Abstract
Primary cilia (PC) function as microtubule-based sensory antennae projecting from the surface of many eukaryotic cells. They play important roles in mechano- and chemosensory perception and their dysfunction is implicated in developmental disorders and severe diseases. The basal body that functions in PC assembly is derived from the mature centriole, a component of the centrosome. Through a small interfering RNA screen we found several centrosomal proteins (Ceps) to be involved in PC formation. One newly identified protein, Cep164, was indispensable for PC formation and hence characterized in detail. By immunogold electron microscopy, Cep164 could be localized to the distal appendages of mature centrioles. In contrast to ninein and Cep170, two components of subdistal appendages, Cep164 persisted at centrioles throughout mitosis. Moreover, the localizations of Cep164 and ninein/Cep170 were mutually independent during interphase. These data implicate distal appendages in PC formation and identify Cep164 as an excellent marker for these structures.
Introduction
The primary cilium (PC) is a microtubule-based structure that protrudes from the surface of most vertebrate cells. It generally comprises a membrane-bound 9 + 0 ciliary axoneme, which consists of nine outer doublet microtubules but lacks both the central microtubule pair and dynein arms. Thus, with few exceptions, PC are nonmotile and instead function as sensory organelles (Pazour and Witman, 2003; Singla and Reiter, 2006; Satir and Christensen, 2007). They play important roles during development, particularly with regard to the establishment of left–right asymmetry, as well as later in life when they are required for the processing of mechanical or chemical signals in many organs (Ibañez-Tallon et al., 2003; Praetorius and Spring, 2005). For instance, in kidney epithelial cells, PC sense fluids flow within the lumen of the nephron, which is critical for normal epithelial development and function. Proteins localizing to the ciliary membrane, known as polycystins, play an important role in mediating this mechanosensory function, and mutations in the corresponding genes cause polycystic kidney disease (Boucher and Sandford, 2004). Similarly, retinal degeneration can be caused by dysfunction of the connecting cilium, a highly specialized PC connecting the inner and outer segments in vertebrate photoreceptors (Badano et al., 2006; Singla and Reiter, 2006). Moreover, recent studies implicate PC in various signal transduction pathways, including sonic hedgehog, platelet-derived growth factor receptor α, and Wnt signaling (Singla and Reiter, 2006; Satir and Christensen, 2007). Ciliary defects have also been causally linked to several pleiotropic disorders, including Bardet-Biedl syndrome (BBS), Alstrom syndrome (ALMS), oral-facial-digital syndrome type I, and nephronophthisis (Badano et al., 2006; Hildebrandt and Zhou, 2007; Zariwala et al., 2007).
The assembly of the PC requires a basal body, which in turn is derived from one of the two centrioles that constitute the centrosome. During ciliogenesis, this basal body is positioned close to the plasma membrane and ciliary microtubules elongate from its distal end. Ciliogenesis requires the assembly of multiple soluble and membranous protein complexes. In particular, the so-called intraflagellar transport (IFT) system is then responsible for moving cargo (IFT particles) to and from the tip of the growing axoneme. IFT, first described in the algae Chlamydomonas reinhardtii (Kozminski et al., 1993), is now known to be mediated by the association of IFT particles with kinesin II and dynein microtubule–based motors for antero- and retrograde movement, respectively (Rosenbaum and Witman, 2002; Scholey, 2003). The signaling networks that control PC function during cell cycle progression remain to be elucidated, but several studies concur to identify a key role for the von Hippel-Lindau tumor suppressor in PC formation (Lutz and Burk, 2006; Schermer et al., 2006; Thoma et al., 2007). Furthermore, Aurora A kinase has recently been implicated in PC resorption (Pugacheva et al., 2007).
In this study, we have sought to identify centrosomal proteins (Ceps) that are required for ciliogenesis. Taking advantage of the fact that PC formation can be induced in cultured cells by serum starvation (Tucker et al., 1979; Vorobjev and Chentsov, 1982), we depleted individual centrosomal proteins by siRNA and examined the consequences on subsequent PC formation. This siRNA screen identified several proteins that affected PC formation, albeit to different degrees. A very strong effect was observed upon depletion of Cep164, a protein that had not previously been studied. Our characterization of Cep164 leads to conclude that this protein is not only required for PC formation but also constitutes an excellent marker for distal appendages on mature centrioles or basal bodies.
