Abstract
Recent advances in high-throughput methods and the application of computational tools for automatic classification of proteins have made it possible to carry out large-scale proteomic analyses. Biological analysis and interpretation of sets of proteins is a time-consuming undertaking carried out manually by experts. We have developed PANDORA (Protein ANnotation Diagram ORiented Analysis), a web-based tool that provides an automatic representation of the biological knowledge associated with any set of proteins. PANDORA uses a unique approach of keyword-based graphical analysis that focuses on detecting subsets of proteins that share unique biological properties and the intersections of such sets. PANDORA currently supports SwissProt keywords, NCBI Taxonomy, InterPro entries and the hierarchical classification terms from ENZYME, SCOP and GO databases. The integrated study of several annotation sources simultaneously allows a representation of biological relations of structure, function, cellular location, taxonomy, domains and motifs. PANDORA is also integrated into the ProtoNet system, thus allowing testing thousands of automatically generated clusters. We illustrate how PANDORA enhances the biological understanding of large, non-uniform sets of proteins originating from experimental and computational sources, without the need for prior biological knowledge on individual proteins.
INTRODUCTION
In recent years, new experimental and computational methods have greatly increased the capability of performing large-scale proteomic and genomic studies. In this line of research, large sets of proteins or genes are being studied simultaneously. There are numerous such studies that reflect experimental as well as computational approaches (1,2). Innovation in high-throughput technologies has led to a flood of data from DNA microarrays, two-hybrid screens, phage displays, 2D gels and advanced mass-spectrometry experiments (3,4). On the computational side, comparative genomics, phylogenetic profiling and numerous methods for a global organization of genes and proteins have led to a large collection of protein sets for which structural and functional understanding is desirable (5,6). The biological analysis of such sets tends to be complicated and time-consuming due to the immense size of the data as well as the necessity of thorough biological knowledge of each protein. This often leads to an insufficient analysis of only a small subset of proteins, which provides very limited biological understanding of the result. However, much effort has been put into annotating protein sequences in recent years (7–9). We define an ‘annotation’ or a ‘keyword’ as a binary property that may be assigned to a protein. Resources such as InterPro (10), Gene Ontology (GO) (11), ENZYME (12) and SCOP (13) provide a wealth of biological information, in the form of annotations. Different annotations offer a whole spectrum of information for each protein of interest. For well-studied proteins, information concerning structure, sequential motifs, cellular localization, association with biochemical pathways and taxonomy is usually provided. Examination of the annotation sources used by PANDORA shows that more than 95% of the proteins are associated with two annotations or more (excluding taxonomical annotations). The average number of annotations per protein is 10.9 and the median is 10. The increasing amount of available annotations allows us to study protein sets without the need of a deeper examination of individual proteins.
The organization of annotations into well-focused dictionaries of keywords enables the usage of computational methods to analyze such annotation data. The simplest way to analyze a set of proteins is based on tallying individual keywords. However, this naïve method can often obscure much of the biological information. Consider for example a set of 100 proteins with 50 appearances of the keyword ‘membrane’ and 50 appearances of the word ‘enzyme’. What can be concluded? The set could consist of 50 proteins that are membrane-localized enzymes, two disjoint sets of membrane proteins and enzymes, or intersecting sets. Naïve tallying is too weak a method to distinguish between these possibilities. It entails a loss of relevant biological information, especially when rich and complex protein-keyword sets are being considered. Therefore it is important to recognize that intersection and inclusion (subset/superset) relations between annotation-specific subsets of proteins possess crucial biological data.
We have developed PANDORA (Protein ANnotation Diagram ORiented Analysis), a web tool based on the SwissProt protein database (14) that allows us to carry out integrative biological annotation analysis of protein sets, using annotations from various sources. PANDORA currently integrates annotations from the following sources: SwissProt keywords, NCBI Taxonomy (15), InterPro, GO, SCOP and ENZYME.
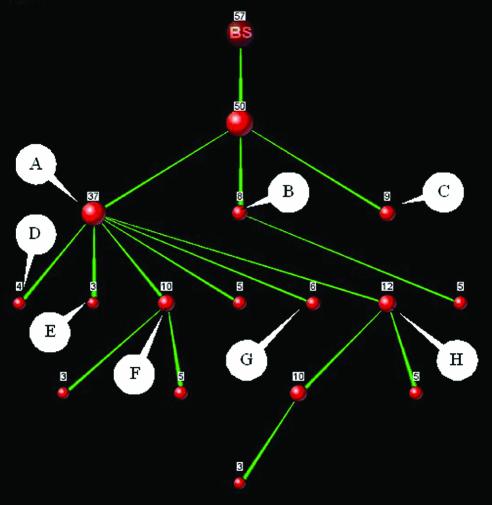
The input to PANDORA is a protein set and a selection of one or more annotation types. The system displays the full protein-keyword relations between the proteins of the set and the keywords of the selected types. This is displayed as an intersection-inclusion Directed Acyclic Graph (DAG). An intersection-inclusion DAG is a hierarchical graph that describes all intersection and inclusion relationships between given sets. In our case, these sets would be protein sets, each protein set sharing a unique mixture of keywords. This allows presentation of the whole collection of protein-keyword relations without loss of the initial information. This concept is demonstrated in Figure 1.
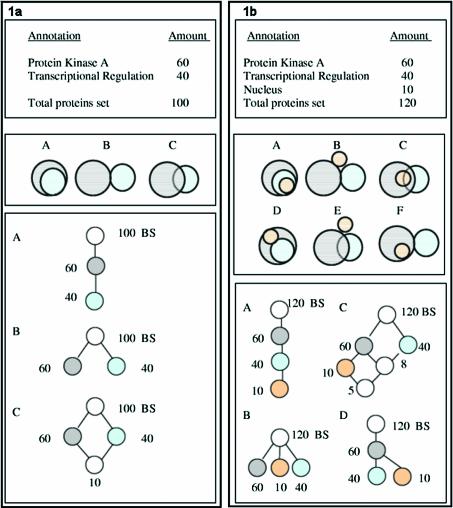
Figure 1.
