Abstract
We report that the Drosophila mind bomb2 (mib2) gene is a novel regulator of muscle development. Unlike its paralogue, mib1, zygotic expression of mib2 is restricted to somatic and visceral muscle progenitors, and their respective differentiated musculatures. We demonstrate that in embryos that lack functional Mib2, muscle detachment is observed beginning in mid stage 15 and progresses rapidly, culminating in catastrophic degeneration and loss of most somatic muscles by stage 17. Notably, the degenerating muscles are positive for apoptosis markers, and inhibition of apoptosis in muscles prevents to a significant degree the muscle defects. Rescue experiments with Mib1 and Neuralized show further that these E3 ubiquitin ligases are not capable of ameliorating the muscle mutant phenotype of mib2. Our data suggest strongly that mib2 is involved in a novel Notch- and integrin-independent pathway that maintains the integrity of fully differentiated muscles and prevents their apoptotic degeneration.
Introduction
In Drosophila, the larval somatic (skeletal) musculature arises from the fusion of two distinct types of myoblasts, the founders and fusion-competent cells (for review see Beckett and Baylies, 2006). Subsequent differentiation programs, including activation of muscle-specific gene expression and asymmetrical cell fusion between the differentially marked founders and fusion-competent myoblasts, are required for the generation of syncytial muscle fibers. Maturation of these syncytia into functional muscle fibers involve additional events, including pathfinding processes and the formation of attachments to the tendon cells, as well as the establishment of neuromuscular junctions (for review see Volk, 1999; Schnorrer and Dickson, 2004). The functional characterization of integrins and downstream effectors of integrin signals has underscored the importance of this pathway in establishing muscle attachment sites (for review see Bokel and Brown, 2002). However, the molecular basis for many other aspects of morphogenesis and maintenance of the mature muscles is still poorly defined.
Herein, we present a functional characterization of Drosophila mind bomb2 (mib2), which shares structural similarities with its paralogue mib1. Unlike mib1, mib2 is prominently expressed in muscle progenitors and differentiated musculatures. We show that loss of mib2 activity leads to muscle detachment and massive muscle degeneration. We also demonstrate that mib2 functions in a novel integrin- and Notch-independent manner to maintain the integrity of the mature somatic musculature.
Results and discussion
Characterization of the Drosophila mind bomb2 gene product
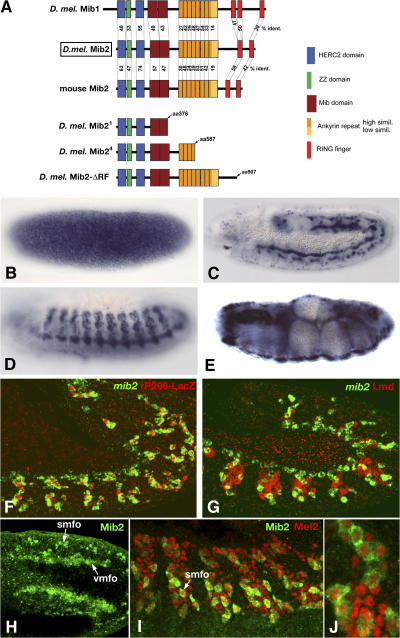
Drosophila mind bomb2 (mib2) encodes a 1,050-amino acid protein with several notable features (Fig. 1 A). A ZZ zinc finger domain within the N-terminal portion is flanked by two regions that share homology with HERC2, a protein that may function in protein trafficking and degradation pathways (Lehman et al., 1998; Ji et al., 1999). The ZZ domain, characterized by Cys-X2-Cys and Asp-Tyr-Asp-Leu motifs, is found in a small number of proteins, including some transcriptional adaptor proteins and Dystrophin/Utrophin, and is implicated in protein–protein interactions (Ponting et al., 1996). Following the ZZ and HERC2-like domains is a repeated sequence that is specific to Mib proteins. Eight Ankyrin repeats are in the middle portion of the protein and two RING fingers at the C-terminal end. Ankyrin repeats are present in a vast number of proteins and their role in protein–protein interactions is well documented (Sedgwick and Smerdon, 1999), while RING fingers proteins are known to participate in protein–protein interactions in the ubiquitination pathway (Joazeiro and Weissman, 2000). The presence of these various domains suggests that Mib2 functions as an adaptor-type of protein and/or as a component of a ubiquitination pathway.
Figure 1.
Protein structure of Mib2 and mib2 mRNA expression in embryos. (A) Top: comparison of homology domains between the D. melanogaster Mib2 protein, its paralogue Mib1, and the murine orthologue Mib2. (A) Bottom: truncated gene products expressed from mutant alleles mib21 and mib24. Mib2-ΔRF is a truncated protein minus both RING fingers. (B) Cleavage-stage embryo with evenly distributed maternal mib2 mRNA. (C) Stage 11 embryo with mib2 expression in founder cells of somatic and visceral muscles. (D) Stage 13 embryo with mib2 expression in somatic muscle precursors. (E) Stage 16 embryo with mib2 expression in somatic, visceral, and pharyngeal muscles. (F) Early stage 12 rP298-LacZ embryo, showing colocalization of mib2 mRNA (green) with founder cell-specific LacZ (red). (G) Early stage 12 wild-type embryo, showing mutually exclusive signals for mib2 mRNA (green) in founder myoblasts and Lame duck (Lmd) protein (red) in fusion-competent myoblasts (occasional yellow signals are due to merged signals from different Z positions). (H) Dorsal view of early stage 12 embryo, showing Mib2 protein in founder cells of visceral muscles (vmfo) and somatic muscles (smfo). (I) Lateral view of late stage 12 embryo (J, high magnification), showing Mib2 protein (green) in founder cells of somatic muscles (smfo). Staining is cytoplasmic and nuclei of cells (stained for Mef2, red) appear spared (J).
