Abstract
Among the goals of RNA structural and functional genomics is determining structures and establishing the functions of a rich repertoire of simple sequence repeats in transcripts. These repeats are present in transcripts from their ‘birth’ in the nucleus to their ‘death’ in cytoplasm and have the potential of being involved in many steps of RNA regulation. The knowledge of their structural features and functional roles will also shed more light on the postulated mechanisms of RNA pathogenesis in a growing list of neurological diseases caused by simple sequence repeat expansions. Here, we discuss several different lines of research to support the hypothesis that the mechanism of RNA pathogenesis may be a more common phenomenon triggered or modulated also by abundant long normal repeats. We propose structures of the repeat regions in transcripts of genes involved in Triplet Repeat Expansion Diseases. We have classified the polymorphic repeat alleles of these genes according to their ability to form hairpin structures in transcripts, and describe the distribution of different structural forms of the repeats in the human population. We have also reported the results of a systematic survey of the human transcriptome to identify mRNAs containing triplet repeats and to classify them according to structural and functional criteria. Based on this knowledge, we discuss the putative wider role of triplet repeat RNA hairpins in human diseases. A hypothetical model is proposed in which long normal RNA hairpins formed by the repeats may also be involved in pathogenesis.
Triplet repeat expansion diseases and possible mechanisms of pathogenesis
Sixteen human neurological and neuromuscular disorders are known to be caused by the expansions of trinucleotide repeats (CUG)n, (CGG)n, (CCG)n, (GAA)n and (CAG)n in single genes (reviewed in 1–5). These diseases include myotonic dystrophy, fragile X syndrome, Friedreich ataxia, spinocerebellar ataxias and Huntington’s disease. Their common feature is genetic anticipation, i.e. an earlier age of the onset and increased severity of clinical symptoms in subsequent generations, correlated with the increasing length of expanded alleles. The repeats are polymorphic in a normal population, and they are believed to be neutral to phenotype until reaching a certain pathogenic length characteristic for each gene. In the majority of the disease-related genes containing the CAG repeats in their coding sequences this pathological threshold is about 40 repeats (2). In some of these genes this length is very close to that of the longest normal alleles (1,2). The highly expanded alleles rarely contain more than 100 CAG repeats. On the contrary, the expansions of triplet repeats located in the non-coding regions of genes are larger and more variable, reaching even thousands of repeats (1).
Possible mechanisms by which the expansions are involved in the pathogenesis of Triplet Repeat Expansion Diseases (TREDs) fall into three categories (4,6). The gain-of-function mechanism, consistent with dominant inheritance patterns, is characteristic for diseases caused by CAG repeat expansions in translated sequences. The abnormal protein products with extended polyglutamine tracts and altered function are thought to be the major factors in pathogenesis (7,8). The loss-of-function mechanism, consistent with recessive inheritance, is represented by the fragile X syndrome and Friedreich ataxia in which the transcriptional silencing of the FMR-1 gene (9,10) and transcriptional interference in the case of X-25 gene (11) result in reduced levels of their protein products. The third category includes the gain-of-function mechanism known for dominantly inherited myotonic dystrophy, which is caused by trinucleotide repeat expansion in a non-coding sequence, and the expanded repeat is pathogenic on the level of RNA.
According to the originally proposed mechanism of RNA pathogenesis in myotonic dystrophy type 1, the transcript with the expanded CUG repeat alters the functioning of RNA binding proteins which results in the disrupted processing of other CUG repeat containing transcripts (12). The expanded transcript was shown to accumulate as nuclear foci (13) and co-localize with proteins of the muscleblind family (14–16) to which it binds in vitro (17). The altered expression and splicing regulatory function of other CUG-binding proteins including CUG-BP (18) and other members of the CELF protein family (19–23) was also demonstrated in DM1 tissues, and several target transcripts for these proteins were identified (23–28). The proteins of the CELF family differ from the proteins of the muscleblind family in their RNA binding specificity. The former bind to single-stranded CUG repeats while the latter bind to double-stranded CUG repeats. Long CUG repeats were shown earlier to form hairpin structures in vitro (29) and the muscleblind proteins were isolated based on this property (17). In agreement with the recently proposed model of RNA role in DM1 pathogenesis (30,31), the binding of the muscleblind proteins to the expanded CUG repeat hairpins is accompanied by the elevated expression of CUG-BP and the altered splicing of the target transcripts. The cellular consequences of muscleblind protein sequestration by the expanded CUG repeat hairpins remain to be determined. A similar mechanism of RNA pathogenesis is proposed for myotonic dystrophy type 2 (30–33); it may also operate in other diseases caused by the repeat expansions in non-coding sequences (4,34–36) and its contribution to the pathogenesis of the so-called polyglutamine diseases cannot be excluded (37). In order to learn more about the details of such mechanisms, and evaluate their significance for human diseases, a better understanding of the normal roles played by triplet repeats in RNA is also required.