Representation of keyword set relationships as an intersection-inclusion DAG. Numbers indicate amount of proteins in each set. BS indicates the ‘Basic Set’ of all proteins. (a) Top panel: tally of keyword appearances which does not reveal the amount of intersection between the two sets; middle panel: all three possibilities of intersection between two sets (A, inclusion; B, disjoint sets; C, partial intersection); bottom panel: intersection-inclusion DAG representation of these three cases. (b) Top panel: tally of three keyword appearances; middle panel: six possible cases of intersection between three sets; bottom panel: intersection-inclusion DAG representation of four out of these six cases.
In cases of large protein sets and very rich information, we offer the user the option of controlled graph simplification, allowing the user to observe the data at varying levels of detailed granularity. Protein clusters obtained by any computational method are a natural test-bed for biological analysis using PANDORA. For such application, PANDORA is currently being integrated into ProtoNet (16), a system that provides hierarchical agglomerative clustering of all SwissProt proteins.
METHODS
Database and source of annotations
We have used annotation sources associated with proteins in SwissProt version 40.28, containing 114 033 proteins. SwissProt is considered to be a highly reliable protein database, and many annotation sources are already mapped to the SwissProt database. Each protein was associated with annotations obtained from six separate sources. Annotation sources used: SwissProt keywords, NCBI Taxonomy (as of September 2002), InterPro version 5.2, SCOP version 1.57, GO (as of July 2002 using EBI mapping to SwissProt) and ENZYME version 27.0. The properties and the amounts of each of the individual sources are listed in Table 1.
Table 1. Annotation sources used by PANDORA.
| Source | Annotation method | Number of annotations | Annotation type | Data structure |
|---|---|---|---|---|
| SwissProt Keywords (release 40.28) | Expert/automatic | 865 | Wide range of annotations from very general to very specific; www.expasy.org/swissprot/ | Unstructured |
| NCBI Taxonomy (Sept 2002) | Automatic | 10 844 | Taxonomical annotations; www.ncbi.nlm.nih.gov/Taxonomy/ | Tree |
| InterPro (version 5.2) | Integration expert/automatic | 5551 | Sequence based annotations. Integration from: TIGRFAMs, SMART, ProDom, Pfam, PROSITE and PRINTS; four categories: Family (4261), Domain (1200), Repeat (82) and PTM (8); www.ebi.ac.uk/interpro/ | Partly unstructured, some trees; additional relations, i.e. ‘found in’ and ‘contains in’ |
| SCOP (version 1.57) | Expert | 2927 | Structural annotations, based on structural domains; http://scop.mrc-lmb.cam.ac.uk/scop/ | Tree, four levels |
| GO (Gene Ontology; July 2002) | Expert/automatic | 5229 | Three categories: Molecular Function (3004), Cellular Component (480) and Biological Process (1745); www.ebi.ac.uk/go/ | Each category is implemented as a Directed Acyclic Graph (DAG) |
| ENZYME (version 27.0) | Expert/automatic | 1959 | Enzyme classification annotations; www.expasy.org/enzyme/ | Tree, four levels; by an established nomenclature |
Recent releases may have additional information (see text).
PANDORA is a web-based tool written in PHP and HTML, thus allowing it to be platform independent. PANDORA is available on the web at: http://www.pandora.cs.huji.ac.il; supporting data: www.pandora.cs.huji.ac.il/supplement/
URLs of resources are listed: EBI GO Annotation: www.ebi.ac.uk/GOA/; ENZYME: www.expasy.org/enzyme/; GO: www.ebi.ac.uk/go/; InterPro: www.ebi.ac.uk/interpro/; NCBI Taxonomy: www.ncbi.nlm.nih.gov/Taxonomy/; ProtoNet: www.protonet.cs.huji.ac.il/; SCOP: http://scop.mrc-lmb.cam.ac.uk/scop/; SwissProt: www.exapsy.org/swissprot/
Generating the intersection-inclusion Directed Acyclic Graph
Given a set of proteins P as input, and a set of related keywords K assigned to those proteins, a K×P binary matrix is created representing a symmetrical binary relation between proteins and keywords. Using the symmetrical property of this relation, a row in the binary matrix would be a bit vector that represents a set of proteins sharing a common keyword. The representation as a bit vector is possible due to the fact that there is a limited amount of keywords possible, thus allowing each keyword to be assigned an indexed bit in the vector. Each of these K bit vectors becomes a node in a DAG (Directed Acyclic Graph). Nodes are added one by one, are placed correctly and are checked for intersections with other relevant nodes in the graph. The implementation of sets as bit vectors allows computationally efficient manipulation of these sets using fast logical bit-wise operations. When comparing two bit vector nodes, five cases may arise, described in the following pseudo-code:
// A and B are bit vectors of proteins, and
each is assigned one
// or more keywords
If (A = = B){
merge A with B; // new node will share A and B
keywords
}
Else {
C = A & B; // ‘bitwise and’
if (C = = 0){
A and B are disjoint sets;
}
elseif (C = = A){
make B descendant of A; // A is subset of B
}
elseif (C = = B){
make A descendant of B; // B is subset of A
}
else{
make a new ‘intersection’ node from C;
inherit A and B keywords;
place as descendant of A and B;
}
}
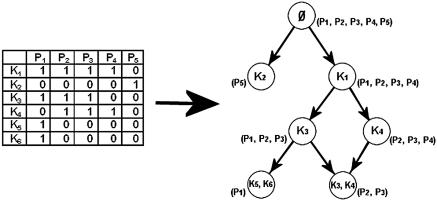
This code is adjusted and implemented recursively, so that it can handle the process of adding the nodes one by one, placing them in their correct position in the graph and checking them for relevant intersections. For ease of use, a node called the ‘Basic Set’, representing all the proteins of the current set, is constructed and placed at the top of the graph. This is the only node whose proteins may not have any keywords in common. Figure 2 shows a schematic example of a protein-keyword binary matrix and the resulting DAG.
