Abstract
The budding yeast regulation of Ace2 and morphogenesis (RAM) network integrates cell fate determination and morphogenesis. Its disruption impairs polarized growth and causes mislocalization of the transcription factor Ace2, resulting in failure of daughter cell–specific transcription required for cell separation. We find that phosphoregulation of the conserved AGC family kinase Cbk1 is critical for RAM network function. Intramolecular autophosphorylation of the enzyme's activation loop is critical for kinase activity but is only partially required for cell separation and polarized growth. In marked contrast, phosphorylation of a C-terminal hydrophobic motif is required for Cbk1 function in vivo but not for its kinase activity, suggesting a previously unappreciated level of control for this family of kinases. Phosphorylation of the C-terminal site is regulated over the cell cycle and requires the transcription factor Ace2 as well as all RAM network components. Therefore, Ace2 is not only a downstream target of Cbk1 but also reinforces activation of its upstream regulator.
Introduction
Eukaryotic cells exhibit exquisite control over their architecture; this is critical for cell motility, division, and polarity. Networks of signaling proteins determine cell morphology by orchestrating cytoskeleton organization, membrane trafficking, and gene expression. Fundamentally important pathways are conserved between metazoans and the budding yeast Saccharomyces cerevisiae (for review see Pruyne et al., 2004). In budding yeast, as in other eukaryotes, cell morphology is closely coupled with cell cycle progress. Bud emergence and growth in late G1 require establishment and maintenance of polarized growth through regulated organization of actin cytoskeleton assembly and membrane traffic. As cells pass into G2/M, buds depolarize to switch from apical to isotropic growth. The cytoskeleton is further reorganized during cytokinesis, when an actomyosin ring forms and contracts to separate mother and daughter cell cytoplasm. During cytokinesis, a septum is deposited between mother and daughter cells; this is destroyed a few minutes later, when cytokinesis is complete, resulting in mother/daughter separation. Temporal coordination of these events requires conserved signaling pathways but remains incompletely understood.
The control of cell morphology is also important for the determination of cell fate. Asymmetric segregation of molecules or structures that influence gene expression links a cell's differentiation status to its underlying structure. Budding yeast cells exhibit cell fate asymmetry that involves segregation of determinants to the daughter cell. The transcription factor Ace2 accumulates specifically in the daughter cell nucleus, where it induces expression of chitinases and glucanases required for septum destruction (Dohrmann et al., 1992; O'Conallain et al., 1999; Colman-Lerner et al., 2001; Weiss et al., 2002). Ace2 asymmetry is also responsible for daughter-specific delay of G1 progression through an as-yet-unknown mechanism (Laabs et al., 2003).
The budding yeast regulation of Ace2 and morphogenesis (RAM) network is a recently discovered signaling pathway that controls cell fate asymmetry and polarized growth. Components of this pathway are conserved in a broad range of eukaryotes and are generally involved in the control of cell architecture (Verde et al., 1998; Gallegos and Bargmann, 2004; He et al., 2005a; Hergovich et al., 2006; Seiler et al., 2006). The yeast network comprises six genes: CBK1, KIC1, HYM1, MOB2, TAO3/PAG1, and SOG2 (Nelson et al., 2003). Cells lacking any of these proteins exhibit two phenotypes: a failure to degrade the septum between mother and daughter, resulting in large groups of connected cells, and poor maintenance of polarized growth. The cell separation defect results from the mislocalization of Ace2 to both mother and daughter nuclei, resulting in the loss of Ace2-dependent transcription (Bidlingmaier et al., 2001; Colman-Lerner et al., 2001). However, defective polarized growth is not attributable to loss of Ace2 function: cells lacking Ace2 can maintain polarized growth (Weiss et al., 2002). Therefore, the RAM network has separate roles in regulation of Ace2 and control of polarized growth.
The localization of RAM network proteins over the cell cycle reflects their dual roles. The proteins concentrate at sites of cell growth, such as the bud tip and the cortex of the expanding bud, and redistribute to the bud neck late in cell division, during telophase (Colman-Lerner et al., 2001; Weiss et al., 2002; Nelson et al., 2003). In addition to cell cortex localization, the Ndr/Lats family protein kinase Cbk1 and its conserved binding partner Mob2 localize to the daughter cell nucleus at the M–G1 transition. This localization requires Ace2, and nuclear localization of Ace2 is similarly dependent on Cbk1 and Mob2 (Colman-Lerner et al., 2001; Weiss et al., 2002). Cbk1 and Mob2 lack canonical nuclear localization sequences, suggesting that the proteins associate with Ace2 and enter the nucleus as a complex.
Cbk1 kinase activity is essential for RAM network function. How is the enzyme controlled? Although Cbk1 protein levels remain constant, kinase activity fluctuates over the cell cycle with maximal specific activity during early bud growth and late mitosis (Weiss et al., 2002). The kinase's activity is low if other RAM network genes are deleted, suggesting that it functions downstream of the other components of the pathway (Nelson et al., 2003).
In addition, Cbk1 has two putative regulatory phosphorylation sites that are conserved among AGC group kinases (for review see Hergovich et al., 2006). Analogous sites are important for the in vivo function of Cbk1-related kinases in budding yeast (such as Dbf2), Drosophila (such as tricornered and warts), and mammals (such as Ndr and Lats; Millward et al., 1999; Mah et al., 2001; Tamaskovic et al., 2003; Wu et al., 2003; Emoto et al., 2004; He et al., 2005b). One site, found in the kinase domain activation loop, is likely autophosphorylated (Tamaskovic et al., 2003). The other site, located in a hydrophobic motif immediately C-terminal to the kinase domain, is phosphorylated by an upstream kinase. Germinal center kinases, which are related to Ste20 and other p21-activated kinases, phosphorylate this site in several cases (Chan et al., 2005; Stegert et al., 2005; Emoto et al., 2006). The role of phosphorylation at each site has been examined in vitro for the mammalian Ndr proteins. In Drosophila, these phosphorylation sites are essential for tricornered function in vivo (Emoto et al., 2004; He et al., 2005b). These findings are consistent with a prevailing model for the regulatory function of phosphorylation at these sites: they increase the kinase's specific activity. This view is supported primarily by enzymatic and structural studies of Akt/protein kinase B (PKB), an AGC family member (Yang et al., 2002a,b; Nolen et al., 2004).
We analyzed the in vivo function of these phosphorylation sites in Cbk1 and found that both were phosphorylated but that their function cannot be explained by simple regulation of the enzyme's catalytic activity. Mutation of one of these sites, which is in Cbk1's activation loop (“T-loop site”), severely compromised in vitro kinase activity but yielded only an intermediate phenotype for cell separation and polarized growth. In contrast, mutation of the C-terminal hydrophobic motif site (“CT-motif”) produced a protein with substantial kinase activity but an entirely null phenotype. Using phosphospecific antibodies, we found that both sites were regulated over the cell cycle; the CT-motif site's phosphorylation exhibited dramatic fluctuation, peaking before bud emergence and during cytokinesis. All RAM network components were essential for CT-motif modification; a subset was essential for T-loop phosphorylation, which we find is an intramolecular reaction in vivo. Cbk1 is extensively phosphorylated at additional sites, and dephosphorylation of these sites requires CT-motif phosphorylation. Interestingly, full phosphorylation of the C-terminal hydrophobic motif required Ace2, a downstream effector of the RAM network; thus, the regulatory target modulates activation of its upstream regulator. This study of Cbk1 phosphoregulation defines a novel role for CT-motif phosphorylation independent of kinase activation. It is this molecular event that bridges RAM network signaling with its phenotypic outputs of cell separation and polarized growth.