Triplet repeat regions in transcripts may form hairpin structures of different architecture
There is a multitude of various hairpins in cellular transcripts but those composed of triplet repeats are special. Their stems containing periodic U/U, A/A, G/G and C/C mismatches are highly regular structures according to the results of in vitro studies (29,38). The G-rich repeat hairpins may also be considered as having the potential to form fold-back quadruplex structures which were demonstrated for DNA (39–42). Expanded (CUG)n repeat hairpins appear in an electron microscope as rod-like structures consistent with the A-form of RNA helix (43). As far as the structural context is concerned the repeated sequence may be hidden in the interior of the transcript structure or exposed and easily accessible for interactions with RNA binding proteins. The structural role of the sequences directly flanking the repeat is important for long normal repeats which are the focus of this survey.
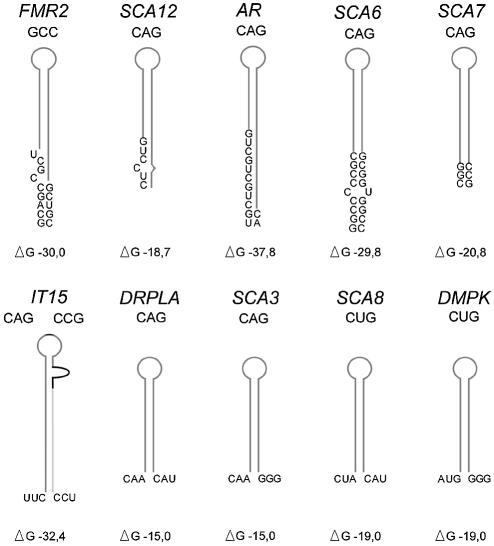
The 10 mRNAs considered here included AR, DRPLA, IT15, SCA3, SCA6, SCA7, in which the repeated sequence is present in ORF, FMR2 and SCA12 with the repeats located in 5′-UTR as well as DMPK and SCA8 in which it occurs in 3′-UTR. The FMR1, SCA1 and SCA2 mRNAs were not considered as the majority of their alleles contain interruptions in the repeat sequences. In agreement with the M-fold (44,45) data, the CAG repeats of the SCA3 and DRPLA mRNAs and CUG repeats of the SCA8 and DMPK mRNAs form hairpins composed of the repeated sequence only (Fig. 1). In IT15 mRNA, two different polymorphic repeated motifs CAG and CCG interact with each other and the excessive CAG repeats may form an extra stem–loop structure. In the other five mRNAs, the sequences flanking the repeat contribute to the hairpin in a way shown in Figure 1.
Figure 1.
Schematic representation of RNA structure modules predicted for trinucleotide repeats and their nearest flanking sequences from 10 TREDs-related transcripts. Repeated sequences (17 repeats) are marked with a gray line. In the case of IT15, an additional 10 CCG repeats (light gray line) and 12 naturally occurring specific nucleotides located between two repeat tracts (black line) were taken into account. The secondary structures were predicted and free energy values calculated using the M-fold program version 3.0 (44).
The predicted free energies of experimentally determined hairpin structures composed of 17 repeats (46) are –15, –19 and –18 kcal/mol for (CAG)17, (CUG)17, and (CCG)17, respectively. Taking into account the contribution from the specific flanking sequences which are shown in Figure 1, these hairpin stabilities rise significantly, and in AR, SCA6 and FMR2 transcripts, they are approximately doubled. Thus, it appears from this analysis that the triplet repeat regions of transcripts related to TREDs are capable of forming hairpin structures of different molecular architectures which may have an impact on their protein binding properties.
Variation in repeat length means variation in hairpin formation ability
Having established the contributions from specific flanking sequences to the predicted hairpin structures formed by the repeats, we then asked the following questions: how may individuals differ in the distribution of their repeat alleles, and how is the natural variation in length of the repeats related to their ability to form hairpin structures? To answer these questions we analyzed the genotyping data collected for the same 10 triplet repeat loci in a normal population.