The Mib2 protein is conserved during evolution. Drosophila Mib2 and its murine orthologue display a similar structural organization and considerable degree of amino acid conservation within all the aforementioned domains (Fig. 1 A; and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200708135/DC1). When compared with Mib2 proteins across species, Drosophila Mib1, an E3 ubiquitin ligase that has been shown to be important in Notch signaling (Itoh et al., 2003; Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005), shows a lower level of homology in most of these domains, indicating that Mib2 is a paralogue of Mib1. In addition, the Mib2 proteins have only two RING finger domains while the Mib1 proteins have three.
mib2 is highly expressed in visceral and somatic mesodermal cells
Maternally derived mib2 transcripts are detected prominently in the fertilized egg (Fig. 1 B). Zygotic expression is first observed at low levels panmesodermally, and beginning at stage 11, high levels of expression appear in progenitors of somatic and visceral muscles (Fig. 1 C) and persist in the differentiated muscles of late stage embryos (Fig. 1, D and E; and unpublished data). mib2 is not detectable in cardiomyocytes. Co-localization of mib2 RNA (cytoplasmic) and LacZ protein (nuclear) in embryos derived from the rP298 enhancer trap line (Nose et al., 1998), which carries a P-LacZ insertion within the dumbfounded (duf) gene that is active in all founders (Ruiz-Gomez et al., 2000), confirmed that mib2 expression is specific for founder myoblasts (Fig. 1 F). Accordingly, mib2 is not detected in Lame duck (Lmd)–positive fusion-competent cells (Duan et al., 2001; Fig. 1 G). Mib2 protein expression is identical to that of mib2 mRNA and appears to be in the cytoplasm of founder cells (Fig. 1, H–J). In contrast, mib1 expression is not detectable in mesodermal cells (unpublished data).
Identification of mib2 mutant alleles
Genetic and molecular analysis in the vicinity of the 37B10 locus identified the lethal complementation group l(2)37Be as a likely candidate for mib2 (CG17492; see Materials and methods). We obtained the two extant alleles, l(2)37Be1 and l(2)37Be4, for further analysis. Sequence analysis of the protein-coding exons showed that the mib2 gene on the l(2)37Be1 mutant chromosome contains a nucleotide change (C to T) that converts Gln377 to a nonsense codon (Fig. 1 A and Fig. S1). On the l(2)37Be4 chromosome, a two-base pair deletion converts Asn587 to a Thr, which is then followed by a nonsense codon. As shown below, expression of wild-type mib2 in l(2)37Be1 mutant embryos can rescue the observed muscle phenotype. We conclude that the l(2)37Be1 and l(2)37Be4 alleles correspond to bona fide mib2 mutations and henceforth designate these alleles as mib21 and mib24, respectively. Based upon our analysis, the mutant Mib21 protein lacks all Ankyrin repeats and RING fingers while the mutant Mib24 protein lacks the RING fingers but retains four out of the predicted eight Ankyrin repeats.
mib2 mutant embryos exhibit detached muscles during later stages of embryogenesis
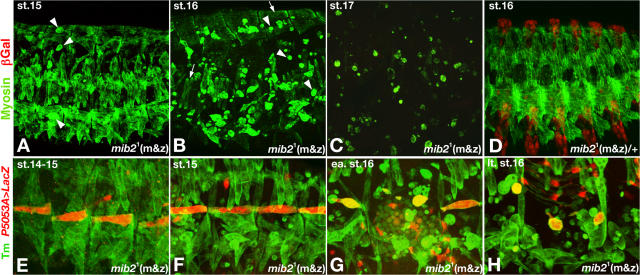
To assess the consequence of loss of mib2 function on muscle development, we stained wild-type and mutant embryos with an antibody against Myosin to visualize the muscle pattern. We focused more on the mib21 allele because the molecular nature of this mutation suggests that it is a stronger mutant allele. As compared with wild-type embryos, stage 15 mutant embryos (derived from mib21 germline clones and zygotically mib21/Df(2L)TW130, termed “mib21 m&z”), which lack both maternal and zygotic mib2 activity, have a well-developed somatic musculature, although a very limited number of detached muscles can already be detected (compare Fig. 2 A with Fig. 2 D). At stage 16, the mutant embryos exhibit a highly deranged muscle pattern that is characterized by a massive number of detached muscles (Fig. 2 B). Many of the rounded muscles have become smaller, followed by rapid muscle degeneration. Consequently, in stage 17 mutant embryos, normal somatic muscles are absent and the size of the rounded muscles decreases dramatically (Fig. 2 C). We observed the same types of muscle deterioration with mib24 mutant embryos that are null for both maternal and zygotic mib2 activity (unpublished data). Unlike the somatic muscles, the midgut muscles do not disintegrate in mib2 mutant embryos; however, the incompletely constricted midgut of these embryos suggests that mib2 also plays a role in visceral muscles (Fig. S3, A and B; available at http://www.jcb.org/cgi/content/full/jcb.200708135/DC1). Cardiac morphology is not affected, as predicted from the absence of mib2 expression in myocardial cells (see Fig. 5 F). Embryos that lack only zygotic mib2 function and homozygous deficiency embryos show similar somatic muscle and gut defects as those derived from germ line clones, although the defects are delayed and less severe (see Fig. 4; and unpublished data). Previous CG17492/mib2 knockdown by RNAi injections also caused some muscle detachments (Estrada et al., 2006).