Results
Two conserved phosphorylation sites are required for Cbk1 function in vivo
Ndr/Lats family kinases share two conserved phosphorylation sites that are important for kinase function (Millward et al., 1999; Tamaskovic et al., 2003; Emoto et al., 2004; He et al., 2005b). One site is found in the T-loop of the kinase domain (serine 570), and the other lies in a short hydrophobic region C-terminal to the kinase domain, the CT-motif (threonine 743). To determine whether the putative phosphorylation sites are important for Cbk1's in vivo function, we mutated these sites to the nonphosphorylatable amino acid alanine and characterized the resulting phenotypes. We made these mutations, as well as substitution of an aspartic acid essential for catalytic activity, in HA-tagged Cbk1 expressed under the control of its endogenous promoter at the native chromosomal locus. We refer to the cbk1-HA-S570A construct as the T-loop allele, to the cbk1-HA-T743A construct as the CT-motif allele, and to the catalytically inactive cbk1-HA-D475A construct as the kinase-dead allele. These proteins are expressed at similar levels (see Fig. 2 A and Fig. 6 D). Cells lacking functional Cbk1 have two distinct phenotypes, which have been reported previously and are shown in Fig. 1 A (Racki et al., 2000; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Weiss et al., 2002; Nelson et al., 2003). These cells fail to degrade the septum between mother and daughter cells after cytokinesis and, as a result, grow as clumps of >100 cells connected by chitin-rich junctions. In addition, cells carrying the kinase-dead allele or cbk1Δ cells are spherical instead of ovoid because they cannot maintain polarized growth (Weiss et al., 2002).
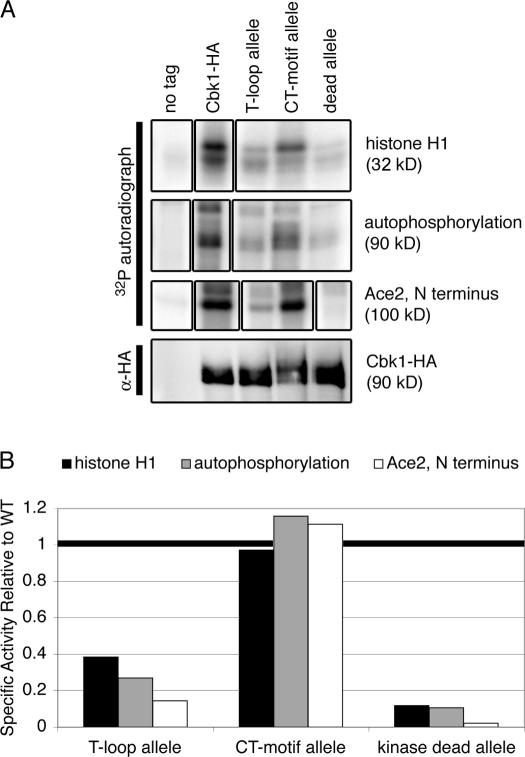
Figure 2.
The Cbk1 T-loop allele lacks detectable kinase activity, but CT-motif phosphorylation is dispensable. (A, top) Immunoprecipitates from strains expressing untagged Cbk1 (ELY126), Cbk1-HA (ELY140), the cbk1 T-loop allele (ELY390), cbk1 CT-motif allele (ELY437), and cbk1 kinase-dead allele (ELY426) were incubated with histone H1 or the N terminus of Ace2 fused to maltose binding protein (MBP) and γ-32P-ATP to assay in vitro kinase activity, as described previously (Weiss et al., 2002). (bottom) Anti-HA blot of the immunoprecipitates imaged using fluorescently labeled secondary for a quantitative measure of protein expression (90 kD). (B) Quantification of histone H1 phosphorylation, autophosphorylation, and Ace2 phosphorylation. The signal from the autoradiograph in A was normalized to Cbk1 expression levels from the anti-HA blot. Phosphorylation of each substrate by wild-type Cbk1 was set as 1, and all values were normalized against this, giving a relative reading of specific activity for each substrate. These experiments were repeated for three biologically independent samples, and the specific activity of the CT-motif allele ranged from ∼50 to 100% of wild-type levels. The T-loop allele kinase activity never exceeded 25% of wild-type levels.
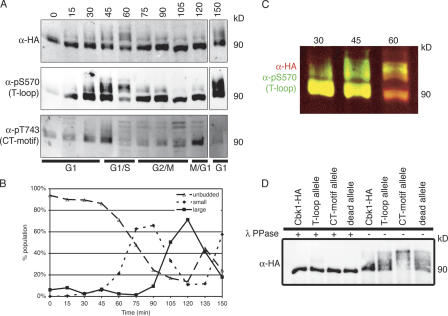
Figure 6.
Cbk1 modification is dynamic over the cell cycle. (A) Cells carrying Cbk1-HA (ELY140) were arrested with α factor for 2 h, until >80% of cells had formed mating projections. They were then washed extensively and released into fresh YPD. Cells were harvested every 15 min, and lysates from each time point were subjected to immunoprecipitation with anti-HA. The resulting proteins were resolved by SDS-PAGE, blotted with the indicated antibodies, and imaged using fluorescently labeled secondary antibodies. Slower migrating forms of Cbk1-HA appear, particularly at the 45-, 60-, and 150-min time points; these are detected by anti-HA and both phosphospecific antibodies. CT-motif phosphorylation peaks at 45 and 120 min. (B) At each time point, cells were fixed in formaldehyde to assess the budding index. Fixed cells were sonicated with a probe to break apart any clumps of cells, and budding morphology was scored (n > 200). (C) Merge of anti-HA and anti-pS570 images in A. Fluorescent detection allows overlay of two color images: red represents anti-HA and green represents anti-pS570. Note bias of T-loop phosphorylation to slower migrating forms of Cbk1-HA in both the 45- and 60-min time points. (D) Proteins were immunoprecipitated with anti-HA from strains expressing Cbk1-HA (ELY140), the cbk1 T-loop allele (ELY390), the cbk1 CT-motif allele (ELY437), and cbk1 kinase-dead allele (ELY426). Half of each immunoprecipitation was treated with λ phosphatase. Proteins were resolved by SDS-PAGE, run longer to enhance separation, and blotted with anti-HA. The resulting bands were imaged using fluorescently conjugated secondary antibodies and subsequently quantified; all proteins are expressed at similar levels, and a pronounced shift is evident under these conditions for the CT-motif allele (Cbk1-HA, 90 kD).
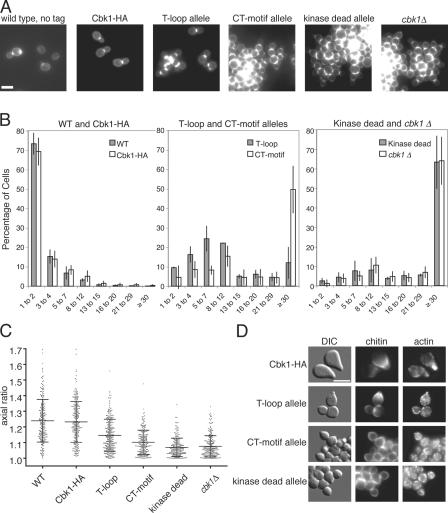
Figure 1.
The T-loop and CT-motif mutant alleles exhibit cell separation and polarized growth defects. (A) Cells expressing untagged Cbk1 (ELY126), Cbk1-HA (ELY140), the cbk1 T-loop allele (ELY390), the cbk1 CT-motif allele (ELY437), cbk1 kinase-dead allele (ELY426), and cbk1Δ (ELY132) were stained for chitin in the cell wall using calcofluor to enhance visualization of cell shape. T-loop and CT-motif refer to the mutant forms of Cbk1 that cannot be phosphorylated at serine 570 and threonine 743, respectively. (B) The number of connected cells in a group was counted in three independent trials (n > 200 groups per strain, per trial). Error bars indicate the standard deviation for the mean of the three trials. (C) Cells from A were measured for length and width in three independent trials (n > 200 cells per strain, per trial). The ratio of length to width gives a measure of the roundness of the cell, with a perfect circle equal to one. Means and standard deviations are shown with data points from only one representative trial, for clarity. (D) Mat a cells expressing Cbk1-HA (ELY140), the cbk1 T-loop allele (ELY461), the cbk1 CT-motif allele (ELY462), or cbk1 kinase-dead allele (ELY463) were arrested in medium containing 10 μM α pheromone. Chitin and F-actin were visualized using chitin and phalloidin staining, respectively. Bars, 5 μm.
