad1 or Smad3, suggesting that Imp8 could directly interact with Smad1 or Smad3 (Fig. 7 D and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200703106/DC1).
One characteristic among importins is that the interaction with their cargoes is regulated by Ran in its GTP-bound form (Gorlich et al., 1996; Mattaj and Englmeier, 1998). To test if this is true between Smad3 and Imp8, we first pulled down HA-Imp8 using GST fusion of full-length Smad3. After washing off the unbound proteins, RanQ69L-GTP (the Q69L mutation locks Ran in its GTP-bound state) or BSA was added to the GST beads for further incubation (Fig. 7 E). Indeed we found that comparing to the BSA control, RanQ69L-GTP caused more release of Imp8 into the supernatant and correspondingly resulted in a decrease of Imp8 remaining bound to GST-Smad3 on the beads (Fig. 7 E). This suggested that association of Smad3 with Imp8 was disrupted upon binding of Ran-GTP, supporting the notion that Smad3 is a nuclear import cargo of Imp8.
Basal state Smads can enter the nucleus via Msk/Imp7/8-independent mechanisms
Without TGF-β stimulation, Smads undergo spontaneous nucleocytoplasmic shuttling and are distributed evenly in both the nucleus and cytoplasm in many types of cells (Inman et al., 2002; Xu et al., 2002; Nicolas et al., 2004). Prompted by the observation that unphosphorylated Smads interact with Imp7 and 8, we examined if Imp7 and 8 are also required for basal state Smads import into the nucleus. In Drosophila S2R+ cells, Flag-Mad was detected throughout the cells without exogenous Dpp (Fig. 8 A). The presence of Flag-Mad in the nucleus is not likely due to autocrine Dpp secreted by the cells, because further blocking any residual Mad phosphorylation by RNAi against punt and tkv did not eliminate the presence of Mad in the nucleus (Fig. 8 A). Treatment with dsRNA targeting msk also had no effect on the presence of Mad in the nucleus at basal state (Fig. 8 A). Because RNAi against punt/tkv and msk have been validated to be highly potent (Fig. 1 C), we concluded that nuclear import of basal state Mad does not rely on Msk, in contrast to Dpp-activated Mad.
Figure 8.
Roles of Msk, Imp7, and Imp8 in nuclear import of Smads at basal state. (A) S2R+ cells transfected with Flag-Mad expression vector were subject to RNAi as indicated. After inducing the expression of Flag-Mad, the cells were analyzed by anti-Flag immunofluorescence staining (green). Bar, 10 μm. (B) HeLa cells were transfected with indicated siRNAs. 3 d later, cells were stained with anti-Smad2/3 antibody (red) without prior TGF-β treatment. Bar, 10 μm. (C) 2 d after HeLa cells were transfected with indicated siRNAs, the cells were further transfected with a Myc-Fox H1 expression vector. Double- immunofluorescence staining with anti-Myc (green) and anti-Smad2/3 (red) was performed 1 d after Fox H1 transfection with no TGF-β stimulation. Bar, 10 μm.
Similar to Drosophila cells, knockdown of Imp7 and Imp8 individually or in combination did not reduce the amount of Smad2/3 in the nucleus of unstimulated HeLa cells (Fig. 8 B). Therefore, in both Drosophila and mammalian cells, although Msk and Imp7/8 interact with basal state Smads, the presence of Smads in the nucleus without TGF-β stimulation is not critically dependent on Msk or Imp7/8.
Overexpressing Fox H1 in HeLa cells is another way to drive endogenous Smad2/3 into the nucleus without TGF-β stimulation (Fig. 8 C). Such nuclear accumulation of Smad2/3 could be due to nuclear sequestration of the shuttling Smad by the nucleus-bound Fox H1, and has been observed with other Smad-interacting transcription factors such as ATF3 (Kang et al., 2003). Transfection of siRNA targeting Imp7 or Imp8, individually or combined, did not alter the “nucleus only” pattern of Fox H1, and did not affect Fox H1–induced nuclear concentration of Smad2/3 (Fig. 8 C). In control siRNA-transfected cells, 82.3% (n = 34) of Fox H1–positive cells contained endogenous Smad2/3 predominantly in the nuclei, whereas in cells with double-knockdown of Imp7 and Imp8, 84.3% (n = 32) of Fox H1–expressing cells have Smad2/3 in the nucleus. Thus, the above observations, from both Drosophila and mammalian cells, led us to conclude that unphosphorylated Smads may be able to enter the nucleus through additional mechanisms, such as those described in previous studies (Xiao et al., 2000; Kurisaki et al., 2001; Xu et al., 2002).