Figure 2.
mib2 is required for maintaining the attachment and integrity of somatic muscles. Muscle phenotypes were analyzed in embryos that were derived from homozygous mib21 mutant germ line clones and have the zygotic genotype of mib21/Df(2L)TW130 (except for embryo in D, which is zygotically mib21/SM6 wg-lacZ). Embryos in E–H also carry P5053A-Gal4 and UAS-lacZ on the Df(2L)TW130 chromosome to highlight muscle 12 (VL1). Color codes for probes are indicated on the left. (A) At stage 15, a few somatic muscles lacking functional Mib2 are detached and rounded up (arrowheads), wheras the majority of muscles are still normal. (B) At stage 16, most somatic muscles (except for some LT and DO muscles, arrows) are rounded up and decreased in size (arrowheads). (C) At stage 17, only small remnants of rounded somatic muscle syncytia remain. (D) Normal somatic muscle pattern in germ line–derived embryo, which is zygotically heterozygous for mib2, shows that maternal products are not required in the presence of functional zygotic mib2. (E–H) Normal attachment and morphogenesis of muscle 12 (E and F; orange-yellow signals) occurs before it detaches and shrinks after stage 16 (G–H). Occasional red signals come from longitudinal gut muscles.
Figure 5.
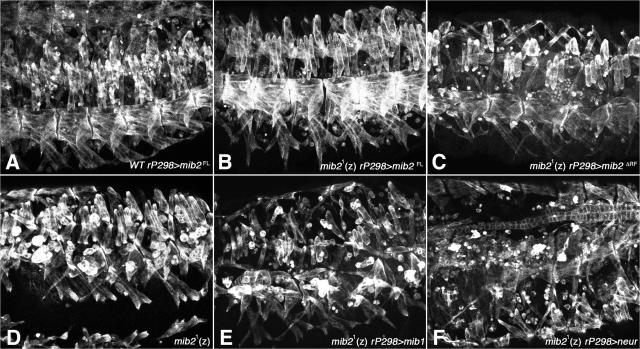
RING finger–deleted Mib2, but not Mib1 and Neuralized, can substitute for Mib2. (A) Stage 16 wild-type embryo, overexpressing full-length Mib2 (Mib2FL) via rP298-Gal4, exhibits a normal somatic muscle pattern. (B) Stage 16 mib21 (zygotic) homozygous mutant embryo with forced Mib2FL expression has normal somatic muscles. (C) Stage 16 mib21 (zygotic) homozygous mutant embryo with forced expression of RING finger–deleted Mib2 (Mib2ΔRF) via rP298-Gal4 shows significant rescue of the mib2 muscle phenotype. (D) Stage 16 (zygotic) homozygous mib21 embryo, shown as control for the rescue experiments, exhibits detached/disintegrating muscles. (E and F) Stage 16 mib21 (zygotic) homozygous mutant embryo with forced expression of Mib1 or Neur via rP298-Gal4. Muscle phenotypes are comparable to mib21 alone.
Figure 4.
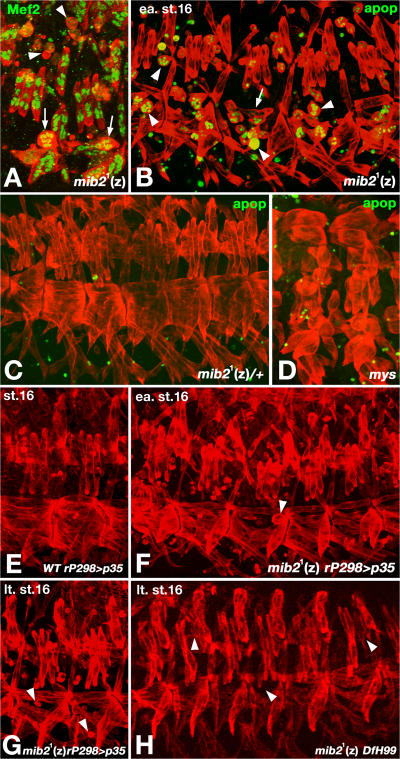
mib2 activity is required for preventing apoptosis of somatic muscles. In all panels, muscles were labeled with anti-Tropomyosin (red). (A) Zygotically (z) mib21 homozygous mutant embryo at stage 16 with disappearance of Mef2-labeled nuclei in shrunken syncytia (arrowheads). Arrows indicate Mef2-positive muscles at an earlier stage of detachment. (B) Embryo as in A but labeled with TUNEL (green), showing strong apoptotic signals in the detached and shrinking muscle syncytia (arrowheads). Arrow indicates detaching muscle with weak apoptotic signals. (C) Stage 16 mib21 heterozygous control embryo without any apoptotic signals in somatic muscles. (D) Stage 16 myospheroid (mys) mutant embryo with an absence of apoptotic signals in detached muscles which are not shrunken. (E) Stage 16 control embryos with p35 overexpression via rP298-Gal4 exhibit normal somatic muscle morphology. (F) Early stage 16 mib21 (z) embryo with forced expression of p35 via rP298-Gal4, showing only a few detached muscles (arrowheads). (G) Late stage 16 embryo as in (F) with only a few detached or missing muscles (arrowheads). (H) Late stage 16 embryo homozygous for mib21 and Df(3R)H99. Most somatic muscles appear normal although a few have aberrant shapes or are missing (arrowheads).
To analyze the temporal progression and cause of the observed muscle phenotype, we recombined P5053A-Gal4, which is a common marker for muscle 12 (or VL1) development (Ritzenthaler et al., 2000; Swan et al., 2004), and UAS-LacZ onto the mib21 chromosome. Wild-type and mutant embryos were double-labeled for Tropomyosin and LacZ expression. At late stage 14, the somatic musculature of mib21 mutant (m&z) embryos, including muscle 12, which is in the final stages of establishing its normal attachments, looks normal (Fig. 2 E). A low degree of muscle detachment becomes detectable at stage 15, although muscle 12 does not seem to be affected immediately, suggesting some differences in susceptibility to loss of mib2 function among the various muscles (Fig. 2 F). However, massive muscle detachments and degeneration, which include muscle 12, occur by stage 16 (Fig. 2, G and H). In the aggregate, this analysis indicates that mib2 function is not needed for the formation of somatic muscles and their initial attachment to tendon cells, but rather it is required at late embryogenesis for maintaining the attachments and the integrity of the mature musculature.