The T-loop and CT-motif mutant alleles showed both phenotypes but to different degrees. Like cells expressing kinase-dead Cbk1, cells expressing the T-loop allele or CT-motif allele proteins grew as groups of cells connected by chitin (Fig. 1 A). We quantified the cell separation defect by counting the number of connected cells per cluster (Fig. 1 B). The separation defects of the CT-motif allele, kinase-dead allele, and cbk1Δ were nearly indistinguishable (49.7 ± 12.1%, 63.3 ± 13.6%, and 64.0 ± 12.3% of cells in clumps containing >30 connected cells; Fig. 1 B, middle and right). In contrast, the T-loop mutant had an intermediate phenotype with most cells in clusters of <12, suggesting that phosphorylation of this site is only partially required for cell separation (Fig. 1 B, middle).
Cells expressing either the cbk1 T-loop or CT-motif alleles were noticeably rounder than wild-type strains, suggesting that phosphorylation of these sites is required for maintenance of polarized growth (Fig. 1 A). To quantify this phenotype in mitotically growing cells, we determined the mean axial ratio by measuring the length and width of >200 cells for each strain in three independent trials (Fig. 1 C). This ratio measures the roundness of a cell, with a perfect circle equal to 1.0 and greater numbers representing deviation toward an ellipse. Wild-type and CBK1-HA cells had a broad distribution of axial ratios, with means of 1.24 ± 0.13 and 1.21 ± 0.12, respectively. In contrast, the mean axial ratios of cells carrying the kinase-dead or cbk1Δ-null alleles were 1.07 ± 0.06 and 1.07 ± 0.07, respectively. Cells with the CT-motif allele have a mean axial ratio of 1.09 ± 0.07, which is similar to both the kinase-dead and null mutants. In contrast, cells carrying the T-loop allele had an intermediate mean axial ratio of 1.13 ± 0.10, which is significantly different from both wild-type and null phenotypes, as well as the CT-motif mutant phenotype (using a nonparametric Mann-Whitney t test, P < 0.0001).
Maintenance of polarized growth can also be assessed by observing mating projection formation in response to pheromone. Cells expressing Cbk1-HA formed mating projections normally, with characteristic chitin deposits at the base of an elongated projection and actin polarized into the tip (Fig. 1 D, top). In contrast, cells expressing the cbk1 T-loop, CT-motif, or kinase-dead alleles formed abortive mating projections, similar to cbk1Δ cells, which are unable to sustain polarized growth (Fig. 1 D; Bidlingmaier et al., 2001). Abortive mating projections are characterized by slight bumps flanked by chitin deposits that lack the elongated projections and pronounced actin polarization of normal mating projections. Collectively, our observations suggest that phosphorylation of the T-loop and CT-motif sites play important yet distinct roles in cell separation and maintenance of polarized growth in vivo.
The in vitro kinase activity of phosphosite mutants does not correlate with in vivo function
The phenotype of kinase-dead Cbk1 shows that the protein's enzymatic activity is critical for its function in both cell separation and polarized growth (Fig. 1), consistent with previous findings (Weiss et al., 2002). Therefore, the phenotypes of the nonphosphorylatable T-loop and CT-motif alleles could be due to reduction of normal Cbk1 kinase activity. To assess this, we characterized the relative specific activity of Cbk1 alleles in vitro, using quantitative detection with fluorescently labeled secondary antibodies to measure relative amounts of the immunoprecipitated enzyme. We evaluated autophosphorylation and histone H1 phosphorylation as previously described (Weiss et al., 2002) and examined phosphorylation of a bacterially produced fragment of the transcription factor Ace2, a likely Cbk1 regulatory target. Surprisingly, wild-type and CT-motif mutant Cbk1 phosphorylated these substrates with comparable efficiency. In contrast, the T-loop mutant protein exhibits minimal kinase activity, only slightly higher than that of the kinase-dead allele (Fig. 2). Cbk1 function and kinase activity requires association with Mob2 (Weiss et al., 2002; Nelson et al., 2003). We therefore tested all of the Cbk1 mutants and found that none were defective for Mob2 association (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200604107/DC1). In sum, these results show that the phenotypic differences between the T-loop and CT-motif alleles is not attributable to differences in kinase activity.
Cbk1 and Ace2 asymmetric nuclear localization requires CT-motif phosphorylation
When expressed under the control of its endogenous promoter, Cbk1 localizes to sites of active cell growth at the cortex (Racki et al., 2000; Colman-Lerner et al., 2001; Weiss et al., 2002). To determine whether the phenotypes of the T-loop, CT-motif, and kinase-dead mutant alleles were attributable to mislocalization, we visualized GFP-tagged versions of these proteins in live cells. All mutant Cbk1-GFP fusion proteins were present at the cell cortex of presumptive bud sites and the growing bud, like wild type (Fig. 3 A, left and middle). The mutant proteins also accumulated normally at the bud neck during cell division (Fig. 3 A, right). Thus, phosphorylation at the T-loop and CT-motif sites are not required for Cbk1's cortical localization.
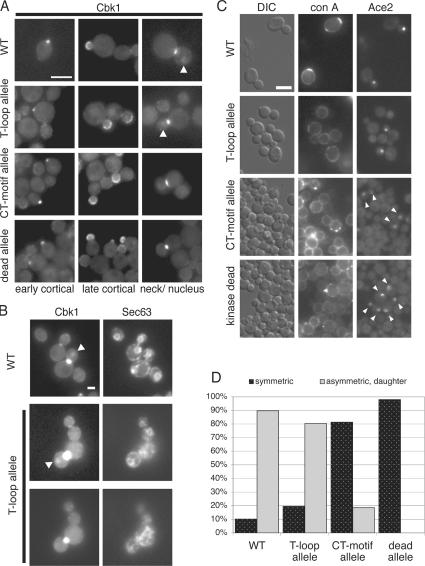
Figure 3.
Mutation of Cbk1 phosphorylation sites is required for Cbk1 and Ace2 nuclear localization. (A) Localization of GFP-tagged Cbk1 (ELY466), the T-loop allele (ELY468), the CT-motif allele (ELY467), and the kinase-dead allele (ELY469) in asynchronous cells. (left) Early cortical localization; (middle) late cortical localization; (right) bud neck localization. Arrowheads indicate daughter cell nuclear localization. Bar, 5 μm. (B) To confirm nuclear localization of both wild-type and the T-loop allele of Cbk1-GFP, the nuclear envelope was visualized with Sec63-tomato (ELY535 and ELY536, respectively). The middle and bottom panels show two different focal planes to better visualize both the mother and daughter nuclei. Arrowheads indicate daughter cell nuclear localization. Bar, 2 μm. (C) Localization of Ace2-GFP in strains expressing untagged Cbk1 (ELY410), the T-loop allele (ELY464), the CT-motif allele (ELY465), and kinase-dead Cbk1 (ELY451). Asynchronously growing cells were briefly labeled with rhodamine-conA, washed, and allowed to continue growth for an additional 90 min. (middle) Mother cells labeled with conA; daughter cells are unlabeled. (right) Ace2-GFP localization in large budded cells. Arrowheads indicate symmetric localization. All exposure times, contrast enhancement, and grayscale values were held constant. Bar, 5 μm. (D) The images in C were used to differentiate between symmetric (localization to mother and daughter nuclei; dark bars) and asymmetric (only daughter; light bars) Ace2-GFP nuclear localization. n ≥ 50 cells per strain.
Cbk1 also localizes to the daughter cell nucleus at the M–G1 transition (Colman-Lerner et al., 2001; Weiss et al., 2002). Some nuclear localization of T-loop mutant was evident (Fig. 3, A [arrows] and B [nucleus delineated by localization of the ER marker Sec63-RFP and Cbk1-GFP nuclear localization indicated by arrows]). In contrast, we could not detect either the kinase-dead or the CT-motif mutant GFP fusion proteins in nuclei.