Results
Identification of centrosomal proteins involved in PC formation
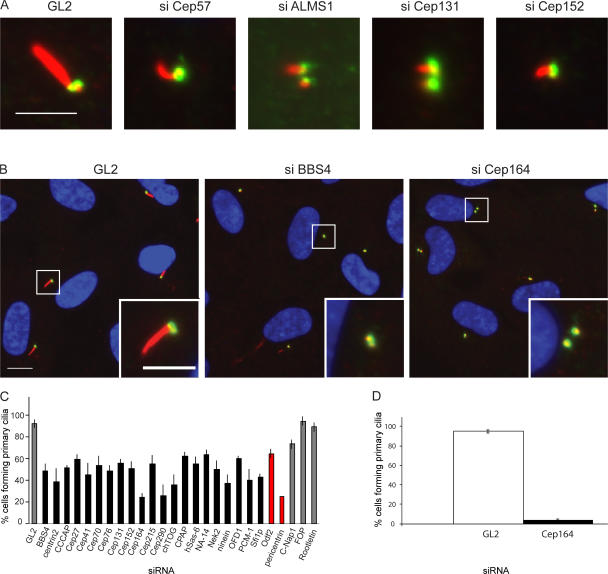
To search for proteins involved in PC formation, an siRNA screen focusing on centrosomal proteins (Andersen et al., 2003) was performed. After the depletion of individual proteins from retinal pigment epithelial (hTERT-RPE1) cells, PC formation was induced by serum starvation (Vorobjev and Chentsov, 1982) and monitored by staining with antibodies against acetylated tubulin (Piperno and Fuller, 1985). Depletion efficiency was assessed by quantitative real-time PCR (qRT-PCR) and, whenever possible, immunofluorescence (IF) microscopy and/or Western blot analysis (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200707181/DC1). Because depletion of outer dense fiber (Odf) 2 and pericentrin had previously been found to impair PC formation (Jurczyk et al., 2004; Ishikawa et al., 2005), these proteins were reexamined to provide points of reference for classifying phenotypes. Of 41 proteins analyzed, depletion of 25 proteins produced clear phenotypes, suggesting that these proteins might be involved in PC formation and/or maintenance (Fig. 1). Depletion of 23 proteins (including Odf2 and pericentrin) considerably reduced the proportion of cells that assembled PCs in response to serum starvation, from 90–95% in controls to 25–60% in depleted cells (Fig. 1, B and C; and not depicted). Furthermore, the depletion of Cep57 or ALMS1 led to the formation of morphologically abnormal, stunted cilia without considerably reducing the efficiency of ciliogenesis (Fig. 1 A), whereas the depletion of Cep131 and Cep152 resulted in both a clear reduction in ciliogenesis and morphological aberrations in those cilia that did form (Fig. 1, A and C). In contrast, no qualitative or quantitative effects on PC formation were observed upon siRNA-mediated depletion of 16 other proteins analyzed, including fibroblast growth factor receptor 1 oncogene partner, rootletin, and centrosomal Nek2-associated protein 1 (C-Nap1; Fig. 1 C, gray; and not depicted).
Figure 1.
siRNA screen for proteins implicated in PC formation. hTERT-RPE1 cells were transfected for 48 h with control (GL2) or protein-specific oligonucleotide duplexes, followed by serum starvation to induce PC formation. Cells were analyzed by IF microscopy using antibodies against acetylated tubulin (red) and C-Nap1 (green), used to mark cilia and centrioles, respectively. (A) Normal cilia in control cells (GL2) and stunted cilia after siRNA targeting Cep57, ALMS1, Cep131, or Cep152. (B) Normal cilia in control cells (GL2) and suppressed cilia formation after siRNA targeting BBS4 or Cep164. Insets show magnifications of the boxed areas. (C) Influence of various siRNA treatments on the percentage of cells showing PC formation. A reduction in PC formation was considered substantial if siRNA-mediated depletion of a particular centrosomal protein showed a comparable effect as depletion of either pericentrin or Odf2, two proteins previously implicated in PC formation (Jurczyk et al., 2004; Ishikawa et al., 2005). No effects were seen upon siRNA treatments targeting fibroblast growth factor receptor 1 oncogene partner (FOP), rootletin, or C-Nap1. (D) hTERT-RPE1 cells were transfected for 48 h with control (GL2) or Cep164-specific oligonucleotide duplexes. PC formation was monitored specifically in those cells that had been efficiently depleted of Cep164 as determined by staining with anti-Cep164 antibodies. This focus on efficiently depleted cells explains the higher impact of Cep164 depletion on PC formation in these experiments, as compared with the screen (C), in which all cells had been analyzed regardless of siRNA efficiency. At least 250 cells were counted for each bar in four experiments and error bars denote SD. Bars: (A) 5 μm; (B) 10 μm; (B, inset) 5 μm.