Loss of mib2 function does not interfere with the localization of known muscle attachment components
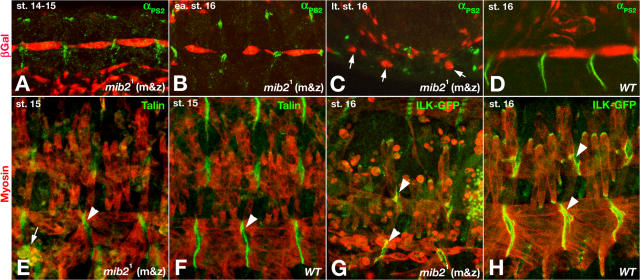
Because of the muscle detachment phenotype, we examined whether loss of mib2 function disrupts the localization of integrin signaling components that are known to establish stable muscle/tendon attachments (for review see Bokel and Brown, 2002). In mib21 (m&z) embryos, αPS2 integrin localizes normally to the attachment sites within the tips of muscle 12 before and during the early stages of their detachment (compare Fig. 3, A and B with Fig. 3 D). Hence, the gradual disappearance of localized αPS2 integrin during later stages (Fig. 3 C) is presumably a consequence of the muscle detachments and deterioration rather than a cause of the detachment. Likewise, all other integrin pathway components examined, including Talin (Fig. 3, E and F), Pinch (not depicted), ILK (Fig. 3, G and H), and Tyr397-phosphorylated FAK (Fig. S3, C and D) are initially localized normally within the muscle ends near the attachment sites in the absence of mib2 activity. Several components on the epidermal side of the attachments were also unaffected (unpublished data). These observations argue against a function of mib2 in establishing stable muscle attachments via integrin signaling components unless there is a yet unknown parallel pathway to ILK that is affected.
Figure 3.
mib2 is not required to localize integrin signaling components to muscle attachments. Except when denoted “WT” (wild type), embryos shown are derived from homozygous mib21 mutant germ line clones and have the zygotic genotype of mib21/Df(2L)TW130. Embryos in A–D also carry P5053A-Gal4 and UAS-lacZ on the Df(2L)TW130 chromosome to highlight muscle 12 (VL1). (A) At stage 14–15, normal localization of αPS2 integrin to the ends of muscle 12 near the attachment sites in the absence of mib2 activity. (B) At early stage 16, mostly normal localization of αPS2 integrin during the early phase of muscle 12 detachment. (C) During late stage 16, no localization of αPS2 integrin in muscle 12 syncytia (arrows) that begin to deteriorate. Small red cells in A–D correspond to longitudinal gut muscles. (D) Stage 16 control embryo, showing αPS2 integrin localized to the attachment sites within muscle 12. (E and F) Stage 15 embryos without and with mib2 activity, showing normal localization of Talin to the attachment sites within the myosin-stained muscles (arrowheads). In E, a few ventral muscles that have detached show evenly distributed Talin (arrows). (G and H) Stage 16 embryos without and with mib2 activity, showing normal localization of ILK-GFP to the attachment sites (arrowheads) except where muscles have detached and were shrinking.
Absence of mib2 function triggers apoptosis in muscles
As shown in Fig. 4 A, Mef2-positive nuclei are still present in the large rounded muscles of zygotic (z) mib21 mutant embryos at stage 16, whereas they are absent in the rounded muscles that are severely decreased in size. Because cell shrinkage and chromatin deterioration are hallmarks of apoptosis, we used TUNEL staining to detect apoptotic cells in mib2 mutant embryos. Indeed, the detaching muscles in mib21 (z) embryos are positive for the apoptotic marker (Fig. 4 B), whereas heterozygous control embryos only show apoptotic signals in the CNS and other nonmuscle tissues (Fig. 4 C). Of note, the detached muscles in myospheroid (mys) mutant embryos, which lack functional β-integrin at their attachment sites, do not show any significant apoptotic signals and do not shrink, indicating that apoptosis is not an automatic consequence of muscle detachment (Fig. 4 D). Hence, we propose that the muscle detachment in mib2 mutants is a consequence of apoptotic events in these muscles.
To test this proposal further, we blocked apoptosis through forced expression of the caspase inhibitor p35 in muscle founders and their derived muscles. Blocking apoptosis in muscles of mib21 (z) mutant embryos leads to a significant reduction of muscle detachment and deterioration at early stage 16 (compare Fig. 4 F with Fig. 4 B; no effects are seen with analogous expression of p35 in a wild-type background [Fig. 4 E]). At late stage 16, some muscle degeneration still occurs in the p35-overexpressing mutant embryos, as evidenced by the slightly larger number of rounded muscles with decreased sizes and missing muscle fibers, although it is much less severe than in mib21 (z) mutants without blocked apoptosis (Fig. 4 G). A large number of muscle fibers are still present at stage 17 in these apoptosis-blocked mib21 (z) mutant embryos (unpublished data). Notably, the muscle degeneration phenotype is rescued in mib21 (z) mutant embryos that are also homozygous for Df3R)H99, which deletes the apoptosis inducers reaper, hid, and grim, (Fig. 4 H). In this background the majority of the muscles appear normal until at least late stage 16, although we do not know whether they change their morphology after cuticle formation. Together, these observations suggest that muscle detachment and degeneration in mib2 mutants are largely a consequence of triggered apoptosis.