Figure 2.
A protein-keyword binary matrix and its matching intersection-inclusion DAG. K1–K6 are keywords, P1–P5 are proteins. Arrows (edges) in the graph represent hierarchical inclusion (subset/superset) relationships. Each node represents a set of proteins that share a unique combination of keywords. The top node of the graph is the ‘Basic Set’, representing all the keywords of the set (it is marked empty due to the fact that there are no keywords common to all proteins in the set). The proteins of each node in the graph appear in parentheses next to it. While the matrix seems cryptic, the graph shows the proteins are divided into two major groups: P1–P4 that have K1 in common, and P5 that has K2. Keywords are passed by inheritance due to the hierarchical inclusion relationships. For example, all the proteins that have the keyword K5 and K6 (in this case only P1—in the node on the bottom left) also have the keywords K3 and K1 due to inheritance. For details of graph construction see Methods.
Input methods
Selection of the basic protein set as input for PANDORA reflects the underlying type of research that is involved.
For a global proteomic protein family research, one might wish to study a set of all proteins that share a keyword of interest as a starting point for study. Note that this feature can be used to study various biological sets, such as all the proteins that are involved in a certain biological process (according to GO) or that are structurally similar (according to SCOP).
For any sets of proteins resulting from external data, whether they are obtained by computational or experimental methods, the user may input the proteins either manually or by file upload.
In case the proteins of interest do not appear in the SwissProt database or the accession numbers are unknown, the user might wish to find similar proteins in SwissProt, and study them with PANDORA. In this case, the sequences of the proteins are submitted, and each sequence is locally aligned using the BLAST (17) search engine against the SwissProt database, and the accession number of the protein with the highest E-score (above a set threshold) is returned. The entire list of proteins found by the BLAST search can be used as input for PANDORA. As PANDORA is not limited to proteomic information, genomic information may also be used as input sequences for BLASTing. A set of nucleotide sequences may be used for alignment with the protein database using BLASTX. It is important to note that a local alignment may be misleading in terms of protein similarity. This problem may be more severe when nucleotide sequences are used as input. Proteins that are considered ‘similar’ by local alignment may often share only a local region, resulting in an incorrect annotation transfer. Changing the E-score threshold to force a more significant alignment will not necessarily resolve this problem. This is a widely known problem (18) which is not addressed in this current version of PANDORA.
PANDORA allows study of protein clusters. Several sources for protein families are known that include SwissProt proteins, i.e. ProtoMap (19) and ProtoNet (16). Other systems are based on additional information from TrEMBL and from complete genomes, i.e. ClusTR (20), Systers (21) and iProClass (22). ProtoNet is a website that provides a hierarchical agglomerative clustering organization of all SwissProt proteins including a rich view on the quality of each protein cluster. PANDORA enables biological annotation analysis of these protein clusters.
RESULTS
Combining all protein-keyword relations, a DAG is constructed (for details on graph construction, see Methods). Each node in the graph represents a number of proteins sharing a unique combination of keywords. The graph shows inclusion and intersection relations between these nodes. The hierarchy in the graph is based on the proteins of each node, so that the keywords are passed by inheritance. This means that each of the proteins of a given node share not only the keywords of that node but also all the keywords of its ancestors. Figure 2 shows a schematic example of a protein-keyword relation DAG.
We begin with an explanation of the mathematical/ computational concepts that underlie PANDORA.
Concepts and principles for PANDORA development
Dealing with complex data. It is important to understand that a DAG constructed contains ALL the information of relationships between keywords and proteins, specifically, all unique keyword combinations with underlying proteins. This necessarily means that complex data will yield a complex graph. A larger set of proteins potentially means more keywords and increased complexity. In the worst possible case, a graph with K keywords will have
nodes (the collection of all possible subsets, also known as the Power Set). This worst-case scenario is clearly unacceptable. For example, a set annotated by merely 20 keywords will have over a million nodes (220 = 1 048 576). Fortunately, instances of such immense complexity, though possible from the pure mathematical perspective, do not tend to manifest themselves in the world of proteins, due to some intrinsic properties of protein-keyword relationships. Still, graphs of complexity that humans find hard to digest do occur. We have developed some computational methods that address this issue.
The first method is based on the notion of ‘resolution’. This essentially is a user-controlled parameter that trades off a graph’s accuracy for simplicity. We define the level of resolution according to the maximal error allowed. The error is measured by the number of proteins by which two nodes may differ and still be considered the same. For example, with a resolution of three proteins, nodes that differ by three proteins or less are considered equal and are merged. Furthermore, two nodes whose intersection is ‘almost’ equal to one of them (i.e. within the set resolution) are shown as parent and child. This concept simplifies the graph and allows the user to control the amount of data lost by this simplification. Based on experience, we set the default resolution value to 1% of the number of proteins in the set.
‘Zooming’ is an additional simplification method introduced in PANDORA. While studying a complex graph at low resolution, i.e. trading accuracy for simplicity, the user sees a simplified global view of the data. However, one may wish to view a biologically interesting subset of proteins at higher accuracy (higher resolution). For this we provide the ‘zooming’ method. Each node in the graph can be selected as a new set of proteins, and studied separately in a new graph. This allows focusing on a specific subset of proteins, studying it at any desired resolution, removing irrelevant nodes and edges and studying different aspects of the chosen subset by including other annotation types.