To exclude the possibility that depletion of centrosomal proteins might have influenced cell cycle profiles, FACS analyses were performed 48 or 72 h after siRNA treatments (unpublished data). These analyses provided no evidence for major deviations from controls, except for an increase in the proportion of G2/M cells upon depletion of Plk4 and other proteins implicated in centriole duplication, as expected (Habedanck et al., 2005). Although the depletion of several centrosomal proteins has recently been reported to induce a p53-dependent G1 arrest (Mikule et al., 2007), we found no evidence to indicate that depletion of any of the centrosomal proteins analyzed here produced a G1 arrest. Interestingly, in the course of the siRNA screen we occasionally observed splitting of centrosomes, concomitant with the impairment of PC formation (Fig. 1 B, right [inset]; note that centrosome splitting is defined here as the separation of parental centrioles by >2 μm). However, this splitting was only seen in serum-deprived cells and not in cycling cells. Furthermore, there was no strict correlation between centrosome splitting and PC formation. The depletion of BBS4 did not induce splitting although this protein was required for ciliogenesis, whereas the depletion of proteins implicated in centrosome cohesion (rootletin and C-Nap1) induced splitting without detectably impairing PC formation. Thus, the functional relationship, if any, between cell cycle–dependent centrosome splitting and the inhibition of PC formation is not presently clear.
This screen points to the involvement of many proteins in the biogenesis and/or maintenance of the PC, which is consistent with the known complexity of this structure (Ostrowski et al., 2002; Avidor-Reiss et al., 2004; Blacque et al., 2005). With regard to several proteins previously implicated in centriole duplication, we note that depletion of hSas-6 considerably reduced PC formation, in agreement with a recent independent paper (Vladar and Stearns, 2007). In contrast, the depletion of other proteins required for centriole biogenesis, notably Plk4, Cep135, or CPAP (Kleylein-Sohn et al., 2007), produced little adverse effects on PC formation, presumably because the limited duration of the present siRNA experiments prevented a marked reduction in centriole numbers. Remarkably, depletion of only three proteins almost completely prevented PC formation in our screen. The three proteins identified here as essential for ciliogenesis were pericentrin, Cep290, and Cep164. A key role of pericentrin in PC formation has previously been reported (Jurczyk et al., 2004) and a requirement for Cep290 in ciliogenesis falls in line with the implication of this protein in cilia-related diseases, including Joubert syndrome, Senior-Loken syndrome, and Meckel syndrome (Valente et al., 2006; Baala et al., 2007; Hildebrandt and Zhou, 2007). In contrast, Cep164, the focus of this paper, has not previously been analyzed.
The availability of anti-Cep164 antibodies (Fig. 2 B) made it possible to monitor the depletion of Cep164 in siRNA experiments. Focusing on cells that were effectively depleted of Cep164, we found that only 3.6% of cells formed a PC compared with 95% of GL2-treated control cells, clearly demonstrating that Cep164 is indispensable for PC formation (Fig. 1 D). Cep164 was originally identified in a proteomic inventory of human centrosomes (Andersen et al., 2003). The corresponding gene maps to human chromosome 11q23.3 (GenBank/EMBL/DDBJ accession nos. NM_014956 and NP_055771) and database analyses suggest the existence of potential isoforms. The Cep164 protein studied here is made up of 1,460 residues, resulting in a predicted molecular mass of 164 kD. As predicted by the SMART protein domain database (Schultz et al., 2000), the protein comprises a putative N-terminal WW domain (57–89) as well as three coiled-coil regions (589–810, 836–1047, and 1054–1200; Fig. 2 A). Orthologues of Cep164 clearly exist in other vertebrates (e.g., mouse [XP_929307] and zebrafish [XP_697015]) and potentially also in Drosophila melanogaster (NP_611787; Gherman et al., 2006). No obvious structural homologues could be identified in C. reinhardtii, Paramecium tetraurelia, or Tetrahymena termophila, but this does not exclude the existence of functional homologues. Considering the importance of flagellated and ciliated single-cell eukaryotes for the study of basal body function (Marshall and Rosenbaum, 2000; Beisson and Wright, 2003; Dutcher, 2003), the identification of Cep164-related proteins in a genetically tractable organism would be of obvious interest.
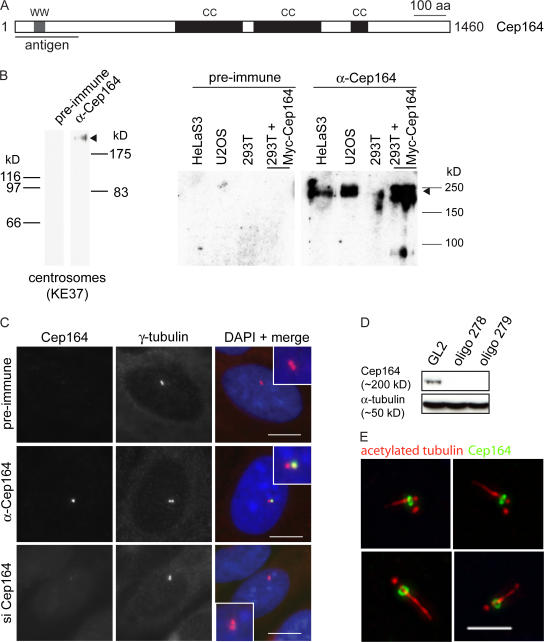
Figure 2.