Ubiquitin ligases in the Notch pathway cannot substitute for Mib2 during muscle development
Because RING fingers are implicated in protein ubiquitination, we sought to test whether the RING fingers of Mib2 are required for its activity and whether Mib1 and Neuralized (Neur), E3 ubiquitin ligases that have been shown to be modulators of the Notch pathway by ubiquitinating Delta (Lai and Rubin, 2001; Pavlopoulos et al., 2001; Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005), could substitute for Mib2. Overexpression of full-length Mib2 in muscle founders and the derived muscles of wild-type embryos does not affect the pattern and stability of the muscles, although there is an increased number of unfused myoblasts at late stages (Fig. 5 A). In the mib21 mutant background, forced expression of full-length Mib2 leads to essentially complete rescue of the muscle detachment and deterioration phenotype (compare Fig. 5 B with Fig. 5 D), although an excessive number of unfused myoblasts is also evident. Notably, forced expression of a Mib2 version lacking both RING fingers (Mib2ΔRF; see Fig. 1 A) in the mutant background also allowed a significant, albeit incomplete, rescue of the muscle defects. In these embryos there is only occasional detachment of muscles and very few signs of apoptotic decay, although the muscles sometimes appear shorter and thicker as compared with normal muscles (compare Fig. 5 C with Fig. 5, A and D). Analogous overexpression of this mutated Mib2 version in a wild-type background does not have any effects on muscles (unpublished data). Based upon the significant degree of rescue with MibΔRF, we conclude that the RING fingers have a less prominent role in promoting muscle integrity as compared with the other domains, and that ubiquitination may not be the main activity of Mib2 that is required for muscle development.
In sharp contrast to full-length Mib2 and Mib2ΔRF, Mib1 and Neur are not able to confer any rescuing activity under similar experimental conditions (compare Fig. 5, E and F with Fig. 5 D), suggesting that Mib2 possesses important targets that are different from those of Mib1 and Neur, and that blocked Notch signaling is not the cause of the observed muscle defects in mib2 mutant embryos. This latter point is underscored by our results from experiments with the Nts allele, which never yield any embryos with muscle phenotypes that are similar to those of mib2 mutants (unpublished data; see also Fuerstenberg and Giniger, 1998). However, we do not exclude the possibility that Mib2 can act in the Notch pathway in other contexts, such as in post-embryonic tissues, which we have not yet examined. It has been shown in cell culture that vertebrate Mib2 is capable of ubiquitinating Delta and Jagged, and Mib2 was also identified as a binding partner of Drosophila Delta in yeast two-hybrid screenings (Formstecher et al., 2005; Koo et al., 2005; Takeuchi et al., 2005; Zhang et al., 2006).
In summary, Mib2 appears to have a unique and Notch- independent role in “protecting” differentiated body wall muscles from entering apoptosis, undergoing detachment, and being subject to degradation. We speculate that Mib2 is required in a yet undefined pathway for the establishment of specific functional features of the sarcomeres or other structures of the myofibers. In this context, it is interesting to note that mouse Mib2 (also known as skeletrophin) was identified as a binding partner of α-actin and is expressed in skeletal muscles (Takeuchi et al., 2003). The disruption of these unknown structural and functional features in the absence of mib2 activity could become detrimental upon stimulation of contractility and trigger entry into apoptosis. Apoptotic degradation of multiple components of the muscle fibers could first result in detachment because the contractile force renders muscle attachment more sensitive to disruptions, and leads to the degradation of the entire syncytia. The identity of the functional target(s) of Mib2 in muscles is currently unknown, as the expression of all markers examined to date, including founder cell markers (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200708135/DC1), muscle attachment proteins (Fig. 3), and differentiation markers (Fig. 3; Fig. 4; and unpublished data), is unaffected in mib2 mutant embryos. Future studies, including the identification of interaction partners or mutations in other genes with similar phenotypes, will help to elucidate the pathway in which Mib2 acts to protect muscle integrity. Whether this pathway is involved in preventing skeletal muscle atrophies in which caspase-3 activation contributes to the breakdown of actomyosin complexes of myofibrils (Du et al., 2004) could also be explored.
Materials and methods
Drosophila stocks
mys1 (Digan et al., 1986), UAS-p35 (made by Bruce Hay, CalTech, Pasadena, CA; Zhou et al., 1997), P5053A-Gal4 (Lopez, 1998), P{neoFRT}40A (Xu and Rubin, 1993), UAS-lacZ (Bg4-1-2; Brand and Perrimon, 1993), CyO wgen11-LacZ (Kassis et al., 1992), SM6 eve-LacZ8.0 (Panzer et al., 1992), TM2 P{lArB}C40.1S3 (Bellen et al., 1989), Df(3R)TW130 (Wright et al., 1976), and the alleles l(2)37Be1 and l(2)37Be4 (Stathakis et al., 1995), which were induced by EMS and EMS plus γ-rays, respectively, were obtained from the Bloomington Drosophila stock center. Other fly stocks used include: mib1EY9780 (Pitsouli and Delidakis, 2005), UAS-neur (Pavlopoulos et al., 2001), UAS-mib1-3 (Wang and Struhl, 2005), rP298-Gal4 (Menon and Chia, 2001), and ILK-GFP (Zervas et al., 2001).
To generate UAS-mib2 and UAS-mib2ΔRF transgenic lines, we subcloned the regions coding for amino acids 1–1,050 and amino acids 1–907, respectively, from the EST clone LP14687 (obtained from the Berkeley Drosophila Genome Project/BDGP) into the pUAST vector and injected the resulting constructs into Drosophila embryos by using standard protocols. Multiple independent insertions were obtained and analyzed for each construct.