An inherent problem of the different types of annotation sources is the multiple usages of similar (or identical) keywords to describe different sets (23,24). For example, 321 proteins are annotated ‘Voltage-gated channel’ by GO ‘molecular function’, but only 312 proteins are annotated ‘Voltage-gated channel’ by SwissProt. It is important to mention the resolution method as a means of dealing with such annotation inconsistency. Lowering graph resolution separates protein sets with a low degree of intersection, and unifies sets that have a high degree of intersection. Thus, protein sets that share similar keywords from different sources and are roughly equal (e.g. the ‘Voltage-gated channel’ sets mentioned above) will be unified.
Integration of multiple annotation types. One of the strengths of PANDORA is the capability of integrating several annotation types. For a given set of proteins, the user may choose a preferable type of annotation. Jointly with the ‘zooming’ option (see above), this provides rich biological data that are not easily accessible. For example, assume a set of proteins is studied through taxonomical annotations. The user then ‘zooms in’ on the subset of proteins that are specific to Drosophila, examining it in a new graph. Now this subset can be further studied through their cellular location information, for example, zooming in on all proteins that are located in the nucleus. This will open a new graph, and so on.
Another helpful method is to look at more than one annotation type on the same graph at the same time. This could show relations between different biological aspects such as structure-function or function-location intersections. PANDORA provides this feature, allowing selection of multiple annotation types simultaneously. However, it is important to remember that graph complexity potentially increases exponentially with the amount of keywords, so this feature should be used while keeping in mind that the graphs may become very complex.
Assessing the quality of a protein set. When studying protein sets that share a keyword, it is often important to be able to quantitatively assess the contents of the set in terms of quality or significance. For example, when studying a random set of 10 proteins sharing a keyword, it might be useful to know if this keyword is assigned to only 10 proteins in the database or to 100 000 proteins. The way PANDORA quantifies the quality of a protein set is by sensitivity, defined as:
where TP(k) are the True-Positives (the amount of proteins in the set and are assigned the keyword k) and FN(k) are the False-Negatives (the amount of proteins that are not in the set and are assigned the keyword k). The sensitivity scores are displayed on the graph, allowing easy evaluation of the protein set (sensitivity is also color-coded in the graph; nodes fill from red to white: red represents false-negatives, white represents true-positives).
In this version of PANDORA, no statistical evaluation is assigned for the keywords of the protein sets. This would require a prior assumption on the source of the data and the properties of the proteins that are used as input (for a null model). We choose not to limit PANDORA to a given method of data input (for details see Methods) and thus application of a statistical model seems inappropriate. However, the sensitivity scoring does supply a quantitative measure of evaluation and can give some indication as to the significance of the results.
Analysis of complex biological examples
All graphs mentioned in the examples are available interactively at the PANDORA supplemental web site (http://www.pandora.cs.huji.ac.il/supplement/). Studying these graphs interactively on-line may enhance the understanding of these examples.
We bring examples of three typical uses of PANDORA: analysis of a protein set resulting from a comparative proteomics experiment, analysis of a protein cluster derived computationally and detection of false annotations by analysis of a protein set sharing an annotation.
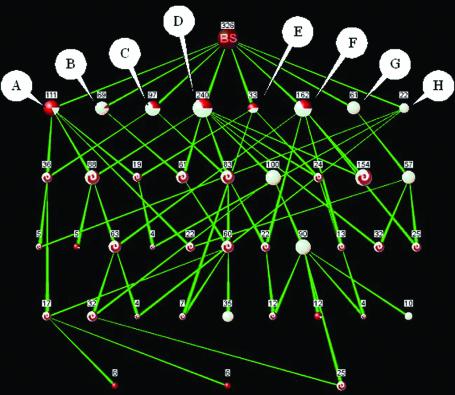
Analysis of a large protein cluster. Large protein clusters resulting from computational methods are natural targets for biological analysis with PANDORA. Such clusters are extremely difficult to analyze manually, especially the identification of ‘biological’ subsets of proteins. We applied PANDORA on a ProtoNet protein cluster (A220629) containing 326 proteins. The cluster contains 61 proteins marked by InterPro as ‘Glucose-inhibited division protein, A family’ (gidA). GidA proteins are highly conserved proteins found in both prokaryotes and eukaryotes, yet the function of these proteins remains unknown. Previous research suggests localization to the periplasm in prokaryotes (25) and to the mitochondria in eukaryotes (26) and binding of FAD (25). Some of the gidA proteins are partially annotated by InterPro, but no annotation of any source is shared by a significant amount of these proteins. Biological interpretation of the 326-protein cluster may provide some further evidence as to these proteins’ function. Looking at the graph of InterPro ‘domain’ and ‘family’ keywords (Fig. 3), the cluster consists of eight main biological subsets.
Figure 3.
PANDORA graph of a ProtoNet protein cluster (A220629), containing 326 proteins. Resolution level is of three proteins. Annotations (InterPro): (A) ‘NAD binding site’; (B) ‘Pyridine nucleotide-disulfide oxidoreductase, class-II’; (C) ‘Adrenodoxin reductase’; (D) ‘FAD-dependent pyridine nucleotide-disulfide oxidoreductase’; (E) ‘Aromatic ring hydroxylase’; (F) ‘Pyridine nucleotide-disulfide oxidoreductase, class-I’; (G) ‘Glucose-inhibited division protein, A family’; (H) ‘Thiamine biosynthesis Thi4 protein’. Sensitivity is reflected by the color of the nodes: red reflects misses and white reflects hits (intersection nodes are marked by a red-white swirl). Nodes A–H appear to be highly interconnected, reflecting the sharing of biological properties. Node D is the largest node, containing 240 proteins out of 331 ‘FAD-dependent pyridine nucleotide-disulfide oxidoreductase’ proteins in the database.