Discussion
Genome-wide RNAi screening in this study offers a genetic approach to uncover new elements in TGF-β signal transduction. Here we identify and validate with in vivo evidence that Msk and its mammalian orthologues Imp7 and 8 are critical components in transporting TGF-β–activated Smads into the nucleus. Biochemical evidence further suggests that Msk/Imp7/8 directly import phospho-Smads as cargoes.
Although there appears to be some discrepancy between these new findings and our previous reports that importins are dispensable for the nuclear import of Smads, these observations can be reconciled (Xu et al., 2002, 2003). Our present and previous studies, based on different approaches, may have revealed different nuclear import mechanisms used by basal and activated Smads to enter the nucleus. There are important differences comparing Smads import with or without TGF-β stimulation. Unphosphorylated Smads are monomers, but phosphorylated Smads are assembled into complexes with Smad4 and are thus much larger in size (Wu et al., 2001; Chacko et al., 2004). Moreover, as phospho-Smads accumulate in the nucleus they have to move across the nuclear pore against an ascending concentration gradient of Smads already in the nucleus, whereas unphosphorylated Smads never reach a higher concentration in the nucleus than in the cytoplasm. Thus, importing phospho-Smad complexes and unphosphorylated Smad monomers may entail different mechanisms, with or without the participation of importins. Indeed, our RNAi data in both Drosophila and mammalian cells suggest that nuclear import of the two forms of Smads is very different regarding the requirement of Msk/Imp7/8. This type of differential requirement for import factors is not unique to Smads. In fact, STATs (signal transducers and activators of transcription) in the interferon pathway are another example in which the latent STATs are imported by an importin-independent mechanism, whereas the phosphorylated STATs depend on importins to accumulate in the nucleus (Meyer et al., 2002; McBride and Reich, 2003; Marg et al., 2004). It is also interesting to note that phospho-Smads were still detected in the nucleus upon RNAi-mediated knockdown of Msk/Imp7/8. Although we cannot rule out the trivial explanation that this may be due to incomplete depletion of the targeted proteins, this observation may also suggest additional import mechanisms for activated Smads. We recognize that our previous finding of importin-independent nuclear import of Smads was largely based on an in vitro reconstituted nuclear import assay (Xu et al., 2002, 2003). Although this in vitro system is widely accepted, it may not fully recapitulate nuclear import of activated Smads in cells (Adam et al., 1992). Based on our RNAi data, regarding the requirement of importins, the conclusion drawn from the in vitro import assay may not apply to phospho-Smads in intact cells. However, the current study does not necessarily contradict the previous suggestions that direct Smad–nucleoporin interaction is critical for nuclear import of Smads (Xu et al., 2002; Sapkota et al., 2007).
Our data showed that Msk/Imp7/8 interacted with Smads regardless of their phosphorylation status; thus, additional factors must be involved to explain why only TGF-β/BMP– activated Smads can accumulate in the nucleus. Because basal- state Smads are actively exported out of the nucleus (Inman et al., 2002; Xu et al., 2002; Kurisaki et al., 2006), it is possible that retaining only phospho-Smads in the nucleus requires blocking Smads nuclear export, a scenario that has been demonstrated for Smad4 (Chen et al., 2005). This hypothesis would be consistent with findings in live cells, in which TGF-β signaling led to reduced mobility of Smad2 in the nucleus (Schmierer and Hill, 2005).
Because Msk, Imp7, and Imp8 are shown to be critical for targeting phospho-Smads into the nucleus, it is conceivable that regulatory inputs to this nuclear import factor would impact TGF-β signaling. Although we did not notice any changes in subcellular localization of Msk or Imp7/8 in response to TGF-β in cultured cells (Fig. 6 B and unpublished data), during Drosophila embryonic development, Msk distribution changed between cytoplasm and nucleus in a dynamic fashion (Lorenzen et al., 2001). Moreover, Msk is phosphorylated on tyrosine residues with yet-unknown functional consequences (Lorenzen et al., 2001). If and how Msk localization is regulated and by what signals are completely open questions at present.