Identification of mib2 mutants
The mib2 gene maps genetically at 37B10, a genomic region that was characterized genetically and molecularly in the context of the Dopa decarboxylase gene (Stathakis et al., 1995). By comparing the data of Stathakis et al. (1995) with those from the BDGP, we determined that mib2 is uncovered by the overlapping deficiencies Df(2L)OD15, Df(2L)hkUC1, and Df(2L)TW130 (Wright et al., 1976). Based upon additional molecular and genetic mapping data, we identified a complementation group, l(2)37Be, of originally five embryonic lethal alleles as the most likely candidate for the mib2 gene (Stathakis et al., 1995; Adams et al., 2000). The supporting evidence includes: (1) genomic rescue experiments done by Stathakis et al. (1995) with a construct, which according to our analysis only contains mib2 and a neighboring gene called catsup (l(2)37Bc), rescued the lethality of l(2)37Be and catsup mutations; and (2) one allele, l(2)37B5, which no longer exists, was shown to be associated with an ∼800-bp deletion of sequences that we now have identified as being part of the Drosophila mib2 gene.
Sequencing of mib2 from wild-type and mutant alleles
EST clone LP14687 was fully sequenced and the derived mib2 ORF was identical to that of CG17492. For allele sequencing, the alleles were balanced with a “blue balancer”. Fixed embryos were stained with an antibody against β-galactosidase, and homozygous l(2)37Be1 or l(2)37Be4 mutant embryos were identified by the absence of LacZ expression from the “blue balancer”. Hand-picked embryos of the appropriate genotype were incubated in a solution of 10 mM Tris-HCl, 1 mM EDTA, 25 mM NaCl, and 200 μg/ml proteinase K. Amplified products were purified and subjected directly to automated sequencing. Specific primers were used for sequencing all exons and exon–intron boundaries. For confirmation, the fragment that contained a sequence aberration was reamplified from genomic DNA and resequenced. The mouse Mib2 sequence data are based on our sequencing of the cDNA clone IMAGE:6516763.
Rescue (and overexpression) experiments
For rescue and overexpression experiments the following stocks were generated and used: UAS-mib2Full Length(FL)-1.UAS-mib2ΔRING(ΔRF)-2. rP298-Gal4; mib21/SM6, eveLacZ8.0. mib21/SM6, eveLacZ8.0;UAS-mib2-1. mib21/SM6, eve-LacZ8.0;UAS-mibΔRF-2. mib21/ CyO, wgen11-LacZ;UAS-mib1-3. mib21/SM6, eve-LacZ8.0;UAS-p35. mib21/SM6, eve-lacZ8.0; Df3R)H99/TM2, P{lArB}C40.1S3. mib21/SM6, eve-LacZ8.0;UAS-neur12.4.
Generation of Drosophila Mib2 antibodies
pET30-Mib2(COOH) was generated by cloning the PCR fragment that code for amino acids 650–1,038 of Mib2 in frame with the 6xHis tag of the pET-30a vector (Novagen). The fusion protein was expressed in Escherichia coli and purified with Ni2+-affinity chromatography under denaturing conditions (QIAGEN). Antiserum production in guinea pigs was done by Covance Research Products and affinity purification was performed against bacterially expressed S-tag (Novagen)-Mib2 fusion protein.
In situ RNA hybridization and immunocytochemistry of whole-mount embryos
The embryonic CG17492 mRNA expression was first described in the BDGP in situ hybridization database (Tomancak et al., 2002). We confirmed and extended these expression data with the use of a digoxygenin-labeled mib2 RNA probe that was generated by using the LP14687 cDNA clone and published protocols (Tautz and Pfeifle, 1989). Embryos were photographed with Nomarski DIC optics on a microscope (AX70; Olympus) with a 20× Uplan Fl/0.5 NA objective and a color camera (5.0 RTV; QImaging). Images were acquired with QImaging software and processed with Adobe Photoshop.
Immunocytochemistry was performed essentially as described (Reim et al., 2003) and the TSA amplification system was used as needed. Cy3 and FITC were used as fluorochromes. Embryo stainings were analyzed on a confocal microscope (TCS-SP 4D; Leica) with a HC Plan Apo20×/0.7 NA and a HCX Plan Apo40×/0.75–1.25 NA oil objective at 20°C. Generally, Z-scans were taken at 1- to 1.5-μm steps and four to eight Z-scans were merged via maximum projection using the Leica TCS software package or Adobe Photoshop CS. Except for the final adjustment of contrast and brightness with Adobe Photoshop CS, no other processing of the imaging was performed.
Antibodies were used as follows: mouse anti-βgalactosidase (1:100; developed by J. Sanes, Washington University, St. Louis. MO, and obtained through DSHB, Iowa University, Iowa City, IA), rabbit anti-β-galactosidase (1:1500; ICN), rat anti-Tropomyosin (1:50; Babraham Tech), mouse anti-Myosin (1:200; Kiehart and Feghali, 1986); rabbit anti-Mef2 (1:700; Bour et al., 1995); rat anti-αPS2 (1:10, TSA; Wilcox et al., 1981); rabbit and mouse anti-Talin (1:750, TSA, and 1:20, TSA, respectively; Brown et al., 2002); rabbit anti-Pinch (1:15,000, TSA; Clark et al., 2003); rabbit anti-FAK[pY397] (1:300; Biosource International); and rabbit anti-GFP (1:10,000, TSA; Molecular Probes). Biotinylated (1:200, Vector Laboratories) and fluorescent (1:100, Jackson ImmunoResearch Laboratories) secondary antibodies were also used. The Apoptag kit (Intergen) was used for detecting apoptotic cells as described in Reim et al. (2003).