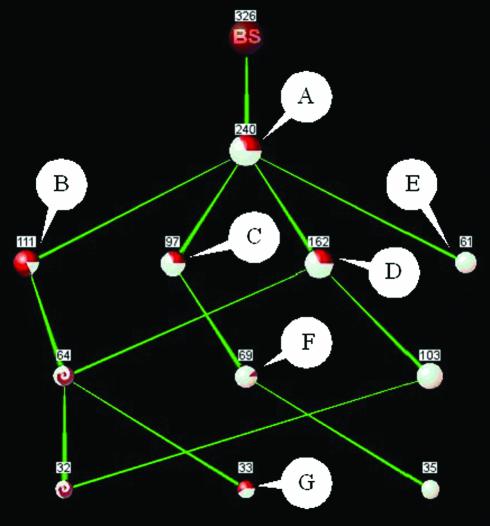
The most prominent set is ‘FAD-dependent pyridine nucleotide-disulfide oxidoreductase’ (FPNDO), with a sensitivity of 73% (color-coded in the graph) and containing 74% of the proteins in the cluster. Additionally these eight main subsets appear highly interconnected, indicating that they share many biological properties, more specifically InterPro annotations. This high degree of interconnection is especially apparent between the group of 240 FPNDO proteins and the other seven main subsets. This can be appreciated by decreasing the resolution, thus lowering the accuracy and removing annotation ‘noise’: the low-resolution graph (Fig. 4) shows the FPNDO set as a superset, containing all other main subsets, indicating a high degree of intersection with these groups.
Figure 4.
PANDORA graph of a ProtoNet protein cluster (A220629), containing 326 proteins. Resolution level is of 30 proteins. Annotations (InterPro): (A) ‘FAD-dependent pyridine nucleotide-disulfide oxidoreductase’; (B) ‘NAD binding site’; (C) ‘Adrenodoxin reductase’; (D) ‘Pyridine nucleotide-disulfide oxidoreductase, class-I’; (E) ‘Glucose-inhibited division protein, A family’; (F) ‘Pyridine nucleotide-disulfide oxidoreductase, class-I’; (G) ‘Aromatic ring hydroxylase’. Sensitivity is reflected by the color of the nodes: red reflects misses and white reflects hits (intersection nodes are marked by a red-white swirl). Node A appears to be a superset of all other protein sets in this highly simplified graph. This low-resolution view of the graph shown in Figure 3 helps appreciate the high degree of intersection between the main subsets (nodes B–G) and the ‘FAD-dependent pyridine nucleotide-disulfide oxidoreductase’ set (node A).
Furthermore, examining the cluster through ‘enzyme’ keywords shows that all 201 proteins that are annotated by ‘enzyme’ are annotated as ‘oxidoreductase’, spanning several different types of oxidoreductases. A graph of SCOP annotations (not shown) reveals 12 proteins are assigned structures, all belonging to the SCOP ‘FAD/NAD(P)-binding domain’ superfamily. None of the gidA proteins are assigned as solved structures, yet a prediction using GenTHREADER (27) fold-recognition server of a gidA protein (GIDA_BUCAI) that has no annotations (except as gidA) showed several statistically significant hits (Table 2), all folds of oxidoreductases.
Table 2. Fold ‘hit list’ for an unannotated gidA protein (GIDA_BUCAI) as predicted by GenTHREADER fold recognition server.
| Description | PDB no. | E-value |
|---|---|---|
| Fumarate reductase | 1qjd | 0.001 |
| Sarcosine oxidase | 1b3m | 0.002 |
| Fumarate reductase | 1qla | 0.002 |
| Adenylylsulfate reductase | 1jnr | 0.002 |
| Thioredoxin reductase | 1trb | 0.003 |
| Phenol hydroxylase | 1foh | 0.003 |
| Flavocytochrome C sulfide dehydrogenase | 1fcd | 0.003 |
| Glutathione reductase | 3grs | 0.003 |
| Dihydrolipoamide dehydrogenase | 1lvl | 0.003 |
| Adrenodoxin reductase | 1cjc | 0.003 |
Ten best matches are shown. All structures are of oxidoreductases.
Despite the large size of the cluster and some degree of inconsistency between various annotation sources, PANDORA provides a global view of this ProtoNet protein cluster, providing further evidence that gidA proteins function as NAD/FAD binding oxidoreductases. The full extent of these annotations, such as the subdivision into main biological subsets and high degree of interconnectivity, would be extremely difficult to appreciate by means of examining the long annotation ‘hit lists’ or other view of individual proteins.
Biological interpretation of experimental results. Proteomic experiments often result in long lists of proteins. Biological analysis of these protein sets is difficult due to the amounts of proteins and their different biological properties. The inability to interpret the full extent of these sets often results in an analysis of a small subset of proteins, undermining the attempt to achieve a global view of complex biological processes. Selenium salts are toxic at high concentrations and have mutagenic effects in several prokaryotes (28). However, selenium is a trace element required for the synthesis of the amino acid selenocysteine. In mammals, selenium is known to have an insulin-like hypoglycemic effect of stimulating glucose uptake and metabolism (29,30). Furthermore, there is also some evidence that selenium possesses anticarcinogenic properties in mammals (31). We chose a set of 57 proteins that were induced by selenium salts in Escherichia coli (32), and used PANDORA in order to obtain a comprehensive biological view of the set. Figure 5 shows the PANDORA graph of these proteins (at a two protein resolution), using GO ‘biological process’ annotations.
Figure 5.
PANDORA graph of 57 proteins induced by selenium salts in E.coli. Resolution is of two proteins. Annotations (GO ‘Biological process’): (A) ‘Metabolism’; (B) ‘Stress response’, ‘Temperature response’; (C) ‘Transport’; (D) ‘Electron transport’; (E) ‘Oxygen and radical metabolism’; (F) ‘Nucleic acid metabolism’; (G) ‘Glucose catabolism’, ‘Glycolysis’; (H) ‘Biosynthesis’. The graph shows a clear division into biological subsets. Induction of the proteins of nodes B and E is obviously stress-related. Node G (six proteins) indicates induction of glycolysis.