Characterization of anti-Cep164 antibodies. (A) Schematic illustrating primary structure of human Cep164. The protein is predicted to comprise an N-terminal WW domain (WW) and three coiled-coil domains (CC). The horizontal line underneath the scheme denotes the region used for antibody production. (B) Antibodies directed against Cep164 and corresponding preimmune serum were tested by Western blotting on centrosome preparations from KE37 cells (left) and total lysates of HeLaS3, U2OS, and 293T cells as well as 293T cells overexpressing myc-Cep164 (right). Affinity-purified antibody was used at 1 μg/ml. Arrowhead indicates Cep164. Note that in the centrosome sample Cep164 shows a retarded electrophoretic mobility caused by a high sucrose concentration. (C) Antibodies directed against Cep164 and corresponding preimmune serum were used in IF microscopy on U2OS cells. The anti-Cep164 antibody (green) recognizes only one of two centrioles, as indicated by colocalization with γ-tubulin (red). This staining was abolished upon siRNA-mediated depletion of Cep164, and preimmune serum showed no specific staining. (insets) Enlarged centrosome areas. (D) Western blot on U2OS cells illustrating efficient depletion of Cep164 by siRNA. Transfections were performed for 48 h using two different Cep164-specific oligonucleotide duplexes (278 and 279) or GL2 for control. Blotting for α-tubulin is shown to control for equal loading. (E) Cep164 localizes to mature centrioles at the base of the PC but not along the length of the axoneme. hTERT-RPE1 cells were serum starved for 48 h to induce PC formation and stained with antibodies against Cep164 (green) and acetylated tubulin (red). The four representative examples show bar- and ringlike staining of Cep164 at the base of the PC in close association with the mature centriole. Bars: (C) 10 μm; (E) 5 μm.
Cep164 localizes specifically to mature centrioles
Antibodies raised against an N-terminal fragment (1–298) of recombinant Cep164 recognized a band of ∼200 kD on Western blots performed on either centrosomes purified from KE37 lymphoblastoid cells or lysates from HeLaS3, hTERT-RPE1, and 293T cells, whereas preimmune sera showed no specific reactivity (Fig. 2 B). The reason for the unexpectedly slow migration of Cep164 is not entirely clear but may relate to the relatively acidic isoelectric point of the protein (5.32). In any case, we emphasize that a myc-tagged Cep164 protein expressed in human 293T cells showed a similarly retarded migration (Fig. 2 B). Moreover, siRNA-mediated depletion of Cep164 (using two different siRNA oligonucleotide duplexes) abolished immunoreactivity (Fig. 2 D), making us confident that the 200-kD protein detected by the anti-Cep164 antibody represents endogenous Cep164. IF microscopy demonstrated that Cep164 localizes to the centrosome (Fig. 2 C), confirming earlier results based on the localization of GFP- and myc-tagged Cep164 constructs (Andersen et al., 2003). Interestingly, when compared with the staining produced by antibodies against γ-tubulin (Fig. 2 C), the anti-Cep164 antibody stained only one of two centrioles (Fig. 2 C, middle, inset). Preimmune serum did not produce any centrosomal staining (Fig. 2 C, top) and Cep164 staining was abolished by siRNA-mediated depletion of Cep164 (Fig. 2 C, bottom), attesting to the antibody's specificity.
To determine whether the localization of Cep164 to only one centriole relates to the known difference in maturity between the two centrioles, we performed costaining with Cep170, an appendage-associated protein and established marker for the mature parental centriole (Guarguaglini et al., 2005). Although Cep164 and Cep170 did not colocalize exactly (a point to which we will return later), the two proteins clearly localized to the same single centrioles (Fig. S1, A and B [GL2 controls], available at http://www.jcb.org/cgi/content/full/jcb.200707181/DC1). This indicates that Cep164, like Cep170, associates specifically with mature centrioles. To corroborate this conclusion, we also examined the localization of Cep164 in hTERT-RPE1 cells that had been induced to form a PC by serum starvation (Fig. 2 E). Costaining of the PC with antibodies against acetylated tubulin revealed that Cep164 was always associated with the one centriole that was located at the base of the PC, in agreement with the fact that only the mature centriole is able to initiate PC formation (Vorobjev and Chentsov, 1982). Interestingly, anti-Cep164 antibodies stained bar- or ringlike Cep164-positive structures at the base of the PC, depending on the angle of viewing (Fig. 2 E), whereas the axoneme itself was unstained.