Online supplemental material
Figure S1 shows protein sequence alignment of D. mel. Mib2 with D. mel Mib1 and mouse Mib2. Figure S2 shows absence of any effects of mib2 mutation on muscle founder marker expression. Figure S3 shows gut phenotype and phospho-FAK staining in mib2 mutant embryos. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708135/DC1.
Supplementary Material
Acknowledgments
We thank Dongyun Wu for technical assistance and Hong Duan for technical advice. We also thank M. Beckerle, N. Brown, C. Delidakis, D. Kiehart, S. Menon, G. Struhl, the Bloomington Drosophila Stock Center, Developmental Studies Hybridoma Bank at the University of Iowa (DSHB, under the auspices of NICHD), and Berkeley Drosophila Genome Project for sharing various antibodies, fly stocks, and cDNA clones. Confocal laser scanning microscopy at the MSSM-Microscopy Shared Resource Facility was supported by NIH-NCI (R24 CA095823) and National Science Foundation (DBI-9724504).
This research was supported by grants from the Muscular Dystrophy Association (to H.T.N) and from the NIH (AR4628 to H.T.N. and DK59406 to M. Frasch).
N. Ezzeddine's present address is Department of Biochemistry and Molecular Biology, The University of Texas Houston Medical School, Houston, TX 77030.
Abbreviations used in this paper: mib2, Drosophila mind bomb2; mys, myospheroid; Neur, Neuralized.
References
- Adams, M.D., S.E. Celniker, R.A. Holt, C.A. Evans, J.D. Gocayne, P.G. Amanatides, S.E. Scherer, P.W. Li, R.A. Hoskins, R.F. Galle, et al. 2000. The genome sequence of Drosophila melanogaster. Science. 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Beckett, K., and M.K. Baylies. 2006. The development of the Drosophila larval body wall muscles. Int. Rev. Neurobiol. 75:55–70. [DOI] [PubMed] [Google Scholar]
- Bellen, H.J., C.J. O'Kane, C. Wilson, U. Grossniklaus, R.K. Pearson, and W.J. Gehring. 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3:1288–1300. [DOI] [PubMed] [Google Scholar]
- Bokel, C., and N.H. Brown. 2002. Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev. Cell. 3:311–321. [DOI] [PubMed] [Google Scholar]
- Bour, B., M. O'Brien, W. Lockwood, E. Goldstein, R. Bodmer, P. Taghert, S. Abmayr, and H. Nguyen. 1995. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9:730–741. [DOI] [PubMed] [Google Scholar]
- Brand, A.H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Brown, N.H., S.L. Gregory, W.L. Rickoll, L.I. Fessler, M. Prout, R.A. White, and J.W. Fristrom. 2002. Talin is essential for integrin function in Drosophila. Dev. Cell. 3:569–579. [DOI] [PubMed] [Google Scholar]
- Clark, K.A., M. McGrail, and M.C. Beckerle. 2003. Analysis of PINCH function in Drosophila demonstrates its requirement in integrin-dependent cellular processes. Development. 130:2611–2621. [DOI] [PubMed] [Google Scholar]
- Digan, M.E., S.R. Haynes, B.A. Mozer, I.B. Dawid, F. Forquignon, and M. Gans. 1986. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev. Biol. 114:161–169. [DOI] [PubMed] [Google Scholar]
- Du, J., X. Wang, C. Miereles, J.L. Bailey, R. Debigare, B. Zheng, S.R. Price, and W.E. Mitch. 2004. Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J. Clin. Invest. 113:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, H., J.B. Skeath, and H.T. Nguyen. 2001. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development. 128:4489–4500. [DOI] [PubMed] [Google Scholar]
- Estrada, B., S.E. Choe, S.S. Gisselbrecht, S. Michaud, L. Raj, B.W. Busser, M.S. Halfon, G.M. Church, and A.M. Michelson. 2006. An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher, E., S. Aresta, V. Collura, A. Hamburger, A. Meil, A. Trehin, C. Reverdy, V. Betin, S. Maire, C. Brun, et al. 2005. Protein interaction mapping: a Drosophila case study. Genome Res. 15:376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerstenberg, S., and E. Giniger. 1998. Multiple roles for notch in Drosophila myogenesis. Dev. Biol. 201:66–77. [DOI] [PubMed] [Google Scholar]
- Itoh, M., C.H. Kim, G. Palardy, T. Oda, Y.J. Jiang, D. Maust, S.Y. Yeo, K. Lorick, G.J. Wright, L. Ariza-McNaughton, et al. 2003. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell. 4:67–82. [DOI] [PubMed] [Google Scholar]
- Ji, Y., M.J. Walkowicz, K. Buiting, D.K. Johnson, R.E. Tarvin, E.M. Rinchik, B. Horsthemke, L. Stubbs, and R.D. Nicholls. 1999. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum. Mol. Genet. 8:533–542. [DOI] [PubMed] [Google Scholar]
- Joazeiro, C.A., and A.M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 102:549–552. [DOI] [PubMed] [Google Scholar]
- Kassis, J.A., E. Noll, E.P. VanSickle, W.F. Odenwald, and N. Perrimon. 1992. Altering the insertional specificity of a Drosophila transposable element. Proc. Natl. Acad. Sci. USA. 89:1919–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart, D.P., and R. Feghali. 1986. Cytoplasmic myosin from Drosophila melanogaster. J. Cell Biol. 103:1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, B.K., H.S. Lim, R. Song, M.J. Yoon, K.J. Yoon, J.S. Moon, Y.W. Kim, M.C. Kwon, K.W. Yoo, M.P. Kong, et al. 2005. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 132:3459–3470. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., and G.M. Rubin. 2001. neuralized functions cell-autonomously to regulate a subset of notch-dependent processes during adult Drosophila development. Dev. Biol. 231:217–233. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., F. Roegiers, X. Qin, Y.N. Jan, and G.M. Rubin. 2005. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 132:2319–2332. [DOI] [PubMed] [Google Scholar]
- Le Borgne, R., S. Remaud, S. Hamel, and F. Schweisguth. 2005. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of Delta and Serrate signaling in Drosophila. PLoS Biol. 3:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, A.L., Y. Nakatsu, A. Ching, R.T. Bronson, R.J. Oakey, N. Keiper-Hrynko, J.N. Finger, D. Durham-Pierre, D.B. Horton, J.M. Newton, et al. 1998. A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc. Natl. Acad. Sci. USA. 95:9436–9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, J. 1998. Embryonic Expression Patterns of GAL4 Enhancer Trap Lines. Personal communication to FlyBase. FBrf0105492:1998.11.24.