The graph shows a clear division into three distinct subsets: nine ‘transport’ proteins, eight ‘stress response’ proteins and 37 ‘metabolism’ proteins (a low degree of intersection between these sets was removed by the resolution threshold). Furthermore, the 37 ‘metabolism’ proteins are subdivided into six major subsets. The authors (32) suggest that selenium toxicity under aerobic conditions may result from superoxide production, due to the induction of superoxide dismutases. The induction of a stress response can be easily recognized in the graph, with eight ‘stress response’ proteins and three ‘oxygen and radical metabolism’ proteins. Yet the graph shows that there are other interesting subsets, especially six proteins annotated as ‘glucose catabolism’. Moreover, using the ‘zoom’ option to view separately the graph of the nine ‘transport’ proteins reveals by annotation two proteins involved in carbohydrate uptake. The induction of six proteins involved in glucose metabolism and two proteins involved in carbohydrate uptake could possibly explain the insulin-like hypoglycemic effects attributed to selenium in mammals. Consistently with our analysis, research suggests aerobic glycolysis as a means of protection against reactive oxygen species (33).
A global view of the results such as the one presented here allows easy perception of the data in a biological context, offering a clearer view of a complex process. All this is achieved without any specific knowledge about the identities of the individual proteins, only through annotations. However, the division into subsets by using lower resolution not only allows a comprehensible view, but also facilitates navigation of the data in order to easily focus research on biologically relevant subsets that are small enough to allow study of individual proteins.
Identification of false annotations. Automatic annotation sources are commonly based on statistical evaluation. As such, they are susceptible to both errors of type I and II (34). Furthermore, incorrect annotations tend to be mistakenly transferred across species via sequence similarity, and once they have penetrated into widely used databases only careful manual screening may trace these false annotations (35).
When examining large protein sets, PANDORA facilitates the detection of protein subsets that share common biological properties. This notion can be applied to a set of proteins that were all given the same annotation by an automatic statistical method (e.g. InterPro). When examining such a set, one would expect to find that the proteins share several properties, especially if looking at annotations that relate to various biological aspects. This would result in a highly interconnected graph. However, separation of the large set into distinct subsets would mean that there are no biological properties shared by these subsets. There are three possible reasons for this: (a) the keyword that was given to the proteins of the set really does describe some aspect that is shared by protein subsets that have no other biological properties in common; (b) the proteins of the set are very poorly annotated, therefore no shared biological properties are found; (c) some of the distinct subsets are incorrectly annotated. Viewing the graph, it is very easy to discern between these three cases, simply by looking at the meanings of the specific annotations.
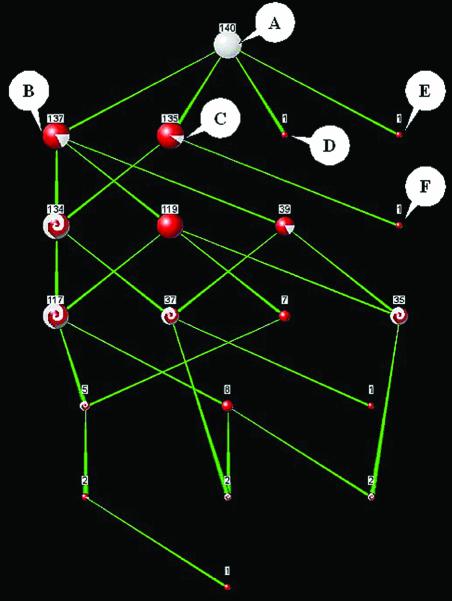
To demonstrate the detection of false annotations, we examined the set of all 140 proteins annotated by InterPro as ‘Homeobox protein, antennapedia type’. According to PROSITE (from which the InterPro annotation was taken), there are three known proteins that are annotated falsely. We examined the PANDORA graph of these 140 proteins using InterPro ‘family’, InterPro ‘domain’ and SwissProt keywords. The graph (Fig. 6) shows two proteins that are absolutely distinct from the rest of the proteins, each sharing a list of unique keywords. A third protein has a list of unique keywords apart from one (‘Developmental Protein’) that is shared with the rest.
Figure 6.
PANDORA graph of 140 proteins annotated by InterPro as ‘Homebox protein, antennapedia type’. Resolution is maximal (no simplification). Select annotations on nodes (InterPro in italics, SwissProt underlined): (A) ‘Homebox protein, antennapedia type’; (B) ‘Homeobox’, ‘DNA-binding’, ‘Nuclear protein’; (C) ‘Developmental protein’; (D) ‘Ribosomal protein’, ‘Mitochondrion’; (E) ‘ABC transporter’, ‘Nitrate assimilation’, ‘Membrane’; (F) ‘Chymotrypsin Serine protease S1’, ‘Zymogen’. Nodes D, E and F appear biologically distinct in the graph, and are falsely annotated as ‘Homebox protein, antennapedia type’ proteins.
This supplies preliminary evidence that these proteins may be falsely annotated. An examination of the keywords of these three proteins and the other interconnected set of 137 proteins confirms this suspicion. While the large set of 137 proteins shares annotations such as ‘Homeobox’, ‘Nuclear Protein’ and ‘DNA-Binding’, the other three proteins appear to be completely different biologically: one is annotated ‘Ribosomal protein’ and ‘Mitochondrion’, the second as ‘ABC transporter’ and ‘Membrane’, and the third as ‘Serine protease’ and ‘Zymogen’. Surely enough, these three proteins were found to be the three known false-positives mentioned by PROSITE.