The nuclear accumulation of Cbk1 both coincides with and depends on Ace2 nuclear localization; in turn, Ace2 asymmetric localization requires Cbk1 kinase activity (Colman-Lerner et al., 2001; Weiss et al., 2002). Thus, it is possible that cells carrying the cbk1 CT-motif and kinase-dead alleles fail to recruit Ace2 to nuclei. Therefore, we examined Ace2-GFP localization in each of the mutant backgrounds (Fig. 3 C). We briefly incubated cells with rhodamine–concanavalin A, which binds stably to the cell wall, and allowed the cells to grow for ∼90 min in the absence of this vital stain. This specifically labeled mother cells and allowed us to distinguish between mother and daughter cells in the mutant strains, which grow as large clusters of connected cells. In wild-type cells, Ace2-GFP was distributed asymmetrically to the daughter cell nucleus, with relatively little cytoplasmic background in ∼90% of cells (Fig. 3, C and D). In the cbk1 T-loop mutant, Ace2-GFP localized to daughter nuclei in ∼80% of cells. In contrast, in CT-motif mutant cells, Ace2-GFP localized faintly to both mother and daughter nuclei in ∼80% of cells (Fig. 3, C and D). This loss of Ace2-GFP asymmetry is similar to that seen in cbk1 kinase-dead and cbk1Δ cells. Ace2 expression levels do not vary appreciably in the mutant backgrounds (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200604107/DC1). Thus, both CT-motif phosphorylation and kinase activity are required for the proper and asymmetric distribution of Cbk1 and Ace2 to the daughter cell nucleus.
Phosphorylation events at the T-loop and CT-motif sites are independent
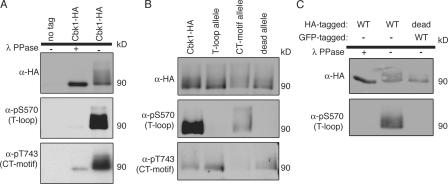
To determine whether Cbk1 is phosphorylated at the T-loop and CT-motif sites in vivo, we generated antibodies against the relevant phosphopeptides for each site. These antibodies strongly detected immunoprecipitated Cbk1-HA but only weakly recognized Cbk1-HA treated with λ phosphatase (Fig. 4 A). The phosphospecific antibodies are not general phosphoserine or phosphothreonine antibodies, as they did not recognize other highly phosphorylated proteins, such as GST-Ste20 (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200604107/DC1) or the mutant alleles for each site (Fig. 4 B). Thus, Cbk1 is phosphorylated in vivo at both the T-loop and CT-motif sites.
Figure 4.
T-loop modification occurs through intramolecular autophosphorylation. (A) Proteins were immunoprecipitated with anti-HA from wild-type (ELY126) and Cbk1-HA (ELY140) strains. Half of the immunoprecipitated proteins was removed and treated with λ phosphatase. The proteins were then blotted with the indicated antibodies. Anti-pS570 was used to detect T-loop site phosphorylation, and anti-pT743 was used to detect CT-motif site phosphorylation (Cbk1-HA; 90 kD). (B) Proteins were immunoprecipitated with anti-HA from strains expressing Cbk1-HA (ELY140), the T-loop allele (ELY390), the CT-motif allele (ELY437), and kinase-dead Cbk1 (ELY426). Proteins were resolved by SDS-PAGE and blotted with the indicated antibodies. (C) Proteins were immunoprecipitated from a haploid strain expressing Cbk1-HA (ELY140) and a diploid strain expressing Cbk1-GFP and kinase-dead Cbk1-HA (ELY537). Immunoprecipitation was done with anti-HA so that only kinase-dead Cbk1 would precipitate from the diploid strain (in addition, GFP-tagged Cbk1 runs at a higher molecular weight than HA-tagged Cbk1). Half the immunoprecipitates from Cbk1-HA were treated with λ phosphatase. The proteins were resolved by SDS-PAGE and probed with the indicated antibodies.
To determine whether phosphorylation of Cbk1's T-loop and CT-motif are independent regulatory inputs, we used the phosphospecific antibodies to detect modification of each site in the absence of the other's phosphorylation (Fig. 4 B). The T-loop mutant protein was detected by anti-pT743, the CT-motif phosphospecific antibody. The CT-motif mutant protein was detected by anti-pS570, the T-loop phosphospecific antibody, although the signal was decreased about fourfold relative to wild type. This suggests that each phosphorylation event can occur in the absence of the other, although CT-motif modification may promote maximal T-loop phosphorylation in vivo.
T-loop autophosphorylation occurs through an intramolecular reaction in vivo
Kinase-dead Cbk1 was phosphorylated at the CT-motif site but not at the T-loop site, suggesting that the T-loop site is autophosphorylated in vivo (Fig. 4 B). This is consistent with the model proposed for other kinases related to Cbk1, such as hNDR (nuclear Dbf2-related kinase; Tamaskovic et al., 2003). Autophosphorylation may occur in cis as an intramolecular reaction or in trans as an intermolecular reaction between Cbk1 molecules. We constructed a diploid strain that expresses both wild-type Cbk1-GFP and the kinase-dead allele of Cbk1-HA to distinguish between these possibilities. As shown in Fig. 4 C, immunoprecipitated kinase-dead protein is not phosphorylated at the T-loop site by wild-type Cbk1 in vivo. Therefore, it is likely that Cbk1 modifies itself in vivo in cis; this is in contrast to the CT-motif site, which must be phosphorylated by a different kinase.
Ace2 and RAM network proteins affect T-loop and CT-motif phosphorylation
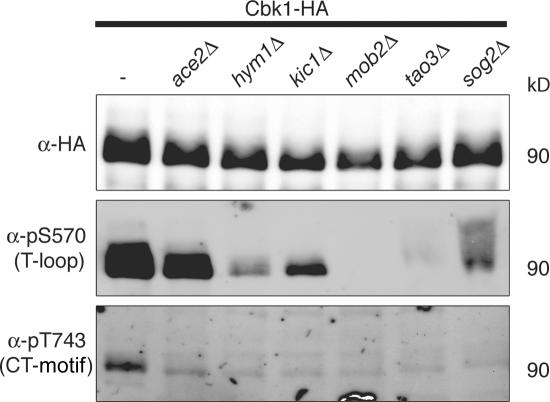
Cbk1 works with other RAM network components to control cell separation and polarized growth. Cbk1 kinase activity is low in cells lacking any other RAM network component, suggesting that activation of Cbk1 is among the final outputs of the pathway (Weiss et al., 2002; Nelson et al., 2003). To determine whether the RAM network is required for phosphorylation of Cbk1's T-loop and CT-motif sites, we used phosphospecific antibodies in quantitative Western blotting to evaluate the modification of Cbk1-HA immunoprecipitated from cells lacking different RAM network genes (Fig. 5). We were able to detect a very low level of phosphorylation at the T-loop site using the anti-pS570 antibody in hym1Δ and kic1Δ strains (∼4 and ∼11% of wild-type phosphorylation). T-loop phosphorylation is detectable on Cbk1-HA immunoprecipitated from sog2Δ cells (∼12% of wild type) but is biased toward forms shifted to higher molecular weights. Phosphorylation of the T-loop site was absent in mob2Δ and tao3Δ backgrounds. The CT-motif–specific anti-pT743 antibody recognized only Cbk1-HA immunoprecipitated from wild-type cells and not from any of the strains lacking RAM network components. Therefore, all RAM network proteins are essential for CT-motif phosphorylation and only a subset is essential for phosphorylation of the T-loop site.
Figure 5.
T-loop and CT-motif phosphorylation in cells lacking Ace2 and RAM network components. Cbk1-HA was immunoprecipitated with anti-HA from the following strains: ace2Δ (ELY397), hym1Δ (ELY395), kic1Δ (ELY398), mob2Δ (ELY399), tao3Δ (ELY396), sog2Δ (ELY436), and wild type (ELY140). The proteins were resolved by SDS-PAGE and imaged with fluorescently labeled secondary antibodies for quantitative Western blotting (Cbk1-HA; 90 kD).
Cbk1 kinase activity is not reduced in ace2Δ cells, suggesting that Ace2 is an endpoint effector of the pathway; our data also support this (Fig. 2). In addition, Ace2 is not required for maintenance of polarized growth (Bidlingmaier et al., 2001; Weiss et al., 2002; Nelson et al., 2003). We used phosphospecific antibodies to detect Cbk1-HA immunoprecipitated from ace2Δ cells to determine whether Ace2 is important for phosphorylation at either the T-loop or CT-motif sites. T-loop phosphorylation is similar in wild-type and ace2Δ backgrounds (Fig. 5). Surprisingly, however, we found substantial reduction of Cbk1 CT-motif phosphorylation in ace2Δ cells (∼43% of wild type, measured by quantitative Western blotting with fluorescent secondary; see Materials and methods). RAM network genes were expressed at similar levels in wild-type and ace2Δ cells, as measured by RT-PCR (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200604107/DC1), indicating that loss of CT-motif modification is not an indirect effect of low RAM network gene expression. Therefore, full modification of Cbk1's critically important CT-motif site is reinforced or protected by the presence of this downstream effector.