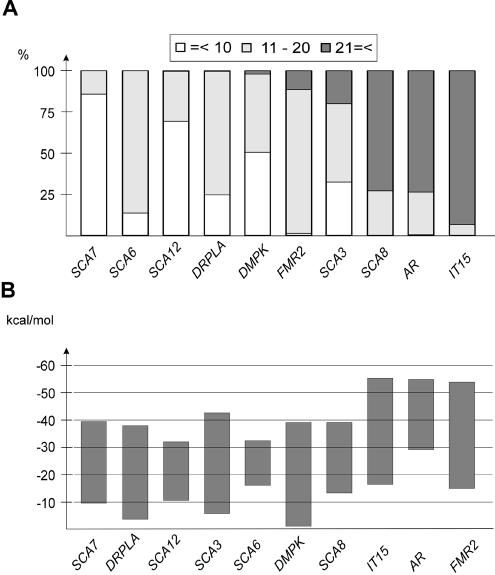
For the purpose of this discussion, the repeats present at the investigated loci were classified according to their length as short, medium and long (Fig. 2A). Transcripts containing 10 or less repeats were shown to be single-stranded, those harboring 11–20 repeats may form quasi-stable hairpins and stable hairpins may occur in transcripts with longer runs of the repeated sequence (17,29,43). From this classification, it turns out that IT15, SCA8 and AR have the highest incidence of long repeat tracts (Fig. 2A). A comparison of the multilocus genotypes between individuals from the studied population revealed that the most frequently observed ratio of short/medium/long repeat alleles at 10 disease loci (20 alleles altogether) was 6:9:5, respectively (A.Jasinska et al., submitted for publication). The number of long alleles ranged from 1 to 8 among the analyzed individuals reflecting a large variation in their distribution.
Figure 2.
Structural interpretation of genotyping data. (A) Distribution of short, medium and long repeat alleles at 10 TREDs loci classified as described in the text. Data are based on the results of genotyping 234 chromosomes from a Polish population using a standard method. In the case of the IT15 gene, the combined number of CAG and adjacent CCG repeats was determined. Three ranges of the repeat length are distinguished by the different shading of the bars as specified in the graph legend. The genes are ordered according to the increasing contribution of long repeat tracts. No association was found between short alleles on the one side and long alleles on the other. (B) Calculated free energy of RNA structures formed by triplet repeats of various lengths in specific flanking sequence context shown in Figure 1. The bars show the M-fold predicted free energy ranges at each locus. The following triplet repeat length ranges which spanned the smallest and the largest repeat alleles found in a normal world population were used in this analysis: 12–31 repeats for SCA3, 7–18 for SCA6, 7–17 for SCA7, 16–37 for SCA8, 7–28 for SCA12, 6–35 for DRPLA, 11–33 for AR, 5–37 for DMPK, 7–35 for FMR2 and 17–51 for IT15 (3,5).
The genotyping data may be interpreted from the perspective of thermodynamics. The allele length ranges observed in a normal population may be expressed as RNA structure stabilities and the obtained ranges of free energy values are presented in Figure 2B. In the majority of transcripts, the predicted free energy difference between structures formed by the shortest and the longest alleles are higher than 30 kcal/mol. The spectra of different repeat alleles in transcripts will therefore form hairpin energy landscapes which, like combinations of genotypes at different loci, will be characteristic for different individuals. This will result in differences in the efficiency of RNA–protein interactions and the efficiency of those cellular processes that require hairpin unwinding, e.g. the initiation step of mRNA translation (47,48).
Are long trinucleotide repeats common in human mRNAs?
In the preceding sections, we have discussed the varieties of RNA structures that may be formed by triplet repeats of different length and their flanks, using the example of TREDs-related transcripts. Among other questions regarding the structures and functions of the repeats in RNAs, the following seem to be of special importance: how many other human mRNAs harbor the repeats in their sequences? At what frequencies do certain types of repeats occur? What are the preferred repeat locations in mRNAs? And, to what functional classes of proteins do products of these mRNAs belong? We have identified a large set of these mRNAs, classified and characterized in the transcriptome-wide survey. Our observations are discussed here from the perspective of RNA structural genomics (49) and the postulated mechanism of RNA pathogenesis in TREDs.