A number of mitogen-induced phosphorylation events in the linker region of Smad have been suggested to inhibit TGF-β–induced nuclear translocation of Smads in Xenopus and mammalian cells (Kretzschmar et al., 1997, 1999; Grimm and Gurdon, 2002; Sapkota et al., 2007). Because part of the Imp7/8 binding was mapped to the linker region of Smad3, it will be interesting to determine if linker phosphorylation would affect the interaction between Smads and Imp7/8 and hence the rate of nuclear import. It is also worth noting that Msk has been genetically implicated in the nuclear import of activated ERK in Drosophila. Such convergence on the same molecule for nuclear import raises the possibility of cross-talk between MAP kinase and TGF-β pathways at the level of nuclear translocation of key signal transducers.
Materials and methods
Whole-genome RNAi screening
The dsRNA library targeting the whole Drosophila genome and the format for screening have been described previously (Armknecht et al., 2005). dsRNAs were deposited in 384-well plates, and in each plate one well is reserved for gfp dsRNA (negative control) and one for combined punt and tkv dsRNA (positive control). 104 S2R+ cells in 10 μl serum-free media were seeded in each well and incubated for 1 h, after which 30 μl of serum-containing media was added. After incubation for 3 d, the cells were induced with 0.5 mM CuSO4 for 3 h followed by fixation with 4% paraformaldehyde in PBS (10 min) and immunofluorescence staining with anti-Flag antibody (Sigma-Aldrich). Automated microscopy was performed using the Discovery1 system (Molecular Devices). The screening was repeated once. The wells containing selected hits were reported to DRSC, which in turn revealed the identities of the dsRNAs contained in those wells. The complete dataset including strong and weak hits in the two screenings is available at http://www.flyrnai.org.
Cell culture, transfection, and immunofluorescent staining
Drosophila S2 and S2R+ cells were cultured in Schneider media with 10% fetal bovine serum (Invitrogen) and transfected with Effectene (QIAGEN). HeLa and 293T cells were cultured in DME with 10% fetal bovine serum and transfected with Lipofectamine 2000 (Invitrogen). Cells with or without treatment (1 nM Dpp, 100 pM TGF-β, or 100 ng/ml BMP2 as indicated; all from R&D Systems) were processed for immunofluorescence staining as described previously (Xu et al., 2002). PS1 was a gift from P. ten Dijke (Leids University, Netherlands). Alexa 488– or Alexa 633–conjugated anti–rabbit secondary antibodies (Invitrogen) were used as indicated, and cells were mounted in Vectorshield (Vector Laboratories). Immunofluorescence microscopy and image acquisition were done with an inverted microscope (20×/0.45, 40×/0.6, 60×/1.40; Eclipse TE2000-S; Nikon) and a digital camera (SPOT RT-KE; Diagnostic Instruments, Inc.) using vendor-provided software. For confocal microscopy, a DMIRE2 inverted microscope and the TCS scanning system from Leica were used. The images were captured wither lasers at: UV (DAPI), 488 nM (Alexa 488 or GFP), and 633 nM (Alexa 633) wavelengths at room temperature using vendor-provided software. 20×/0.70, 40×/(1.25–0.75), and 63×/1.40 oil immersion objectives were used for low and high magnification images. Adobe Photoshop was used to adjust the brightness and contrast of the entire images if necessary. Final figures were assembled using Adobe Photoshop. For quantitation purposes, confocal sections with the strongest signal were selected, and the staining intensity in the nucleus and cytoplasm was measured using NIH ImageJ. Only cells with unsaturated signals were chosen for such analysis.
Drosophila strains and immunostaining of imaginal discs
y w ey:FLP;P{w +mC=Ubi:GFP} P{neoFRT}80B and msk5 P{neo FRT}80B (gifts from A. Vrailas, Emory University, Atlanta, GA) were used to generate msk5 clones in the developing eye imaginal disc as described previously (Baker et al., 2002; Vrailas et al., 2006). Third instar stage larvae were dissected and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. The imaginal discs were permeabilized with 0.3% Triton X-100/PBS for 30 min at room temperature and blocked with 5% normal horse serum/0.3% Triton X-100/2%BSA/PBS for 5 h at room temperature. PS1 was used at 1:2,000 dilution in the blocking buffer and the incubation lasted overnight at 4°C. After washing, the discs were stained with Alexa 633–conjugated anti–rabbit secondary antibody (2 μg/ml; Invitrogen) for 2 h at room temperature. The imaginal discs were mounted in Vectorshield/PBS (1:1; Vector Laboratories) and examined with confocal microscopy as described above.