Dynamic regulation of Cbk1 phosphorylation in vivo
Cbk1 functions in both polarized growth and cell separation, two temporally regulated processes. To assess the cell cycle–dependent control of phosphorylation at the T-loop and the CT-motif sites, we used the phosphospecific antibodies to monitor modification of these sites in synchronized cells (Fig. 6 A). T-loop phosphorylation fluctuated only slightly over the course of cell division: phosphorylation levels were generally high but diminished as cells entered S phase (Fig. 6, A and C). Before bud emergence, slower migrating forms of Cbk1 were highly phosphorylated at the T-loop site (Fig. 6, A and C). CT-motif phosphorylation is more dramatically regulated in a cell cycle–dependent manner (Fig. 6 A). Phosphorylation of this site peaked early, 45 min after release from G1 arrest, just before bud emergence. Phosphorylation then remained low until late in the cell cycle, as cytokinesis occurred. When the majority of cells had passed from M to G1, CT- motif phosphorylation decreased again. The peaks in CT-motif phosphorylation coincide with times of polarized growth and cell separation. In summary, Cbk1 is highly phosphorylated at the T-loop site over most of the cell cycle, whereas CT-motif phosphorylation coincides with periods of polarized growth and cell separation.
Gel mobility of Cbk1-HA immunoprecipitated at different times after release from pheromone arrest shifted markedly over the course of the experiment (Fig. 6, A and C, anti-HA signal). These changes in mobility suggest that the protein is dynamically phosphorylated at multiple sites as the cell cycle progresses. Both phosphospecific antibodies recognized multiple forms of Cbk1, indicating that these shifts are not solely attributable to phosphorylation of the T-loop and CT-motif sites. Consistent with this finding, phosphorylation site mutants and the kinase-dead allele were also shifted considerably relative to wild-type Cbk1 when separated by SDS-PAGE (Fig. 6 D). The CT-motif allele was biased most dramatically to slower migrating forms. These shifts were abolished by treatment with λ phosphatase, indicating that they are due to hyperphosphorylation. As indicated by the mobility shifting seen in both the mutants and over the cell cycle, there are likely additional sites of phosphorylation that are dynamically modified across the cell cycle; these as-yet-unknown sites may represent additional regulatory mechanisms for Cbk1 function.
Discussion
Functional differences between Cbk1's regulatory sites
We have shown that Cbk1 is controlled by phosphorylation of two highly conserved sites. Analogous sites have been characterized in other AGC kinases, most notably Akt/PKB, as well as more closely related enzymes, such as human Ndr and Lats and Drosophila tricornered (Millward et al., 1999; Tamaskovic et al., 2003; Emoto et al., 2004; He et al., 2005b; Hergovich et al., 2005). The prevailing view, based largely on studies of in vitro kinase activity, is that these modifications promote AGC kinase signaling primarily by increasing the target enzyme's catalytic activity. In contrast, we find that the relative functional importance of these sites in vivo is not directly related to their importance for Cbk1's enzymatic activity. Thus, our findings argue that this enzyme activation model does not fully explain the role of these regulatory inputs for Cbk1.
We find that the Cbk1 CT-motif allele has relatively normal kinase activity but exhibits severely defective polarized growth and failed cell separation. In contrast, the Cbk1 T-loop allele has extremely low kinase activity but a moderate phenotypic effect. How does this compare with the existing in vitro characterization of mammalian Ndr? Mammalian enzyme mutated at the T-loop site is similarly compromised for in vitro activity, suggesting that this modification functions similarly in yeast and metazoan enzymes (Millward et al., 1999; Tamaskovic et al., 2003; Stegert et al., 2004). An Ndr CT-motif mutant retains some activity, but this is 7–10-fold lower than the wild-type enzyme (Millward et al., 1999; Tamaskovic et al., 2003; Stegert et al., 2004). Furthermore, CT-motif phosphorylation clearly increases Ndr's in vitro activity (Tamaskovic et al., 2003; Hergovich et al., 2005; Stegert et al., 2005). It is possible that Cbk1 has inherently higher basal activity than Ndr in the absence of this modification, and our studies do not preclude the possibility that Cbk1's catalytic activity is increased when the enzyme is phosphorylated at the CT-motif. However, the relatively mild phenotype of the severely catalytically compromised T-loop allele indicates that Cbk1's in vivo function requires only minimal levels of the enzyme's catalytic activity. Therefore, our data strongly favor a revision of the prevailing hypothesis for regulation of Cbk1 homologues: that CT-motif phosphorylation has distinct regulatory functions in addition to promoting optimal catalytic conformation of the kinase domain.
What are these additional functions of Cbk1 CT-motif phosphorylation? Both T-loop and CT-motif mutant proteins localize normally to the cell cortex, indicating that neither modification is required to recruit the enzyme to its site of action; similarly, both mutant proteins interact with the conserved Cbk1 binding partner Mob2. Cbk1 CT-motif phosphorylation is, however, required for asymmetric localization of both the kinase and Ace2 to the daughter nucleus late in the cell cycle (Fig. 3). Asymmetric nuclear localization of Cbk1 and Ace2 is interdependent, and the proteins may enter the nucleus as a complex (Colman-Lerner et al., 2001; Weiss et al., 2002; Nelson et al., 2003). Thus, we propose that CT-motif phosphorylation is necessary for the formation of a productive Cbk1–Ace2 complex.
Intriguingly, the Cbk1 CT-motif mutant protein is dramatically hyperphosphorylated in vivo. This suggests that phosphorylation of the CT-motif site triggers dephosphorylation of Cbk1 at other sites, perhaps by recruiting phosphatases. Given the null phenotype of the CT-motif mutant, it is possible that Cbk1 hyperphosphorylation inhibits the kinase's interaction with downstream targets in vivo. Mammalian Ndr kinases are also phosphorylated at a third site immediately N-terminal to the kinase domain. This modification is important for Mob protein association and stimulation of kinase activity in vivo (Tamaskovic et al., 2003), although mutation of the site does not dramatically affect basal in vitro kinase activity or Mob binding (Tamaskovic et al., 2003; Bichsel et al., 2004). We have mutated Cbk1 at the corresponding site, serine 339, and find no phenotypic effects (unpublished data). Thus, modification of this site is not critical for Cbk1's in vivo function, although it remains possible that it is phosphorylated in vivo as part of a redundant regulatory mechanism. We are currently pursuing identification of the modified sites in hyperphosphorylated Cbk1 and investigating their functional significance.
The in vivo function of mammalian Ndr kinases is unknown; therefore, it is not presently possible to characterize the phenotypes of T-loop and CT-motif alleles of these enzymes. The physiological role of tricornered, the Drosophila homologue of Cbk1, is better understood: it functions in morphogenesis of actin-rich projections as well as branching and tiling of sensory neuron dendrites (Geng et al., 2000; Emoto et al., 2004). Mutation of both T-loop and CT-motif sites renders tricornered, which is essential for embryonic development, nonfunctional (He et al., 2005b). Overexpression of the mutant alleles exerts a dominant-negative phenotype, with the single site mutants having a less extreme wing hair phenotype than the double mutant (He et al., 2005b). The dominant-negative phenotype of tricornered loss-of-function alleles is complex: intriguingly, kinase-inactive tricornered is dominant negative in neuron morphogenesis but not in wing hair development (Emoto et al., 2004; He et al., 2005b). Although these overexpression studies are fundamentally different from our analysis of endogenously expressed alleles, it is clear that the regulatory phosphorylation sites are functionally important. We find that a Cbk1 double-mutant allele has a null phenotype equivalent to the single-mutant CT-motif allele; we do not observe dominant-negative effects in heterozygotes for any cbk1 alleles at endogenous expression levels (unpublished data). If our analysis of the precise roles of Cbk1's regulatory sites applies more generally, we predict that T-loop alleles of the Ndr family kinases will have a less severe phenotype than CT-motif alleles when expressed under endogenous conditions. Furthermore, we speculate that, under these conditions, CT-motif modification of other Cbk1-related kinases may promote formation of functional complexes or dephosphorylation of other regulatory sites.