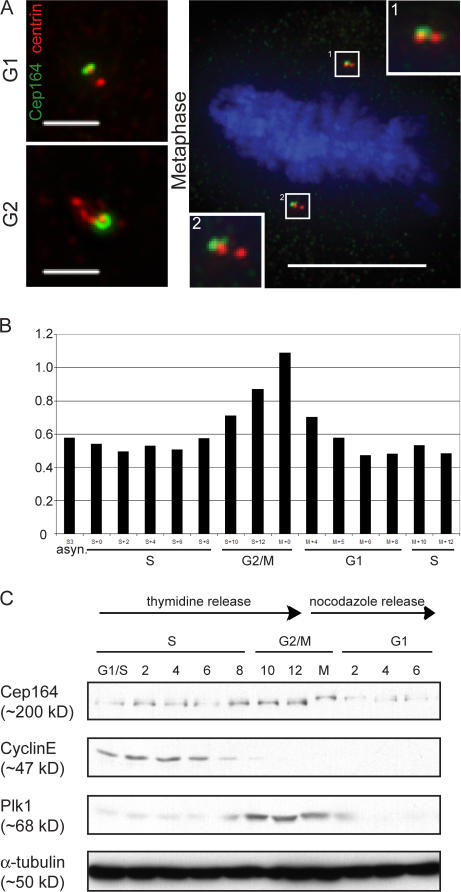
Next, we examined Cep164 localization during cell cycle progression. The protein was detectable at centrioles at all stages, including mitosis, although staining was more intense during interphase (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200707181/DC1). When compared with centrin, which is a marker for individual centrioles (Salisbury et al., 2002), Cep164 was restricted to only one centriole within both G1 and duplicated G2 centrosomes as expected (Fig. 3 A). Increased staining of a second parental centriole could then be detected at the onset of centrosome separation during prophase (Fig. S2 A, prophase), supporting the conclusion that Cep164 associates with structures that form concomitant with centriole maturation. During subsequent stages of mitosis, Cep164 was associated at similar levels with one centriole at each spindle pole (Figs. 3 A and S2 A). This is in stark contrast to Cep170 and ninein, both of which were displaced from centrioles during mitosis (Fig. S2, B and C).
Figure 3.
Cell cycle profile of Cep164 expression. (A) U2OS cells were costained with antibodies against Cep164 (green) and centrin as a marker for individual centrioles (red). Note that Cep164 (green) localizes to only one centriole in both G1 centrosomes (top left) and duplicated G2 centrosomes (bottom left). During mitosis, one of the two centrioles present at each spindle pole is positive for Cep164 (right). (insets) Magnifications of the spindle poles. Bars: (interphase cells) 2 μm; (metaphase cell) 10 μm. (B) Cep164 mRNA levels across the cell cycle were determined in synchronized HeLaS3 cells using qRT-PCR. (C) Cep164 protein levels across the cell cycle. HeLaS3 cells were arrested at the G1/S boundary by a double thymidine block or in M phase by a thymidine block followed by nocodazole treatment and released into fresh medium. Samples were harvested at the indicated time points and subjected to immunoblotting using the indicated antibodies.
We also examined Cep164 expression during the cell cycle using biochemical approaches. As determined by qRT-PCR, Cep164 mRNA levels showed little change throughout G1 and S phase, but levels increased beginning in G2 and peaked in mitotic cells, reaching a twofold increase compared with interphase (Fig. 3 B). A similar fluctuation was also seen at the protein level. Whereas Cep164 protein was present at a low level throughout interphase, it became slightly more abundant in mitosis (Fig. 3 C). Furthermore, in mitotic samples Cep164 displayed a retarded electrophoretic mobility, suggestive of a mitotic modification (most likely phosphorylation).