RNAi in Drosophila and mammalian cells
General procedures for generating dsRNA and RNAi in Drosophila S2 and S2R+ cells were as described previously (Clemens et al., 2000). Two amplicons corresponding to different coding regions of msk, DRSC11340 and DRSC23929, were used to generate nonoverlapping dsRNA targeting msk. Sequence information for the two msk amplicons can be found at the DRSC website (http://www.flyrnai.org). For HeLa and HaCaT cells, siRNA was transfected using HiPerFect at a final concentration of 40 nM (QIAGEN). siRNA targeting Imp7: GAUGGAGCCCUG CAUAUGA dTdT (a gift from M. Stevenson, University of Massachusetts Medical School, Worcester, MA); siRNA targeting Imp8: GAGAUCTTCCGAACUAUUAdGdT. To generate the rescue constructs, the coding sequences targeted by the siRNAs were mutated into Imp7: GATGGAGCCT TACACATGA; Imp8: GAGATCTTTAGGACAATAA. The residues changed by mutagenesis are underlined.
S2 cell fractionation
S2 cells were suspended in 20 mM Hepes, pH 7.6, 5 mM MgCl2, 10 mM KCl, 1 mM EGTA, 1 mM EDTA, 250 mM sucrose, and 0.025% digitonin (EMD Biosciences). Cells were passed through 181/2 G syringes three times and incubated on ice for 5 min. The homogenate was centrifuged at 800 g for 10 min. The supernatant was collected as the cytoplasmic fraction. The pellet was further extracted with 20 mM TrisCl, pH 7.5, 250 mM NaCl, and 0.5% NP-40 to yield the nuclear fraction.
Protein–protein interaction assay
For coimmunoprecipitation experiments, Drosophila or 293T cells were lysed in 20 mM Tris Cl, pH 7.4, 200 mM NaCl, 5 mM Mg2Cl, 20 mM NaF, 20 mM Na4P2O7, 20 mM β-glycerolphosphate, 0.5% NP-40, and 2 mM DTT supplemented with protease inhibitors. Cell extracts were incubated with anti-Flag conjugated to agarose beads (Sigma-Aldrich) at 4°C for 4–16 h, followed by 3× wash in the lysis buffer before immunoblotting. Anti-Msk was a gift from L. Perkins (Harvard Medical School, Boston, MA).
For GST pull-down experiments, Hela or 293T cells were lysed as above and incubated with 10 μg purified GST fusion protein on glutathione beads at 4°C for 4–16 h. The beads were then washed 3× in the lysis buffer. For RanQ69L-GTP elution, GTP-loaded His-RanQ69L or BSA as the control (both at ∼0.4 μg/μl) were added to the washed beads and further incubated at room temperature for 45 min. At indicated time points, an aliquot of the supernatant was taken for immunoblotting. At the end point, the beads were washed once in the lysis buffer and the bound proteins were analyzed by anti-HA or anti-Imp7 (Imgenex).
Full-length human Imp8, Smad1, and Smad3 were produced in E. coli and purified as GST fusions. GST-Smad1 (a gift from F. Liu, Rutgers University, NJ) and GST-Smad3 were digested with thrombin to remove the GST moiety (Novagen). 10 μg of GST-Imp8 on beads was incubated with Smad1 or Smad3 (0.5–1 μg/μl) at 4°C with rotation for 4 h. The buffer contained 20 mM TrisCl, pH 7.5, 200 mM NaCl, 0.05% NP-40, 5% glycerol, and PMSF. The beads were washed 3× in the same buffer and the bound proteins were analyzed by immunoblotting.
Online supplemental material
Fig. S1 shows the controls for the RNAi experiments in Fig. 2. Fig. S2 shows the SDS-PAGE of purified recombinant proteins used in Fig. 7 D. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200703106/DC1.
Supplementary Material
Acknowledgments
We thank B. Mathey-Prevot (Harvard Medical School) for the screening facility and suggestions; D. Brower (University of Arizona, Tucson, AZ), M. Brodsky (University of Massachusetts Medical School), R. Padgett (Rutgers University, NJ), and D. Gorlich (ZMBH, Heidelberg, Germany) for reagents; and D. Wingate for her contributions to the study. We also thank Drs. R. Davis and Q. Xu for critical review of the manuscript.
This work was funded by National Institutes of Health grants to L. Xu (RO1 CA108509) and Y.T. Ip (RO1 GM53269).
Abbreviations used in this paper: BMP, bone morphogenetic protein; dad, daughters against decapentaplegic; Dpp, Decapentaplegic; Mad, Mothers against decapentaplegic; Msk, Moleskin; Tkv, Thickvein.