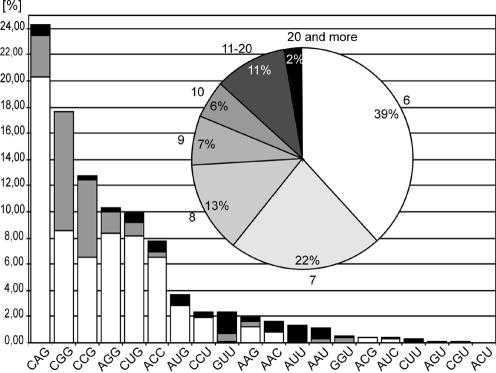
The GenBank nucleotide sequence database (January 2003) was searched using the BLAST program to identify transcripts containing triplet repeat tracts composed of at least six repeats. Single interruptions were allowed in the repeated sequences that were longer than six repeats. Twenty different trinucleotide repeat motifs were analyzed and the gene functions were assigned according to OMIM and GenBank. A total of 718 triplet repeat tracts were found in 619 different mRNAs. Their list is available as Supplementary Material. The triplet repeat distribution in mRNAs (Fig. 3) shows that six types of the repeated motif (CAG)n, (CGG)n, (CCG)n, (CUG)n, (AGG)n and (ACC)n account for 83% of all occurrences. Different triplet repeats are distributed unevenly in the human transcripts. Most of the repeats (67%) are located in ORFs, 24% in 5′-UTRs and only 9% in 3′-UTRs. Taking into account the median length of these three sections in human mRNAs 240 nt for 5′-UTR, 1100 nt for ORF and 400 nt for 3′-UTR (50), the repeats are relatively over-represented in 5′-UTR and under-represented in 3′-UTR. The three motifs (CGG)n, (CCG)n and (CAG)n, account for 78% of all repeats in 5′-UTR and the (CAG)n repeats are the most frequent in ORFs. On the other hand, the U-rich repeat motifs, (GUU)n, (AUU)n and (CUU)n, and A-rich motifs, (AAC)n and (AAU)n, are slightly more prevalent than (CAG)n in 3′-UTRs. The localization of the repeated sequence in mRNA may hint at its possible function. 5′-UTRs are usually involved in translation regulation, while 3′-UTRs play an important role in mRNA stability, transport and cellular localization. The prevalence of structure-forming (CNG)n repeats in 5′-UTRs and ORFs, and preferentially single-stranded U-rich and A-rich repeats in 3′-UTRs, agrees well with this scenario.
Figure 3.
Trinucleotide repeats in human mRNAs according to the GenBank nucleotide sequence database. The graph shows the distribution of repeats according to the type of repeated motif and its localization in mRNA: ORF, white bars; 5′-UTR, gray bars; 3′-UTR, black bars (100% = 718 tracts from 619 mRNAs). Inset, frequencies of triplet repeat tracts of different length among a total of 718 tracts.
All identified human mRNAs containing triplet repeats were also classified according to the repeat length (Fig. 3, inset). It appears that the medium length repeats, reiterated 11–20 times, make up 11% (77/718) of the cases, and there is only a small proportion, about 2% (19/718) of mRNAs, with 21 or more repeats. Besides the TREDs related mRNAs there were only seven other mRNAs with long repeats identified (gene symbols and OMIM or GenBank numbers: TRIM9 *606555, MAB21L1 *601280, HRIHFB2206 XM_043054, TFIID *313650, MN1 *156100, MLLT3 *159558, KCNN3 *602983). This surprisingly low number of mRNAs with long repeats may be because of several reasons. First is the under-representation of genes with long repeats in the presently available human genome sequence. The alleles with long repeats are prone to deletion when cloned in bacteria as well as difficult to sequence, and they are likely to be hidden in the remaining sequence gaps. Second, the repeat polymorphism has not yet been extensively studied and relevant information is available for only 7% of the 718 repeat tracts reported here. A much higher rate of polymorphic repeats in human transcriptome should be expected, as it was shown earlier that about 50% of the analyzed genes were polymorphic (51–53). If this low number of mRNAs containing long repeats is not strongly underestimated this would mean that the contribution of TREDs loci to the global cellular balance of long alleles forming stable hairpins in transcripts is significant.
Of the 619 mRNAs identified in this study, 54% have an assigned biological function. Each mRNA of known function was classified as belonging to one of the following eight functional categories: transcription/translation, cell–cell communication, intracellular signaling, metabolism, defense/immunity, cytoskeletal/structural, cellular processes, DNA replication/modification. We observed a functional bias when this set of triplet repeat containing mRNAs was compared with a much larger set of over 17 000 human genes with a known function (50): (i) over-representation of proteins involved in intracellular and extracellular signaling (increase from 28 to 39%); (ii) over-representation of proteins involved in transcription and translation (increase from 24 to 34%); and (iii) under-representation of proteins involved in metabolism (decrease from 19 to 9%). Our results confirm the high frequency of transcription factors among the proteins encoded by (CAG)n repeat-containing mRNAs reported earlier (54) and reveal the preponderance of triplet repeats in mRNAs encoding signaling proteins.
From the perspective of RNA pathogenesis in TREDs, it is important to identify all those transcripts the function of which may be compromised by the expanded repeat. Our survey provides a long list of transcripts, which considerably extends the number of previously described human genes with triplet repeats (51,55,56). In a recent study (57), 26 triplet repeat tracts longer than 20 repeats have been identified and not many of them occurred in introns. The functional classification of the transcripts identified in our survey shows which cellular functions could be among those altered by the mutant transcript. It should be emphasized, however, that both the list of transcripts and their classification suffer from being incomplete. Nevertheless, they give the first comprehensive insight into this group of mRNAs.