This example suggests a novel method of spotting false-positive annotations, through the examination of the degree of annotation connectivity of protein sets. This is based on the notion that related proteins usually share a number of different properties of various biological aspects.
DISCUSSION
At present already ∼1000 viruses, over 100 microbial genomes and 10 multi-cellular organisms have been fully sequenced. Methods such as comparative genomics, chromosomal localization, phylogenetic profiles, structural predictions and remote homology search are applied to gain information on new proteins and on their cellular and biochemical function. While it is expected that the huge body of knowledge will accelerate the success in inferring structure and function, the truth of the matter is that a substantial fraction (∼50% in eukaryotes) of all predicted open reading frames (ORFs) are still orphans with no assigned function (36). Several attempts were introduced for evaluating the power of prediction methods at a genomic scale (37–40). We provide a new notion that attempts to analyze proteins not at the level of individual proteins but rather in the context of a protein collection. Such collections may be derived from experimental or computation sources. We have developed PANDORA, a web-based tool that allows integrative biological analysis of protein sets, based on protein annotations derived from multiple annotation sources, including information regarding function, structure, cellular location, biological process and taxonomy. PANDORA offers description of protein-keyword intersection and inclusion relations, controlled graph simplification, easy manipulation of relevant protein sets, multiple annotation-type integration and flexible protein set input methods. The impact of PANDORA is expected to increase by the availability of additional sources of information and by including sources of annotation with high quality and accuracy.
Annotation inconsistency is a potential pitfall for annotation-based studies (35,41,42), especially when integrating multiple annotation sources. As such, efforts have been made to tackle such inconsistency between annotation sources. PANDORA helps overcome such inconsistencies either by showing the degree of intersection between sets or by using the resolution method to simplify the graph via unification of sets that have a high degree of intersection and separation of sets that have a low degree of intersection.
It is important to recognize the limits of an annotation-based approach. Many annotations are transferred to new proteins automatically, often based on statistical methods. It should be taken into consideration that such automatic methods are prone to some instances of false-positive and false-negative annotations. We suggest a novel method of detecting false-positive annotations by utilizing the ability to easily recognize distinct biological subsets that share common biological properties in the graphs produced by PANDORA. This may lead to an improved automatic identification of false-positive annotations (N.Kaplan and M.Linial, in preparation).
Because of the generic way PANDORA is written, the underlying database can be easily substituted with another protein database such as TrEMBL (14) or even with a complete translated genome database. This would of course require sufficient annotation of the data in these databases. Based on the impressive growth in annotation efforts and the addition of new systematic annotation systems (43), we expect PANDORA to cope with these advances in the near future. At present, InterPro annotation already covers 85% of all proteins in TrEMBL. At the same time, extensively studied model organisms such as that of Saccharomyces cerevisiae already include rich genome-wide functional information regarding gene lethality, protein–protein interactions, coordinated gene expression and more. We designed PANDORA to be flexible in terms of the annotation sources, and new annotation sources can be easily added.
PANDORA shares sources with the ProtoNet system, and as such will be updated every few months to include the newest versions of both the SwissProt protein database and the various annotation sources. Future plans include the addition of more annotation sources to provide richer biological analysis, statistical evaluation for specific input types (such as single organism proteomic experiments) and educated annotation transfer to provide more accurate analysis of input sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Hillel Fleischer and Uri Inbar for excellent database management and Alexander Savenok for HTML help and for the integration PANDORA into the ProtoNet website. We would like to thank Nati Linial for valuable discussions. This study is partially supported by the Israeli Ministry of Defense and the Sudarsky Center for Computational Biology in the Hebrew University.
REFERENCES
- 1.Yao T. (2002) Bioinformatics for the genomic sciences and towards systems biology. Japanese activities in the post-genome era. Prog. Biophys. Mol. Biol., 80, 23–42. [DOI] [PubMed] [Google Scholar]
- 2.Dongre A.R., Opiteck,G., Cosand,W.L. and Hefta,S.A. (2001) Proteomics in the post-genome age. Biopolymers, 60, 206–211. [DOI] [PubMed] [Google Scholar]
- 3.Chanda S.K. and Caldwell,J.S. (2003) Fulfilling the promise: drug discovery in the post-genomic era. Drug Discov. Today, 8, 168–174. [DOI] [PubMed] [Google Scholar]
- 4.Burbaum J.J. and Sigal,N.H. (1997) New technologies for high-throughput screening. Curr. Opin. Chem. Biol., 1, 72–78. [DOI] [PubMed] [Google Scholar]
- 5.Baxter S.M. and Fetrow,J.S. (2001) Sequence- and structure-based protein function prediction from genomic information. Curr. Opin. Drug Discov. Dev., 4, 291–295. [PubMed] [Google Scholar]
- 6.Wilson C.A., Kreychman,J. and Gerstein,M. (2000) Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J. Mol. Biol., 297, 233–249. [DOI] [PubMed] [Google Scholar]