Cep164 and known appendage proteins localize to distinct structures
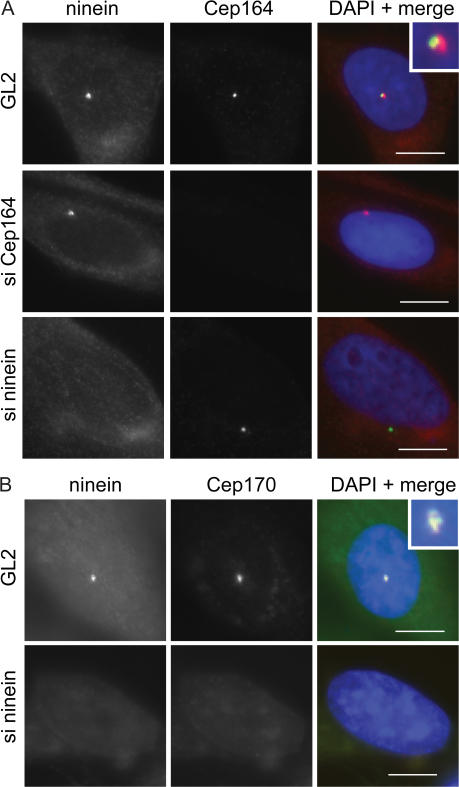
Most proteins that specifically localize to mature centrioles, including Cep170 (Guarguaglini et al., 2005), ninein (Mogensen et al., 2000), centriolin/CEP110 (Ou et al., 2002; Gromley et al., 2003), ɛ-tubulin (Chang et al., 2003), and Odf2 (Ishikawa et al., 2005), are known to associate with appendage structures. To examine whether Cep164 might similarly localize to appendages, we first asked to what extent Cep164 colocalizes with some of these proteins. Although Cep164 and ninein stained the same centriole, they did not colocalize exactly (Fig. 4 A). As already noted in the previous section, the same was true for Cep164 and Cep170 (Fig. S1). In contrast, ninein and Cep170 showed extensive colocalization as expected (Fig. 4 B). Next, we asked whether these proteins depend on each other for correct localization. siRNA-mediated depletion of Cep164 did not detectably affect the localization of ninein, Cep170, or ninein-like protein (Figs. 4 A and S1 A; and not depicted). Conversely, depletion of ninein did not affect Cep164, although it caused the loss of Cep170 from the mature centriole (Fig. 4 B), and depletion of Cep170 did not detectably influence Cep164 localization (Fig. S1 B). These data suggest that Cep164 does not directly interact with any of the appendage proteins investigated here, raising the possibility that it localizes to a distinct appendage structure altogether.
Figure 4.
Localizations and mutual dependencies of appendage proteins. (A) U2OS cells were transfected with control (GL2), ninein-specific, or Cep164-specific siRNA duplexes and costained for ninein (red) and Cep164 (green). (inset) Enlarged centrosome area. (B) U2OS cells were transfected with control (GL2) or ninein-specific siRNA duplexes and costained for ninein (red) and Cep170 (green). (inset) Enlarged centrosome area. Bars, 10 μm.
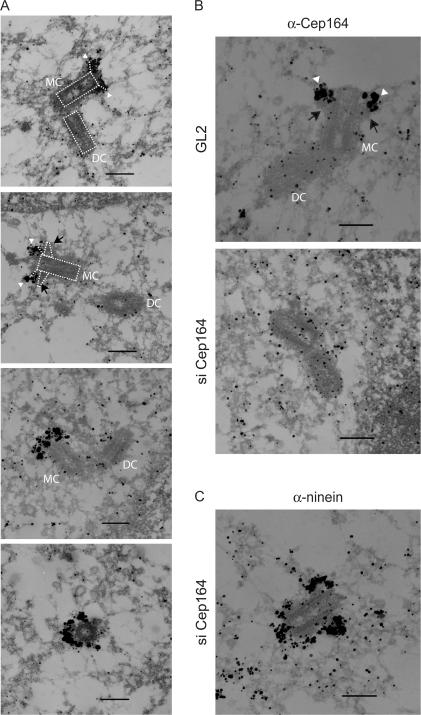
To explore this possibility, immunogold electron microscopy (immuno-EM) was performed. This study revealed that Cep164 localizes specifically to the appendages of mature parental centrioles, whereas immature parental centrioles as well as progeny centrioles were unstained (Fig. 5 A), which is consistent with the IF results. In the past, two distinct sets of appendages, distal and subdistal, have been described (Bornens, 2002). To the best of our knowledge, most appendage proteins characterized so far localize primarily to subdistal appendages, although depletion of Odf2 was found to abolish distal as well as subdistal appendages (Ishikawa et al., 2005). In contrast, our data suggest that Cep164 is a genuine component of distal rather than subdistal appendages. In support of this interpretation, immunogold labeling of Cep164 decorated structures at the very tip of centrioles (Fig. 5, A and B, arrowheads), whereas electron-dense material possibly representing subdistal appendages could occasionally be seen at the proximal side of Cep164-positive structures (Fig. 5, A and B, arrows). Furthermore, whereas siRNA-mediated depletion of Cep164 abolished Cep164 staining, attesting to its specificity (Fig. 5 B), the immunolocalization of ninein was not detectably affected in Cep164-depleted cells (Fig. 5 C; compare with Bouckson-Castaing et al. [1996] and Mogensen et al. [2000]). Collectively, these data suggest that Cep164 is indispensable for PC formation and represents a genuine marker for distal appendages.