Are long normal alleles neutral or pathogenic?
It appears from our analysis that about 2% of human mRNAs contain trinucleotide motifs repeated six and more times, and additional triplet repeats occur in introns (57). It is likely that these repeats are regulatory sites functioning by interactions with repeat binding proteins (21,22,31). By analogy with the CUG repeats (17), one might expect that other repeats of the CNG family also have their specific ssRNA and dsRNA binding proteins (46). Such repeat-type and structure-type specific proteins could be involved in the co-regulation of specific transcripts. Expanded CUG repeats bind proteins of the muscleblind family in a length-dependent manner (17). The threshold length for binding is about (CUG)20 in HeLa cell extract. This shows that these proteins may bind not only to the expanded but also to the normal transcripts that form CUG repeat hairpin structures of sufficient length (17). This may suggest that hairpin structure composed of 20 CUG repeats is not easily unwound by cellular helicases and other proteins present in cell extract. Thus, there is most likely no qualitative difference in the type of RNA structure and type of repeat binding protein between long normal alleles and the expanded alleles. This raises an intriguing question: whether only the giant mutant hairpins can play the role of cellular offenders and normal length hairpins, depleted from dsRNA-binding proteins, are their victims?
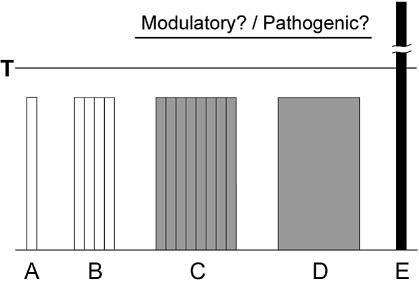
It seems feasible that in the absence of mutant expanded transcripts, one or several long normal repeat-containing transcripts showing abnormally increased expression or the collective action of multiple long but non-expanded hairpins may trigger similar effects in cells as proposed in the model presented in Figure 4. The mechanism of pathogenesis may involve the sequestration or activation of double-stranded RNA binding proteins. If so, ‘sporadic’ cases of diseases may occur with clinical symptoms similar to TREDs but without mutations in the known loci and without clear anticipation. Many such examples of myotonic dystrophies and ataxias have been described previously (58–61). A similar mechanism of pathogenesis may also be considered for common neurodevelopmental and psychiatric diseases such as autism, schizophrenia and bipolar disorder, for which the search for expanded triplet repeat alleles at numerous single loci was conducted but the results remain ambiguous (62–65). Whether these unexplained cases are caused by cumulative contributions from different normal genes or overexpressed genes, as shown in the presented model (Fig. 4), or they result from mutations in yet undiscovered genes remains to be established. Even if the combination of the putative pathogenic effects of long non-expanded RNA hairpins from multiple loci or the effects of overexpressed loci do not cause diseases directly, they may play the role of a risk-modifying factor influencing the time of disease onset and its severity. Clearly, all these questions need to be formally addressed in order to determine the scale on which the mechanism of RNA pathogenesis, shown for two types of myotonic dystrophy so far, may operate in cells.
Figure 4.
A model depicting the putative involvement of long normal RNA repeat hairpins in the pathogenesis. Bars A, B, C, respectively, represent the low, average and high frequency of transcripts with long normal repeat hairpins (represented by the number of columns within bars). Bar D, strong over-expression of a single transcript of that type; Bar E, repeat expansion in a single transcript. Lane T separates normal and expanded repeat length. Note that situations D and E occur in the background of either A, B or C. White, gray and black colors symbolize normal physiological, potentially pathological and pathological states, respectively.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the State Committee for Scientific Research, grant nos 6P04B03118, PBZ/KBN/040/P04/11 and PBZ/KBN/040/P04/12, and the Foundation for Polish Science, grant no. 8/2000.