References
- Adam, S.A., R. Sterne-Marr, and L. Gerace. 1992. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 219:97–110. [DOI] [PubMed] [Google Scholar]
- Armknecht, S., M. Boutros, A. Kiger, K. Nybakken, B. Mathey-Prevot, and N. Perrimon. 2005. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol. 392:55–73. [DOI] [PubMed] [Google Scholar]
- Baker, S.E., J.A. Lorenzen, S.W. Miller, T.A. Bunch, A.L. Jannuzi, M.H. Ginsberg, L.A. Perkins, and D.L. Brower. 2002. Genetic interaction between integrins and moleskin, a gene encoding a Drosophila homolog of importin-7. Genetics. 162:285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut, J., M. Howell, and C.S. Hill. 2007. Kinesin-mediated transport of smad2 is required for signaling in response to tgf-beta ligands. Dev. Cell. 12:261–274. [DOI] [PubMed] [Google Scholar]
- Chacko, B.M., B.Y. Qin, A. Tiwari, G. Shi, S. Lam, L.J. Hayward, M. De Caestecker, and K. Lin. 2004. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol. Cell. 15:813–823. [DOI] [PubMed] [Google Scholar]
- Chen, H.B., J.G. Rud, K. Lin, and L. Xu. 2005. Nuclear targeting of transforming growth factor-beta-activated Smad complexes. J. Biol. Chem. 280:21329–21336. [DOI] [PubMed] [Google Scholar]
- Chen, H.B., J. Shen, Y.T. Ip, and L. Xu. 2006. Identification of phosphatases for Smad in the BMP/DPP pathway. Genes Dev. 20:648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, J.C., C.A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B.A. Hemmings, and J.E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 97:6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, P., L.L. Maduzia, H. Wang, A.L. Finelli, S.H. Cho, M.M. Smith, and R.W. Padgett. 1998. The Drosophila gene Medea demonstrates the requirement for different classes of Smads in dpp signaling. Development. 125:1519–1528. [DOI] [PubMed] [Google Scholar]
- Dean, K.A., O. von Ahsen, D. Gorlich, and H.M. Fried. 2001. Signal recognition particle protein 19 is imported into the nucleus by importin 8 (RanBP8) and transportin. J. Cell Sci. 114:3479–3485. [DOI] [PubMed] [Google Scholar]
- Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J.M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman, N.D., and K.R. Yamamoto. 2004. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol. Biol. Cell. 15:2276–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., N. Pante, U. Kutay, U. Aebi, and F.R. Bischoff. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., M. Dabrowski, F.R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, O.H., and J.B. Gurdon. 2002. Nuclear exclusion of Smad2 is a mechanism leading to loss of competence. Nat. Cell Biol. 4:519–522. [DOI] [PubMed] [Google Scholar]
- Inman, G.J., F.J. Nicolas, and C.S. Hill. 2002. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol. Cell. 10:283–294. [DOI] [PubMed] [Google Scholar]
- Jakel, S., and D. Gorlich. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel, S., W. Albig, U. Kutay, F.R. Bischoff, K. Schwamborn, D. Doenecke, and D. Gorlich. 1999. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18:2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y., C.R. Chen, and J. Massagué. 2003. A self-enabling TGFbeta response coupled to stress signaling. Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 11:915–926. [DOI] [PubMed] [Google Scholar]
- Kretzschmar, M., J. Doody, and J. Massagué. 1997. Opposing BMP and EGF signalling pathways converge on the TGFβ family mediator Smad1. Nature. 389:618–622. [DOI] [PubMed] [Google Scholar]
- Kretzschmar, M., J. Doody, I. Timokhina, and J. Massagué. 1999. A mechanism of repression of TGFβ/Smad signaling by ongenic ras. Genes Dev. 13:804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki, A., S. Kose, Y. Yoneda, C.H. Heldin, and A. Moustakas. 2001. Transforming growth factor-beta induces nuclear import of Smad3 in an importin-beta1 and Ran-dependent manner. Mol. Biol. Cell. 12:1079–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki, A., K. Kurisaki, M. Kowanetz, H. Sugino, Y. Yoneda, C.H. Heldin, and A. Moustakas. 2006. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol. Cell. Biol. 26:1318–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen, J.A., S.E. Baker, F. Denhez, M.B. Melnick, D.L. Brower, and L.A. Perkins. 2001. Nuclear import of activated D-ERK by DIM-7, an importin family member encoded by the gene moleskin. Development. 128:1403–1414. [DOI] [PubMed] [Google Scholar]