RAM network control of CT-motif and T-loop phosphorylation
We have shown that all RAM network proteins are required for phosphorylation of Cbk1's CT-motif, suggesting that this regulatory event is a critical output of the pathway. Which kinase phosphorylates Cbk1's CT-motif site? Recent evidence suggests that germinal center kinases, which are related to p21-activated kinases, are responsible for phosphorylation of this site in other Ndr/Lats family kinases (Chan et al., 2005; Stegert et al., 2005; Emoto et al., 2006). Kic1, a germinal center kinase–family kinase in the RAM network, is a good candidate for a CT-motif kinase, and we find that CT-motif phosphorylation is undetectable in cells lacking Kic1. However, as CT-motif modification depends on all RAM network proteins, Kic1's role may be indirect. The activity of endogenous Kic1 isolated from yeast is extremely low, and definitive in vitro assays have not yet been possible (unpublished data). The p21-activated kinase Ste20 does not phosphorylate Cbk1 in vitro (unpublished data), suggesting that Cbk1 is not a general target of this class of kinases. Furthermore, yeast cells lacking p21-activated kinase function do not exhibit defective polarized growth maintenance or septum degradation (Weiss et al., 2000). Therefore, phosphorylation of the CT-motif site by Kic1 may serve as the critical output of RAM network signaling that results in cell separation and polarized growth.
We find that Cbk1 most likely autophosphorylates itself in vivo at its T-loop site through an intramolecular reaction. In other AGC group kinases, such as Akt/PKB, the analogous modification is performed in trans by an upstream regulatory kinase. Although this is the first demonstration of such behavior for an Ndr/LATS kinase in vivo, it is consistent with previous in vitro observations. Ndr kinases clearly autophosphorylate at this site in vitro (Tamaskovic et al., 2003; Stegert et al., 2004), and biochemical data suggest that this modification can occur in cis (Millward et al., 1995). We also find that T-loop autophosphorylation is only completely abolished in cells lacking Mob2 and Tao3. Because these are the only two RAM network proteins thus far found to coimmunoprecipitate with Cbk1, we suggest that Tao3 and Mob2 directly interact with Cbk1 to either help the kinase adopt a proper conformation for cis autophosphorylation or protect it from phosphatases that reverse the modification (Du and Novick, 2002; Weiss et al., 2002). Consistent with this idea, previous reports have shown that Mob proteins are required for autophosphorylation of the T-loop site in Ndr (Hergovich et al., 2005; Stegert et al., 2005). Tao3 is also conserved in metazoans; we speculate that it plays a similar role in the regulation of Cbk1 homologues in these species.
Interdependency between control of Cbk1 and its regulatory target Ace2
The RAM network's control of the activity and asymmetric localization of Ace2 ensures that Ace2-driven genes involved in septum destruction are only highly expressed once in a cell's life: when it is a daughter. More specifically, Ace2 segregation and function depend on Cbk1's kinase activity (Colman-Lerner et al., 2001; Weiss et al., 2002), and we have shown that Cbk1 phosphorylates the transcription factor in vitro. We are currently investigating the in vivo relevance of this modification. Our in vitro kinase assays show that Cbk1 CT-motif modification does not simply turn on the kinase's ability to phosphorylate Ace2. Rather, this suggests that phosphorylation at the CT-motif site promotes formation of a functional regulatory complex in vivo.
It is interesting that full phosphorylation of Cbk1's CT-motif requires Ace2; this suggests that the downstream target participates in activation of its upstream regulator. Likely, the remaining pool of CT-motif phosphorylated Cbk1 is required for Ace2-independent function in the maintenance of polarized growth. We speculate that Ace2-dependent protection of CT-motif phosphorylated Cbk1 involves either directly blocking phosphatase access or shuttling the protein into a compartment with low phosphatase activity. This phenomenon could help create a reinforcement mechanism that promotes rapid accumulation of CT-motif modified Cbk1 during the M–G1 transition.
The logic of this system likely reflects the importance of temporally controlling Ace2 function. Septum destruction is among the final events of cell division, and Ace2-driven transcription of genes involved in the process is tightly regulated. Ace2 is likely negatively regulated through phosphorylation by mitotic Cdk (O'Conallain et al., 1999; Archambault et al., 2004), and its nuclear entry requires activation of the mitotic exit network (Weiss et al., 2000). Cbk1's CT-motif phosphorylation is delayed until late M/G1. We speculate that Ace2's Cdk phosphorylation is reversed upon mitotic exit and that this coincides with modification of Cbk1. The co-occurrence of these events may create a short temporal window in which Cbk1 can act on Ace2 to promote its rapid accumulation and function in daughter cell nuclei.
Cell cycle control of Cbk1 phosphorylation
Cbk1's phosphorylation is dynamic across the cell cycle. T-loop autophosphorylation is relatively constant, except a short period after bud emergence. In contrast, modification of the CT-motif fluctuates markedly, peaking just before bud emergence and again late in the cell cycle (Fig. 6 A). This coincides with periods of active polarized growth, as well as with cytokinesis and septum degradation after mitotic exit. Because the CT-motif phosphorylation requires all RAM network components, it is likely that this cell cycle–regulated modification is one of the key downstream outputs of this signaling pathway.
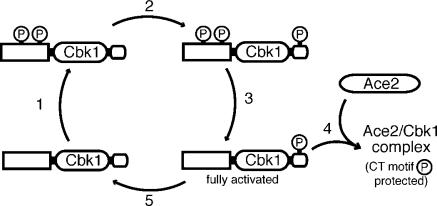
Based on our findings, we propose a speculative model for Cbk1's cycle of activation during M–G1 that consists of a priming phase followed by activation-dependent dephosphorylation. We propose that Cbk1 is initially phosphorylated at multiple priming sites, other than the T-loop and CT-motif, in a RAM network–dependent manner (Fig. 7, 1). We suggest that these modifications both prevent Cbk1's action on downstream targets and promote phosphorylation of the kinase's CT-motif site, most likely by Kic1. This CT-motif modification may then trigger dephosphorylation of the initial priming sites, perhaps by recruiting phosphatases, producing Cbk1 that is fully active (Fig. 7, 2 and 3). This form of Cbk1 may then either productively interact with Ace2, which preserves CT-motif phosphorylation (Fig. 7, 4), or become fully dephosphorylated and inactivated (Fig. 7, 5). This putative kinase/target synergy may work to protect an activated pool of Cbk1 with CT-motif phosphorylation until Ace2's function is performed, ensuring maximal activation of both Cbk1 and Ace2 at the proper place and time.
Figure 7.
Model for Cbk1 activation and Ace2-dependent protection of CT-motif phosphorylation. (1) Cbk1 is primed for activation by RAM network–dependent phosphorylation of as-yet-unknown sites. (2) This modification is permissive for phosphorylation of the CT-motif by an upstream kinase, possibly Kic1. (3) CT-motif phosphorylation promotes removal of priming phosphorylations, making Cbk1 fully able to associate with regulatory targets. (4) Active CT-motif phosphorylated Cbk1 associates with Ace2; this protects the CT-motif from dephosphorylation. (5) In the absence of Ace2, the Cbk1 CT-motif is dephosphorylated.
Materials and methods
Strains, growth conditions, and protein purification
Yeast strains were derived from S288C (Table I) and cultured in YPD media (1% yeast extract, 2% peptone, and 2% dextrose). Overnight cultures were diluted to OD600 0.2 and grown 5 h at 30°C to OD600 0.8 for use in all assays. Deletion mutants and C-terminal fluorescent tags (a gift from R. Tsien, University of California, San Diego, La Jolla, CA) and myc tags were generated using the primers listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200604107/DC1; Longtine et al., 1998; Shu et al., 2006). Cbk1 point mutants were generated using two fragment PCR based on genomic DNA from ELY140. The overlapping fragments were integrated at the endogenous locus into ELY132 and screened by PCR and sequencing.
Table I.