REFERENCES
- 1.Richards R.I. and Sutherland,G.R. (1997) Dynamic mutation: possible mechanisms and significance in human disease. Trends Biochem. Sci., 22, 432–436. [DOI] [PubMed] [Google Scholar]
- 2.Cummings C.J. and Zoghbi,H.Y. (2000) Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet., 9, 909–916. [DOI] [PubMed] [Google Scholar]
- 3.Bowater R.P. and Wells,R.D. (2001) The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol., 66, 159–202. [DOI] [PubMed] [Google Scholar]
- 4.Ranum L.P. and Day,J.W. (2002) Dominantly inherited, non-coding microsatellite expansion disorders. Curr. Opin. Genet. Dev., 12, 266–271. [DOI] [PubMed] [Google Scholar]
- 5.Wilmot G.R. and Warren,S.T. (1998) A new mutational basis for disease. In Wells,R.D. and Warren,S.T. (eds), Genetic Instabilities and Hereditary Neurological Diseases. Academic Press, San Diego, London, Boston, New York, Sydney, Tokyo, Toronto. [Google Scholar]
- 6.Timchenko L.T. and Caskey,C.T. (1996) Trinucleotide repeat disorders in humans: discussions of mechanisms and medical issues. FASEB J., 10, 1589–1597. [DOI] [PubMed] [Google Scholar]
- 7.La Spada A.R. and Taylor,J.P. (2003) Polyglutamines placed into context. Neuron, 38, 681–684. [DOI] [PubMed] [Google Scholar]
- 8.Perutz M.F. (1999) Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem. Sci., 24, 58–63. [DOI] [PubMed] [Google Scholar]
- 9.Pieretti M., Zhang,F.P., Fu,Y.H., Warren,S.T., Oostra,B.A., Caskey,C.T. and Nelson,D.L. (1991) Absence of expression of the FMR-1 gene in fragile X syndrome. Cell, 66, 817–822. [DOI] [PubMed] [Google Scholar]
- 10.Bardoni B. and Mandel,J.L. (2002) Advances in understanding of fragile X pathogenesis and FMRP function and in identification of X linked mental retardation genes. Curr. Opin. Genet. Dev., 12, 284–293. [DOI] [PubMed] [Google Scholar]
- 11.Campuzano V., Montermini,L., Molto,M.D., Pianese,L., Cossee,M., Cavalcanti,F., Monros,E., Rodius,F., Duclos,F., Monticelli,A. et al. (1996) Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science, 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- 12.Wang J., Pegoraro,E., Menegazzo,E., Gennarelli,M., Hoop,R.C., Angelini,C. and Hoffman,E.P. (1995) Myotonic dystrophy: evidence for a possible dominant-negative RNA mutation. Hum. Mol. Genet., 4, 599–606. [DOI] [PubMed] [Google Scholar]
- 13.Taneja K.L., McCurrach,M., Schalling,M., Housman,D. and Singer,R.H. (1995) Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol., 128, 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fardaei M., Larkin,K., Brook,J.D. and Hamshere,M.G. (2001) In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res., 29, 2766–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mankodi A., Urbinati,C.R., Yuan,Q.P., Moxley,R.T., Sansone,V., Krym,M., Henderson,D., Schalling,M., Swanson,M.S. and Thornton,C.A. (2001) Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet., 10, 2165–2170. [DOI] [PubMed] [Google Scholar]
- 16.Fardaei M., Rogers,M.T., Thorpe,H.M., Larkin,K., Hamshere,M.G., Harper,P.S. and Brook,J.D. (2002) Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet., 11, 805–814. [DOI] [PubMed] [Google Scholar]
- 17.Miller J.W., Urbinati,C.R., Teng-Umnuay,P., Stenberg,M.G., Byrne,B.J., Thornton,C.A. and Swanson,M.S. (2000) Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J., 19, 4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timchenko L.T., Timchenko,N.A., Caskey,C.T. and Roberts,R. (1996) Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum. Mol. Genet., 5, 115–121. [DOI] [PubMed] [Google Scholar]
- 19.Lu X., Timchenko,N.A. and Timchenko,L.T. (1999) Cardiac elav-type RNA-binding protein (ETR-3) binds to RNA CUG repeats expanded in myotonic dystrophy. Hum. Mol. Genet., 8, 53–60. [DOI] [PubMed] [Google Scholar]
- 20.Philips A.V., Timchenko,L.T. and Cooper,T.A. (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science, 280, 737–741. [DOI] [PubMed] [Google Scholar]
- 21.Good P.J., Chen,Q., Warner,S.J. and Herring,D.C. (2000) A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J. Biol. Chem., 275, 28583–28592. [DOI] [PubMed] [Google Scholar]