- Marg, A., Y. Shan, T. Meyer, T. Meissner, M. Brandenburg, and U. Vinkemeier. 2004. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 165:823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj, I.W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265–306. [DOI] [PubMed] [Google Scholar]
- McBride, K.M., and N.C. Reich. 2003. The ins and outs of STAT1 nuclear transport. Sci. STKE. 2003:RE13. [DOI] [PubMed]
- Meyer, T., A. Begitt, I. Lodige, M. van Rossum, and U. Vinkemeier. 2002. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 21:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast, N., and L.F. Pemberton. 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14:547–556. [DOI] [PubMed] [Google Scholar]
- Nakao, A., M. Afrakhte, A. Morén, T. Nakayama, J.L. Christian, R. Heuchel, S. Itoh, M. Kawabata, N.E. Heldin, C.H. Heldin, and P. ten Dijke. 1997. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signaling. Nature. 389:631–635. [DOI] [PubMed] [Google Scholar]
- Nicolas, F.J., K. De Bosscher, B. Schmierer, and C.S. Hill. 2004. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 117:4113–4125. [DOI] [PubMed] [Google Scholar]
- Raftery, L.A., and D.J. Sutherland. 1999. TGF-beta family signal transduction in Drosophila development: from Mad to Smads. Dev. Biol. 210:251–268. [DOI] [PubMed] [Google Scholar]
- Raftery, L.A., V. Twombly, K. Wharton, and W.M. Gelbart. 1995. Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics. 139:241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly, T., and J.L. Wrana. 2003. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol. 13:216–220. [DOI] [PubMed] [Google Scholar]
- Sapkota, G., C. Alarcon, F.M. Spagnoli, A.H. Brivanlou, and J. Massague. 2007. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell. 25:441–454. [DOI] [PubMed] [Google Scholar]
- Schmierer, B., and C.S. Hill. 2005. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor {beta}-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 25:9845–9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., and J. Massagué. 2003. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 113:685–700. [DOI] [PubMed] [Google Scholar]
- Stewart, M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8:195–208. [DOI] [PubMed] [Google Scholar]
- Tanimoto, H., S. Itoh, P. ten Dijke, and T. Tabata. 2000. Hedgehog creates a gradient of DPP activity in Dosophila wing imaginal discs. Mol. Cell. 5:59–71. [DOI] [PubMed] [Google Scholar]
- Tsuneizumi, K., T. Nakayama, Y. Kamoshida, T.B. Kornberg, J.L. Christian, and T. Tabata. 1997. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 389:627–631. [DOI] [PubMed] [Google Scholar]
- Vrailas, A.D., D.R. Marenda, S.E. Cook, M.A. Powers, J.A. Lorenzen, L.A. Perkins, and K. Moses. 2006. smoothened and thickveins regulate Moleskin/Importin 7-mediated MAP kinase signaling in the developing Drosophila eye. Development. 133:1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersdorff, V., T. Lecuit, S.M. Cohen, and M. Mlodzik. 1996. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development. 122:2153–2162. [DOI] [PubMed] [Google Scholar]
- Wu, J.W., M. Hu, J. Chai, J. Seoane, M. Huse, C. Li, D.J. Rigotti, S. Kyin, T.W. Muir, R. Fairman, et al. 2001. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol. Cell. 8:1277–1289. [DOI] [PubMed] [Google Scholar]
- Xiao, Z., X. Liu, and H.F. Lodish. 2000. Importin beta mediates nuclear translocation of Smad 3. J. Biol. Chem. 275:23425–23428. [DOI] [PubMed] [Google Scholar]
- Xu, L., Y.G. Chen, and J. Massagué. 2000. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFb-dependent phosphorylation. Nat. Cell Biol. 2:559–562. [DOI] [PubMed] [Google Scholar]
- Xu, L., Y. Kang, S. Col, and J. Massagué. 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. Mol. Cell. 10:271–282. [DOI] [PubMed] [Google Scholar]
- Xu, L., C. Alarcon, S. Col, and J. Massagué. 2003. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J. Biol. Chem. 278:42569–42577. [DOI] [PubMed] [Google Scholar]
- Zawel, L., J.L. Dai, P. Buckhaults, S. Zhou, K.W. Kinzler, B. Vogelstein, and S.E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell. 1:611–617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.