Genotypes of all strains used and generated in this study
| Name | Genotype | Source |
|---|---|---|
| ELY126 (FLY95) | MATα ura3-52 leu2-3,112 trp1Δ1 his3Δ200 (S288C) | F. Luca (University of Pennsylvania, Philadelphia, PA) |
| ELY402 (FLY93) | MAT a ura3-52 leu2-3,112 trp1Δ1 his3Δ200 (S288C) | F. Luca |
| ELY140 (DDY2362) | MAT a CBK1-3xHA∷HIS3 | D. Drubin (University of California, Berkeley, Berkeley, CA) |
| ELY390 | MATα cbk1S570A-3xHA∷HIS3 | |
| ELY461 | MAT a cbk1S570A-3xHA∷HIS3 | |
| ELY437 | MATα cbk1T743A-3xHA∷HIS3 | |
| ELY462 | MAT a cbk1T743A-3xHA∷HIS3 | |
| ELY426 | MATα cbk1D475A-3xHA∷HIS3 | |
| ELY463 | MAT a cbk1D475A-3xHA∷HIS3 | |
| ELY132 | MATα cbk1Δ∷KanMX | |
| ELY447 | MATα MOB2-13xMyc∷TRP1 | |
| ELY417 | MAT a CBK1-3xHA∷HIS3 MOB2-13xMyc∷TRP1 | |
| ELY418 | MATα cbk1S570A-3xHA∷HIS3 MOB2-13xMyc∷TRP1 | |
| ELY448 | MATα cbk1T743A-3xHA∷HIS3 MOB2-13xMyc∷TRP1 | |
| ELY449 | MATα cbk1D475A-3xHA∷HIS3 MOB2-13xMyc∷TRP1 | |
| ELY398 | MAT a CBK1-3xHA∷HIS3 kic1Δ∷TRP1 | |
| ELY395 | MAT a CBK1-3xHA∷HIS3 hym1Δ∷TRP1 | |
| ELY396 | MAT a CBK1-3xHA∷HIS3 tao3Δ∷TRP1 | |
| ELY397 | MAT a CBK1-3xHA∷HIS3 ace2Δ∷TRP1 | |
| ELY399 | MAT a CBK1-3xHA∷HIS3 mob2Δ∷TRP1 | |
| ELY436 | MAT a CBK1-3xHA∷HIS3 sog2Δ∷TRP1 | |
| ELY410 | MATα ACE2-GFP∷KanMX | |
| ELY464 | MAT a cbk1S570A-3xHA∷HIS3 ACE2-GFP∷KanMX | |
| ELY465 | MAT a cbk1T743A-3xHA∷HIS3 ACE2-GFP∷KanMX | |
| ELY451 | MATα cbk1D475A-3xHA∷HIS3 ACE2-GFP∷KanMX | |
| ELY466 (FLY869) | MAT a CBK1-GFP∷KanMX | F. Luca |
| ELY468 | MATα cbk1S570A-GFP∷TRP1 | |
| ELY467 | MATα cbk1T743A-GFP∷KanMX | |
| ELY469 | MATα cbk1D475A-GFP∷KanMX | |
| ELY535 | MAT a CBK1-GFP∷KanMX SEC63-tomato∷HIS3 | |
| ELY536 | MATα cbk1S570A-GFP∷TRP1 SEC63-tomato∷HIS3 | |
| ELY537 | MAT a /MATα CBK1-GFP∷KanMX cbk1D475A-HA∷HIS3 |
All strains are derived from ELY126 and ELY402.
To generate lysates for immunoprecipitation, ∼160 OD600 equivalents were resuspended in 2 ml ice-cold yeast lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.4, 1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 60 mM β-glycerophosphate, 2 mM sodium orthovanadate, 10 mM sodium molybdate, 3 mM benzamidine, 1 mM AeBSF, 1 μg/ml pepstatin, 0.5 mM leupeptin, and 2 μg/ml chymostatin). The cell slurry was split into two 15-ml conical tubes, and 3-ml ice-cold glass beads were added to each. Cells were lysed by vortexing on ice seven times for 1 min, until >85% of cells were broken by microscopy. Lysates were cleared by centrifugation at 13.2 g for 30 min, and the concentration of the resulting supernatant was obtained through protein assay (Bio-Rad Laboratories), using BSA for the standard curve. Protein concentrations were normalized to 35 μg/μl, and 2 ml of normalized lysates were incubated for 30 min on ice with 5.76 μg anti-HA (12CA5; a gift from R. Lamb, Northwestern University, Evanston, IL) or anti-myc (9E10; a gift from H. Folsch, Northwestern University, Evanston, IL) monoclonal antibodies. The lysates were then rotated 1.5 h at 4°C with 50 μl of 1:1 protein G bead slurry (Sigma-Aldrich), equilibrated in yeast lysis buffer. After incubation, beads were washed three times with yeast lysis buffer and three times with yeast wash buffer (omit Triton X-100, glycerol, and phosphatase inhibitors from lysis buffer). For phosphatase treatment, samples were washed once with λ phosphatase buffer and incubated at 37°C for 30 min in 50 μl λ phosphatase buffer with 1 μl λ phosphatase (New England Biolabs, Inc.) added. Samples were subsequently washed once in ice-cold yeast wash buffer and resuspended immediately in 2× SDS-PAGE sample buffer.
Western blotting with phosphospecific antibodies
For all Western blots, proteins were resolved by SDS-PAGE and transferred as described previously (Weiss et al., 2002). Phosphospecific antibodies (Open Biosystems) were generated against the relevant phosphopeptides for the T-loop serine and the CT-motif threonine: CRLMAY(pS)TVGTPD and CPFIGY(pT)YSRFD, respectively. Western blotting was performed using BioTraceW polyvinylidene difluoride (Pall). Membranes were blocked for 30 min at room temperature with 10% BSA in Tris-buffered saline with 0.1% Tween (TBST). The phosphospecific antibodies were diluted 1:250 (anti-pS570) or 1:100 (anti-pT743) in 1% BSA TBST and incubated overnight at 4°C. The diluted anti-pT743 solution also contained the nonphosphorylated peptide at 1:250 (CPFIGYTYSRFD, from a stock solution of ∼0.6 mg/ml in 40% DMSO) to increase specificity for the phosphorylated protein. Blots were subsequently washed three times for 3 min with TBST. They were then incubated with Alexa 680– conjugated goat anti–rabbit secondary (Invitrogen) at 1:5,000 in TBST for 1 h at room temperature. After incubation, blots were washed six times for 5 min in TBST, and both were imaged and quantified on the Odyssey (v2.0; Li-Cor, Inc.). Images were processed using Photoshop (Adobe).
Kinase assays
For kinase reactions, immunoprecipitates were stored overnight at 4°C in yeast lysis buffer with 35% glycerol. Beads were washed three additional times with kinase reaction buffer (20 mM Tris, pH 8.0, 150 mM NaCl, and 5 mM MnCl2). Either 2.5 μg of histone H1 or the N terminus of Ace2 fused to maltose binding protein (a gift from D. Stillman, University of Utah, Salt Lake City, UT) was added in 30 μl kinase reaction buffer containing 20 μM cold ATP and 0.33 μCi/μL γ-32P-ATP. Kinase reactions were allowed to proceed at room temperature for 60 min and stopped by addition of 7 μl of 5× SDS-PAGE sample buffer and 10-min incubation at 85°C. Proteins were separated by SDS-PAGE, and kinase activity was quantified using a Storm 860 and ImageQuant (Molecular Dynamics). Values obtained for kinase activity from the autoradiograph were normalized against anti-HA Western blot of the same immunoprecipitates so that kinase activity could be compared between samples.
Cell synchronization
Log phase cells from 4.8 liters of culture were arrested with 10 μM α factor (GenScript, Inc.). After incubation at 30°C for 2 h, >80% of cells exhibited mating projections. The cells were then harvested by centrifugation for 5 min at 25°C. Cells were released from arrest by washing twice with fresh media and then resuspended in fresh media. At each time point, cells were harvested by centrifugation and immediately frozen in liquid nitrogen. Samples of cells from each time point were also fixed by addition of formaldehyde to 5%, and these cells were imaged to determine budding index, providing an indication of cell cycle progression.