- 22.Ladd A.N., Charlet,N. and Cooper,T.A. (2001) The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol., 21, 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki H., Jin,Y., Otani,H., Yasuda,K. and Inoue,K. (2002) Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells, 7, 133–141. [DOI] [PubMed] [Google Scholar]
- 24.Savkur R.S., Philips,A.V. and Cooper,T.A. (2001) Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nature Genet., 29, 40–47. [DOI] [PubMed] [Google Scholar]
- 25.Seznec H., Agbulut,O., Sergeant,N., Savouret,C., Ghestem,A., Tabti,N., Willer,J.C., Ourth,L., Duros,C., Brisson,E. et al. (2001) Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet., 10, 2717–2726. [DOI] [PubMed] [Google Scholar]
- 26.Buj-Bello A., Furling,D., Tronchere,H., Laporte,J., Lerouge,T., Butler-Browne,G.S. and Mandel,J.L. (2002) Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum. Mol. Genet., 11, 2297–2307. [DOI] [PubMed] [Google Scholar]
- 27.Charlet-B. N., Savkur,R.S., Singh,G., Philips,A.V., Grice,E.A. and Cooper,T.A. (2002) Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell, 10, 45–53. [DOI] [PubMed] [Google Scholar]
- 28.Mankodi A., Takahashi,M.P., Jiang,H., Beck,C.L., Bowers,W.J., Moxley,R.T., Cannon,S.C. and Thornton,C.A. (2002) Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell, 10, 35–44. [DOI] [PubMed] [Google Scholar]
- 29.Napierala M. and Krzyzosiak,W.J. (1997) CUG repeats present in myotonin kinase RNA form metastable ‘slippery’ hairpins. J. Biol. Chem., 272, 31079–31085. [DOI] [PubMed] [Google Scholar]
- 30.Timchenko L.T., Tapscott,S.J., Cooper,T.A. and Monckton,D.G. (2002) Myotonic dystrophy: discussion of molecular basis. Adv. Exp. Med. Biol., 516, 27–45. [DOI] [PubMed] [Google Scholar]
- 31.Faustino N.A. and Cooper,T.A. (2003) Pre-mRNA splicing and human disease. Genes Dev., 17, 419–437. [DOI] [PubMed] [Google Scholar]
- 32.Liquori C.L., Ricker,K., Moseley,M.L., Jacobsen,J.F., Kress,W., Naylor,S.L., Day,J.W. and Ranum,L.P. (2001) Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science, 293, 864–867. [DOI] [PubMed] [Google Scholar]
- 33.Tapscott S.J. and Thornton,C.A. (2001) Reconstructing myotonic dystrophy. Science, 293, 816–817. [DOI] [PubMed] [Google Scholar]
- 34.Hagerman R.J., Leehey,M., Heinrichs,W., Tassone,F., Wilson,R., Hills,J., Grigsby,J., Gage,B. and Hagerman,P.J. (2001) Intention tremor, parkinsonism and generalized brain atrophy in male carriers of fragile X. Neurology, 57, 127–130. [DOI] [PubMed] [Google Scholar]
- 35.Hagerman R.J., Greco,C., Chudley,A., Leehey,M., Tassone,F., Grigsby,J., Hills,J., Wilson,R., Harris,S.W. and Hagerman,P.J. (2001) Neuropathology and neurodegenerative features in some older male permutation carriers of fragile X syndrome. Am. J. Hum. Genet., 69, 177. [Google Scholar]
- 36.Bardoni B. and Mandel,J.L. (2002) Advances in understanding of fragile X pathogenesis and FMRP function and in identification of X linked mental retardation genes. Curr. Opin. Genet. Dev., 12, 284–293. [DOI] [PubMed] [Google Scholar]
- 37.Peel A.L., Rao,R.V., Cottrell,B.A., Hayden,M.R., Ellerby,L.M. and Bredesen,D.E. (2001) Double-stranded RNA-dependent protein kinase, PKR, binds preferentially to Huntington’s disease (HD) transcripts and is activated in HD tissue. Hum. Mol. Genet., 10, 1531–1538. [DOI] [PubMed] [Google Scholar]
- 38.Tian B., White,R.J., Xia,T., Welle,S., Turner,D.H., Mathews,M.B. and Thornton,C.A. (2000) Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA, 6, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fry M. and Loeb,L.A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F.M. (1995) Acid-facilitated supramolecular assembly of G-quadruplexes in d(CGG)4. J. Biol. Chem., 270, 23090–23096. [DOI] [PubMed] [Google Scholar]
- 41.Kettani A., Kumar,R.A. and Patel,D.J. (1995) Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 254, 638–656. [DOI] [PubMed] [Google Scholar]
- 42.Woodford K.J., Howell,R.M. and Usdin,K. (1994) A novel K(+)-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem., 269, 27029–27035. [PubMed] [Google Scholar]
- 43.Michalowski S., Miller,J.W., Urbinati,C.R., Paliouras,M., Swanson,M.S. and Griffith,J. (1999) Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res., 27, 3534–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology. NATO ASI Series, Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 11–43. [Google Scholar]