- 7.Xie H., Wasserman,A., Levine,Z., Novik,A., Grebinskiy,V., Shoshan,A. and Mintz,L. (2002) Large-scale protein annotation through gene ontology. Genome Res., 12, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan N., Montelione,G.T. and Gerstein,M. (2003) Ontologies for proteomics: towards a systematic definition of structure and function that scales to the genome level. Curr. Opin. Chem. Biol., 7, 44–54. [DOI] [PubMed] [Google Scholar]
- 9.Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) Predicting function: from genes to genomes and back. J. Mol. Biol., 283, 707–725. [DOI] [PubMed] [Google Scholar]
- 10.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camon E., Magrane,M., Barrell,D., Binns,D., Fleischmann,W., Kersey,P., Mulder,N., Oinn,T., Maslen,J., Cox,A. et al. (2003) The Gene Ontology Annotation (GOA) Project: implementation of GO in SWISS-PROT, TrEMBL and InterPro. Genome Res., 13, 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bairoch A. (2000) The ENZYME database in 2000. Nucleic Acids Res., 28, 304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard T.J., Ailey,B., Brenner,S.E., Murzin,A.G. and Chothia,C. (1999) SCOP: a Structural Classification of Proteins database. Nucleic Acids Res., 27, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledge base and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasson O., Vaaknin,A., Fleischer,H., Portugaly,E., Bilu,Y., Linial,N. and Linial,M. (2003) ProtoNet: hierarchical classification of the protein space. Nucleic Acids Res., 31, 348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 18.Henikoff S. (1996) Scores for sequence searches and alignments. Curr. Opin. Struct. Biol., 6, 353–360. [DOI] [PubMed] [Google Scholar]
- 19.Yona G., Linial,N. and Linial,M. (2000) ProtoMap: automatic classification of protein sequences and hierarchy of protein families. Nucleic Acids Res., 28, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriventseva E.V., Fleischmann,W., Zdobnov,E.M. and Apweiler,R. (2001) CluSTr: a database of clusters of SWISS-PROT+TrEMBL proteins. Nucleic Acids Res., 29, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause A., Stoye,J. and Vingron,M. (2000) The SYSTERS protein sequence cluster set. Nucleic Acids Res., 28, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C.H., Yeh,L.S., Huang,H., Arminski,L., Castro-Alvear,J., Chen,Y., Hu,Z., Kourtesis,P., Ledley,R.S., Suzek,B.E. et al. (2003) The Protein Information Resource. Nucleic Acids Res., 31, 345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanai I. and DeLisi,C. (2002) The society of genes: networks of functional links between genes from comparative genomics. Genome Biol., 3, res0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Copley R.R., Doerks,T., Letunic,I. and Bork,P. (2002) Protein domain analysis in the era of complete genomes. FEBS Lett., 513, 129–134. [DOI] [PubMed] [Google Scholar]
- 25.White D.J., Merod,R., Thomasson,B. and Hartzell,P.L. (2001) GidA is an FAD-binding protein involved in development of Myxococcus xanthus. Mol. Microbiol., 42, 503–517. [DOI] [PubMed] [Google Scholar]
- 26.Colby G., Wu,M. and Tzagoloff,A. (1998) MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem., 273, 27945–27952. [DOI] [PubMed] [Google Scholar]
- 27.Jones D.T. (1999) GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol., 287. [DOI] [PubMed] [Google Scholar]
- 28.Noda M., Takano,T. and Sakurai,H. (1979) Mutagenic activity of selenium compounds. Mutat. Res., 66, 175–179. [DOI] [PubMed] [Google Scholar]
- 29.Furnsinn C., Englisch,R., Ebner,K., Nowotny,P., Vogl,C. and Waldhausl,W. (1996) Insulin-like vs. non-insulin-like stimulation of glucose metabolism by vanadium, tungsten and selenium compounds in rat muscle. Life Sci., 59, 1989–2000. [DOI] [PubMed] [Google Scholar]
- 30.Stapleton S.R. (2000) Selenium: an insulin-mimetic. Cell. Mol. Life Sci., 57, 1874–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ip C. (1998) Lessons from basic research in selenium and cancer prevention. J. Nutr., 128, 1845–1854. [DOI] [PubMed] [Google Scholar]
- 32.Bebien M., Lagniel,G., Garin,J., Touati,D., Vermeglio,A. and Labarre,J. (2002) Involvement of superoxide dismutases in the response of Escherichia coli to selenium oxides. J. Bacteriol., 184, 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brand K.A. and Hermfisse,U. (1997) Aerobic glycolysis by proliferating cells: a protective strategy against reactive oxygen species. FASEB J., 11, 388–395. [DOI] [PubMed] [Google Scholar]
- 34.Devos D. and Valencia,A. (2001) Intrinsic errors in genome annotation. Trends Genet., 17, 429–431. [DOI] [PubMed] [Google Scholar]
- 35.Linial M. (2003) How incorrect annotation evolved—The case of short ORFs. Trends Biotechnol., 21, 298–300. [DOI] [PubMed] [Google Scholar]
- 36.Galperin M.Y. and Koonin,E.V. (2000) Who’s your neighbor? New computational approaches for functional genomics. Nat. Biotechnol., 18, 609–613. [DOI] [PubMed] [Google Scholar]
- 37.Bilu Y. and Linial,M. (2002) The advantage of functional prediction based on clustering of yeast genes and its correlation with non-sequence based classifications. J. Comput. Biol., 9, 193–210. [DOI] [PubMed] [Google Scholar]
- 38.Rost B. (2001) Review: protein secondary structure prediction continues to rise. J. Struct. Biol., 134, 204–218. [DOI] [PubMed] [Google Scholar]
- 39.Jensen L.J., Gupta,R., Staerfeldt,H.H. and Brunak,S. (2003) Prediction of human protein function according to Gene Ontology categories. Bioinformatics, 19, 635–642. [DOI] [PubMed] [Google Scholar]
- 40.Del Sol Mesa A., Pazos,F. and Valencia,A. (2003) Automatic methods for predicting functionally important residues. J. Mol. Biol., 326, 1289–1302. [DOI] [PubMed] [Google Scholar]
- 41.Yanai I. and DeLisi,C. (2002) The society of genes: networks of functional links between genes from comparative genomics. Genome Biol., 3, research0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Copley R.R., Doerks,T., Letunic,I. and Bork,P. (2002) Protein domain analysis in the era of complete genomes. FEBS Lett., 513, 129–134. [DOI] [PubMed] [Google Scholar]
- 43.Pouliot Y., Gao,J., Su,Q.J., Liu,G.G. and Ling,X.B. (2001) DIAN: a novel algorithm for genome ontological classification. Genome Res., 11, 1766–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]