Cytology, microscopy, and image analysis
Staining of F-actin with rhodamine-phalloidin, cell wall chitin with calcofluor, and cell wall sugars with rhodamine-conjugated concanavalin A were done as described previously (Adams and Pringle 1991; Pringle 1991). Fluorescence/differential interference contrast microscopy was performed using an Axiovert 200M (Carl Zeiss MicroImaging, Inc.) with either a Coolsnap HQ or Cascade II-512B camera (PhotoMetrics, Inc.). Images were obtained with an oil-immersion α plan-fluar 100×/1.45 NA (Carl Zeiss MicroImaging, Inc.), imaged in synthetic complete medium with dextrose. In scoring all phenotypes, cells were sonicated in a water bath four times for 15 s. The cell separation defect was scored as any cluster of more than two cells. Axial ratio was measured by hand using OpenLab (v4.0.4; Improvision) measurement recording program. Nuclear localization of Ace2-GFP was quantified as symmetric or asymmetric, using rhodamine conA labeling to distinguish mother and daughter cells. All subsequent statistical analysis was performed using Excel (Microsoft), Prism (GraphPad), or InStat (GraphPad).
Online supplemental material
Fig. S1 shows the coimmunoprecipitation of Mob2-13xmyc with Cbk1-HA, both wild type and mutants. Fig. S2 demonstrates that Ace2–protein A is expressed at similar levels across the Cbk1 mutants. Fig. S3 shows that the phosphospecific antibodies do not recognize other heavily phosphorylated proteins, such as GST-Ste20, purified from yeast. Fig. S4 demonstrates that deletion of Ace2 does not affect the transcript levels of RAM network proteins. Table S1 shows primers used for the construction of strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200604107/DC1.
Supplementary Material
Acknowledgments
We gratefully thank H. Folsch, L. Hicke, C. LaBonne, R. Lamb, and R. Tsien for the generous gift of reagents; F. Luca and D. Drubin for yeast strains; and D. Stillman for Ace2 expression constructs. We would also like to thank J. Brickner, A. Cuppia, E. Mazanka, S. Salvador, J. Brace, and Y. Lin for critical reading of the manuscript and many useful discussions.
This work was supported by a grant from the American Cancer Society, Illinois Division, to E. Weiss (05-43). J. Jansen is supported by the National Institutes of Health Cellular and Molecular Basis of Disease Training Grant (NIH 5 T32 GM008061-24) and the Malkin Family Cancer Center Fellowship. E. Weiss is a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
Abbreviations used in this paper: PKB, protein kinase B; RAM, regulation of Ace2 and morphogenesis.
References
- Adams, A.E., and J.R. Pringle. 1991. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 194:729–731. [DOI] [PubMed] [Google Scholar]
- Archambault, V., E.J. Chang, B.J. Drapkin, F.R. Cross, B.T. Chait, and M.P. Rout. 2004. Targeted proteomic study of the cyclin-cdk module. Mol. Cell. 14:699–711. [DOI] [PubMed] [Google Scholar]
- Bichsel, S.J., R. Tamaskovic, M.R. Stegert, and B.A. Hemmings. 2004. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by hMOB1 protein. J. Biol. Chem. 279:35228–35235. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier, S., E.L. Weiss, C. Seidel, D.G. Drubin, and M. Snyder. 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, E.H., M. Nousiainen, R.B. Chalamalasetty, A. Schafer, E.A. Nigg, and H.H. Sillje. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 24:2076–2086. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner, A., T.E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter specific genetic programs to induce asymmetric cell fates. Cell. 107:739–750. [DOI] [PubMed] [Google Scholar]
- Dohrmann, P.R., G. Butler, K. Tamai, S. Dorland, J.R. Greene, D.J. Thiele, and D.J. Stillman. 1992. Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6:93–104. [DOI] [PubMed] [Google Scholar]
- Du, L.L., and P. Novick. 2002. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto, K., Y. He, B. Ye, W.B. Grueber, P.N. Adler, L.Y. Jan, and Y.N. Jan. 2004. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 119:245–256. [DOI] [PubMed] [Google Scholar]
- Emoto, K., J.Z. Parrish, L.Y. Jan, and Y.-N. Jan. 2006. The tumor suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 443:210–213. [DOI] [PubMed] [Google Scholar]
- Gallegos, M.E., and C.I. Bargmann. 2004. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 44:239–249. [DOI] [PubMed] [Google Scholar]
- Geng, W., B. He, M. Wang, and P.N. Adler. 2000. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 156:1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., K. Emoto, X. Fang, N. Ren, X. Tian, Y.N. Jan, and P.N. Adler. 2005. a. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol. Biol. Cell. 16:4139–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., X. Fang, K. Emoto, Y.N. Jan, and P.N. Adler. 2005. b. The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with furry during Drosophila wing hair development. Mol. Biol. Cell. 16:689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich, A., S.J. Bichsel, and B.A. Hemmings. 2005. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol. Cell. Biol. 25:8259–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich, A., M.R. Stegert, D. Schmitz, and B.A. Hemmings. 2006. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7:253–264. [DOI] [PubMed] [Google Scholar]
- Laabs, T.L., D.D. Markwardt, M.G. Slattery, L.L. Newcomb, D.J. Stillman, and W. Heideman. 2003. ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 100:10275–10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mah, A.S., J. Jang, and R.J. Deshaies. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 98:7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward, T., P. Cron, and B.A. Hemmings. 1995. Molecular cloning and characterization of a conserved nuclear serine (threonine) protein kinase. Proc. Natl. Acad. Sci. USA. 92:5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward, T.A., D. Hess, and B.A. Hemmings. 1999. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274:33847–33850. [DOI] [PubMed] [Google Scholar]
- Nelson, B., C. Kurischko, J. Horecka, M. Mody, P. Nair, L. Pratt, A. Zougman, L.D. McBroom, T.R. Hughes, C. Boone, and F.C. Luca. 2003. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell. 14:3782–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen, B., S. Taylor, and G. Ghosh. 2004. Regulation of protein kinases: controlling activity through activation segment conformation. Mol. Cell. 15:661–675. [DOI] [PubMed] [Google Scholar]
- O'Conallain, C., M.T. Doolin, C. Taggart, F. Thornton, and G. Butler. 1999. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R. 1991. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194:732–735. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., A. Legesse-Miller, L. Gao, Y. Dong, and A. Bretscher. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559–591. [DOI] [PubMed] [Google Scholar]
- Racki, W.J., A.M. Becam, F. Nasr, and C.J. Herbert. 2000. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19:4524–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., N. Vogt, C. Ziv, R. Gorovits, and O. Yarden. 2006. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol. Biol. Cell. 17:4080–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, X., N.C. Shaner, C.A. Yarbrough, R.Y. Tsien, and S.J. Remington. 2006. Novel chromophores and buried charges control in mFruits. Biochemistry. 45:9639–9647. [DOI] [PubMed] [Google Scholar]
- Stegert, M.R., R. Tamaskovic, S.J. Bischel, A. Hergovich, and B.A. Hemmings. 2004. Regulation of the NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J. Biol. Chem. 279:23806–23812. [DOI] [PubMed] [Google Scholar]
- Stegert, M.R., A. Hergovich, R. Tamaskovic, S.J. Bichsel, and B.A. Hemmings. 2005. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell. Biol. 25:11019–11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaskovic, R., S.J. Bichsel, H. Rogniaux, M.R. Stegert, and B.A. Hemmings. 2003. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J. Biol. Chem. 278:6710–6718. [DOI] [PubMed] [Google Scholar]
- Verde, F., D.J. Wiley, and P. Nurse. 1998. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA. 95:7526–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E.L., A.C. Bishop, K.M. Shokat, and D.G. Drubin. 2000. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2:677–685. [DOI] [PubMed] [Google Scholar]
- Weiss, E.L., C. Kurischko, C. Zhang, K. Shokat, D.G. Drubin, and F.C. Luca. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., J. Huang, J. Dong, and D. Pan. 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 114:445–456. [DOI] [PubMed] [Google Scholar]
- Yang, J., P. Cron, V.M. Good, V. Thompson, B.A. Hemmings, and D. Barford. 2002. a. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 9:940–944. [DOI] [PubMed] [Google Scholar]
- Yang, J., P. Cron, V. Thompson, V.M. Good, D. Hess, B.A. Hemmings, and D. Barford. 2002. b. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol. Cell. 9:1227–1240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.