Figure 5.
Immuno-EM localization of Cep164. (A) U2OS cells were subjected to preembedding immuno-EM. Cells were labeled with anti-Cep164 antibody R171 followed by Nanogold-coupled secondary antibody. Note that Cep164 localizes to very distal appendage structures on parental centrioles (arrowheads, also see B), and sometimes electron-dense material (presumably reflecting subdistal appendages) can be seen proximal to the Cep164 staining (arrows in second panel and in B [top]). The schematic diagrams (dotted lines) in the two top panels illustrate the positions of “mother” (MC) and “daughter” centrioles (DC), along with the presumed positions of distal and subdistal appendages. (B and C) U2OS cells were transfected for 48 h with control (GL2) or Cep164-specific siRNA duplexes and subjected to preembedding immuno-EM. Cells were labeled with either anti-Cep164 or anti-ninein antibody followed by Nanogold-coupled secondary antibody. Bars, 250 nm.
Discussion
The present study identified several proteins whose depletion interfered with PC formation, confirming the view that ciliogenesis is a complex process (Ostrowski et al., 2002; Avidor-Reiss et al., 2004; Blacque et al., 2005). In addition to proteins identified in independent studies, notably pericentrin, Odf2, PCM-1, and BBS4 (Jurczyk et al., 2004; Ishikawa et al., 2005; Nachury et al., 2007), we observed a role in PC assembly for ninein, a protein involved in microtubule nucleation and anchoring (Delgehyr et al., 2005), chTOG, a protein required for stabilizing microtubules and spindle pole organization (Gergely et al., 2003), and Cep290 (also called NPHP6), mutations in which have been linked to several human disease syndromes (den Hollander et al., 2006; Valente et al., 2006; Baala et al., 2007; Hildebrandt and Zhou, 2007). The exact roles of these proteins in ciliogenesis remain to be elucidated, but they are likely to relate to intracellular transport processes that depend on functional centrosome–microtubule interactions. In addition, we identified several proteins, notably Cep57, Cep131, Cep152, and ALMS1, whose depletion resulted in the formation of short or stunted cilia (Li et al., 2007). Little is presently known about the functions of Cep57, Cep131, and Cep152, but ALMS1 is attracting considerable interest because of its implication in ALMS (Collin et al., 2002; Hearn et al., 2002). What precise molecular defects lead to stunted cilia remains to be elucidated, but it is plausible that the functions of Cep57, Cep131, Cep152, and ALMS1 relate to IFT. In support of this proposal, we emphasize that truncated cilia have frequently been seen in C. reinhardtii and C. elegans mutants carrying defects in the IFT machinery (Rosenbaum and Witman, 2002; Scholey, 2003).
The main focus of the present study was the characterization of Cep164, a novel protein of previously unknown function whose depletion severely impaired PC formation. By both IF microscopy and immuno-EM we were able to show that Cep164 localizes specifically to very distally located appendage structures on the mature centriole. On the basis of morphological analyses, centriolar appendages have in the past been classified as distal or subdistal (Vorobjev and Chentsov, 1982). From this perspective, it is remarkable that Cep164 did not colocalize exactly with proteins previously shown to associate with subdistal appendages, notably ninein and Cep170. Furthermore, no reciprocal dependencies for centriolar localization could be observed between Cep164 and ninein or Cep170 and, unlike ninein and Cep170, Cep164 persisted at the mature centriole throughout mitosis. Collectively, these data strongly indicate that Cep164 constitutes a genuine component of distal appendages.
A priori, Cep164 could be required for PC assembly or maintenance. Considering that Cep164 does not localize to the axoneme, it is unlikely to play a direct role in IFT, although it is possible that it contributes to intracellular transport between the interior of the cell and the cilium. Perhaps more likely, Cep164 may form part of a terminal plate (Anderson, 1972), which mediates the interaction between the basal body and the plasma membrane, or provide a docking site for the formation of transition fibers. Further insight into the molecular function of Cep164 will require identification of its interaction partners. In particular, it will be interesting to search for proteins binding to the WW domain that is present within the N-terminal end domain of the protein. One possibility is that Cep164 might contribute to the mediation of interactions with the cytoskeleton. Such interactions may in turn be critical for basal body positioning during PC formation. Alternatively, Cep164 may be indispensable for the formation of distal appendages. Careful ultrastructural analyses of Cep164-depleted centrioles will be required to explore this intriguing possibility.