- Menon, S.D., and W. Chia. 2001. Drosophila Rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev. Cell. 1:691–703. [DOI] [PubMed] [Google Scholar]
- Nose, A., T. Isshiki, and M. Takeichi. 1998. Regional specification of muscle progenitors in Drosophila: the role of the msh homeobox gene. Development. 125:215–223. [DOI] [PubMed] [Google Scholar]
- Panzer, S., A. Fong, and S.K. Beckendorf. 1992. Genetic Notes: New lacZ-marked balancer. Drosoph. Inf. Serv. 72:172. [Google Scholar]
- Pavlopoulos, E., C. Pitsouli, K.M. Klueg, M.A. Muskavitch, N.K. Moschonas, and C. Delidakis. 2001. neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell. 1:807–816. [DOI] [PubMed] [Google Scholar]
- Pitsouli, C., and C. Delidakis. 2005. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 132:4041–4050. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P., D.J. Blake, K.E. Davies, J. Kendrick-Jones, and S.J. Winder. 1996. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 21:11–13. [PubMed] [Google Scholar]
- Reim, I., H.H. Lee, and M. Frasch. 2003. The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development. 130:3187–3204. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, S., E. Suzuki, and A. Chiba. 2000. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat. Neurosci. 3:1012–1017. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., N. Coutts, A. Price, M. Taylor, and M. Bate. 2000. Drosophila Dumbfounded: a myoblast attractant essential for fusion. Cell. 102:189–198. [DOI] [PubMed] [Google Scholar]
- Schnorrer, F., and B.J. Dickson. 2004. Muscle building; mechanisms of myotube guidance and attachment site selection. Dev. Cell. 7:9–20. [DOI] [PubMed] [Google Scholar]
- Sedgwick, S.G., and S.J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311–316. [DOI] [PubMed] [Google Scholar]
- Stathakis, D.G., E.S. Pentz, M.E. Freeman, J. Kullman, G.R. Hankins, N.J. Pearlson, and T.R. Wright. 1995. The genetic and molecular organization of the Dopa decarboxylase gene cluster of Drosophila melanogaster. Genetics. 141:629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan, L.E., C. Wichmann, U. Prange, A. Schmid, M. Schmidt, T. Schwarz, E. Ponimaskin, F. Madeo, G. Vorbruggen, and S.J. Sigrist. 2004. A glutamate receptor-interacting protein homolog organizes muscle guidance in Drosophila. Genes Dev. 18:223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, T., H.H. Heng, C.J. Ye, S.B. Liang, J. Iwata, H. Sonobe, and Y. Ohtsuki. 2003. Down-regulation of a novel actin-binding molecule, skeletrophin, in malignant melanoma. Am. J. Pathol. 163:1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, T., Y. Adachi, and Y. Ohtsuki. 2005. Skeletrophin, a novel ubiquitin ligase to the intracellular region of Jagged-2, is aberrantly expressed in multiple myeloma. Am. J. Pathol. 166:1817–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 98:81–85. [DOI] [PubMed] [Google Scholar]
- Tomancak, P., A. Beaton, R. Weiszmann, E. Kwan, S. Shu, S.E. Lewis, S. Richards, M. Ashburner, V. Hartenstein, S.E. Celniker, and G.M. Rubin. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3:research0088.1–0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk, T. 1999. Singling out Drosophila tendon cells: a dialogue between two distinct cell types. Trends Genet. 15:448–453. [DOI] [PubMed] [Google Scholar]
- Wang, W., and G. Struhl. 2005. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 132:2883–2894. [DOI] [PubMed] [Google Scholar]
- Wilcox, M., D.L. Brower, and R.J. Smith. 1981. A position-specific cell surface antigen in the Drosophila wing imaginal disc. Cell. 25:159–164. [DOI] [PubMed] [Google Scholar]
- Wright, T.R., R.B. Hodgetts, and A.F. Sherald. 1976. The genetics of dopa decarboxylase in Drosophila melanogaster. I. Isolation and characterization of deficiencies that delete the dopa-decarboxylase-dosage-sensitive region and the alpha-methyl-dopa-hypersensitive locus. Genetics. 84:267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., and G.M. Rubin. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237. [DOI] [PubMed] [Google Scholar]
- Zervas, C.G., S.L. Gregory, and N.H. Brown. 2001. Drosophila integrin-linked kinase is required at sites of integrin adhesion to link the cytoskeleton to the plasma membrane. J. Cell Biol. 152:1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Q. Li, and Y.J. Jiang. 2006. Zebrafish Mib and Mib2 are mutual E3 ubiquitin ligases with common and specific Delta substrates. J. Mol. Biol. 366:1115–1128. [DOI] [PubMed] [Google Scholar]
- Zhou, L., A. Schnitzler, J. Agapite, L.M. Schwartz, H. Steller, and J.R. Nambu. 1997. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. USA. 94:5131–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.