- 45.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 46.Sobczak K., de Mezer,M., Michlewski,G., Krol,J. and Krzyzosiak,W.J. (2003) RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res., 31, 5469–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raca G., Siyanova,E.Y., McMurray,C.T. and Mirkin,S.M. (2000) Expansion of the (CTG)(n) repeat in the 5′-UTR of a reporter gene impedes translation. Nucleic Acids Res., 28, 3943–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 49.Doudna J.A. (2000) Structural genomics of RNA. Nat. Struct. Biol., 7 (suppl.), 954–956. [DOI] [PubMed] [Google Scholar]
- 50.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 51.Wren J.D., Forgacs,E., Fondon,J.W.,III, Pertsemlidis,A., Cheng,S.Y., Gallardo,T., Williams,R.S., Shohet,R.V., Minna,J.D. and Garner,H.R. (2000) Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet., 67, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Margolis R.L., Abraham,M.R., Gatchell,S.B., Li,S.H., Kidwai,A.S., Breschel,T.S., Stine,O.C., Callahan,C., McInnis,M.G. and Ross,C.A. (1997) cDNAs with long CAG trinucleotide repeats from human brain. Hum. Genet., 100, 114–122. [DOI] [PubMed] [Google Scholar]
- 53.Grierson A.J., van Groenigen,M., Groot,N.P., Lindblad,K., Hoovers,J.M., Schalling,M., de Belleroche,J. and Baas,F. (1999) An integrated map of chromosome 18 CAG trinucleotide repeat loci. Eur. J. Hum. Genet., 7, 12–19. [DOI] [PubMed] [Google Scholar]
- 54.Gerber H.P., Seipel,K., Georgiev,O., Hofferer,M., Hug,M., Rusconi,S. and Schaffner,W. (1994) Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science, 263, 808–811. [DOI] [PubMed] [Google Scholar]
- 55.Riggins G.J., Lokey,L.K., Chastain,J.L., Leiner,H.A., Sherman,S.L., Wilkinson,K.D. and Warren,S.T. (1992) Human genes containing polymorphic trinucleotide repeats. Nature Genet., 2, 186–191. [DOI] [PubMed] [Google Scholar]
- 56.Stallings R.L. (1994) Distribution of trinucleotide microsatellites in different categories of mammalian genomic sequence: implications for human genetic diseases. Genomics, 21, 116–121. [DOI] [PubMed] [Google Scholar]
- 57.Subramanian S., Madgula,V.M., George,R., Mishra,R.K., Pandit,M.W., Kumar,C.S. and Singh,L. (2003) Triplet repeats in human genome: distribution and their association with genes and other genomic regions. Bioinformatics, 19, 549–552. [DOI] [PubMed] [Google Scholar]
- 58.Mankodi A. and Thornton,C.A. (2002) Myotonic syndromes. Curr. Opin. Neurol., 15, 545–552. [DOI] [PubMed] [Google Scholar]
- 59.Meola G. (2000) Clinical and genetic heterogeneity in myotonic dystrophies. Muscle Nerve, 23, 1789–1799. [DOI] [PubMed] [Google Scholar]
- 60.Schols L., Szymanski,S., Peters,S., Przuntek,H., Epplen,J.T., Hardt,C. and Riess,O. (2000) Genetic background of apparently idiopathic sporadic cerebellar ataxia. Hum. Genet., 107, 132–137. [DOI] [PubMed] [Google Scholar]
- 61.Abele M., Burk,K., Schols,L., Schwartz,S., Besenthal,I., Dichgans,J., Zuhlke,C., Riess,O. and Klockgether,T. (2002) The aetiology of sporadic adult-onset ataxia. Brain, 125, 961–968. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H., Liu,X., Zhang,C., Mundo,E., Macciardi,F., Grayson,D.R., Guidotti,A.R. and Holden,J.J. (2002) Reelin gene alleles and susceptibility to autism spectrum disorders. Mol. Psychiatry, 7, 1012–1017. [DOI] [PubMed] [Google Scholar]
- 63.Vincent J.B., Paterson,A.D., Strong,E., Petronis,A. and Kennedy,J.L. (2000) The unstable trinucleotide repeat story of major psychosis. Am. J. Med. Genet., 97, 77–97. [DOI] [PubMed] [Google Scholar]
- 64.Antonarakis S.E., Blouin,J.L., Lasseter,V.K., Gehrig,C., Radhakrishna,U., Nestadt,G., Housman,D.E., Kazazian,H.H., Kalman,K., Gutman,G. et al. (1999) Lack of linkage or association between schizophrenia and the polymorphic trinucleotide repeat within the KCNN3 gene on chromosome 1q21. Am. J. Med. Genet., 88, 348–351. [PubMed] [Google Scholar]
- 65.Pulver A.E., Mulle,J., Nestadt,G., Swartz,K.L., Blouin,J.L., Dombroski,B., Liang,K.Y., Housman,D.E., Kazazian,H.H., Antonarakis,S.E. et al. (2000) Genetic heterogeneity in schizophrenia: stratification of genome scan data using co-segregating related phenotypes. Mol. Psychiatry, 5, 650–653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.