In conclusion, our study identifies a novel protein, Cep164, as being critically required for PC formation. Furthermore, our results indicate that Cep164 provides an excellent marker for distal (rather than subdistal) appendages, strengthening the conclusion that centriolar appendage proteins are crucial for ciliogenesis (Ishikawa et al., 2005). Finally, considering the rapid emergence of data emphasizing the importance of PC formation and function for human health, it will be interesting to explore a possible relationship between Cep164 and ciliary disease syndromes. In this context, it is intriguing that the Cep164 gene locus (11q23.3) maps close to a region implicated in Jacobsen syndrome (Grossfeld et al., 2004). Thus, it may be rewarding to explore a possible connection between impaired functionality of Cep164 and diseases possibly related to defective ciliogenesis.
Materials and methods
Plasmid preparation and recombinant proteins
PCR was used to amplify a full-length human Cep164 cDNA from clone KIAA1052 (Kazusa DNA Research Institute). The cDNA was subcloned into a mammalian expression vector providing a C-terminal myc tag and the construct was verified by sequencing. For expression of a recombinant Cep164 protein fragment, bp 1–894 of the coding sequence were amplified by PCR, inserted into the expression vector pET28b+ (Novagen), and verified by sequencing. His6-tagged N-terminal Cep164 was expressed in E. coli strain BL21 (DE3) and purified under denaturing conditions according to standard protocols (QIAexpressionistsystem; QIAGEN).
Antibody production
Rabbit anti-Cep164 antisera (R171 and R172) were raised against an N-terminal fragment spanning aa 1–298 (Charles River Laboratories). Similar results were obtained with both sera but R171 was used for most experiments. Antibody R171 was affinity purified using Affigel according to standard protocols (Affigel-10; Bio-Rad Laboratories). The anti-Cep170 mAb (77-419-2) and anti-ninein mAb (79–160) were produced by immunization of BALB/c mice with recombinant fragments of human Cep170 (aa 15–754) and ninein (aa 1,110–2,662) purified from E. coli. Spleen cells were fused with PAIB3Ag81 mouse myeloma cells, and positive hybridoma clones were subcloned by limiting dilution. The anti-Cep170 mAb is an IgG1 and the anti-ninein mAb is an IgG2a.
Cell culture and transfections
Cells were grown at 37°C under 5% CO2. U2OS, HeLaS3, and 293T cells were grown in DME and supplemented with 100 IU/ml of 10% FCS and 100 μg/ml penicillin-streptomycin. hTERT-RPE1 cells were grown in DME nutrient mixture, Ham's F12 (Sigma-Aldrich) supplemented with 10% FCS, penicillin-streptomycin, 2 mM glutamine, and 0.348% sodium bicarbonate. 293T cells were transfected using the calcium phosphate precipitation method (Krek and Nigg, 1991).
Immuno-EM
For preembedding immuno-EM, U2OS cells were grown on coverslips, fixed with 4% formaldehyde for 10 min, and permeabilized with PBS + 0.5% Triton X-100 (Roth) for 2 min. Blocking and primary antibody incubations were then performed as described for IF microscopy followed by incubation with goat anti–rabbit IgG-Nanogold (1:50; Nanoprobes). Nanogold was silver enhanced with HQ Silver (Nanoprobes) and cells were further processed as described previously (Fry et al., 1998; Bahe et al., 2005).
IF microscopy and immunoblotting
Cells were prepared for IF microscopy as described previously (Bahe et al., 2005). The primary antibodies used were 1 μg/ml affinity-purified rabbit anti-Cep164 IgG (R171) and anti–C-Nap1 IgG (Mayor et al., 2000), mouse anti-Cep170 mAb (77-419-2), anti–γ-tubulin mAb (1:1,000, GTU-88; Sigma-Aldrich), anti-centrin mAb (1:3,000, 20H5; provided by J.L. Salisbury, Mayo Clinic, Rochester, MN; Salisbury et al., 2002), and anti-acetylated tubulin mAb (1:2,000; 6-11B-1; Sigma-Aldrich). Secondary antibodies were Alexa Fluor 488/555–conjugated (1:1,000; Invitrogen) and cy2/cy3-conjugated donkey IgGs (1:1,000; Dianova). DNA was stained with 0.2 μg/ml DAPI.
Cells were analyzed using a microscope (Axioskop-2; Carl Zeiss MicroImaging, Inc.) equipped with a 63× NA 1.4 plan apochromat oil immersion objective and standard filter sets (Carl Zeiss MicroImaging, Inc.), a 1,300 × 1,030 pixel cooled charge-coupled device camera (CCD-1300-Y; Princeton Instruments), and Metavue software (Visitron Systems). Alternatively, for the data shown in Figs. 2 E and 3 A, a microscope (Deltavision) on a base (Olympus IX71; Applied Precision) equipped with an apo 100× 1.35 oil immersion objective, a camera (CoolSnap HQ; Photometrics), and a 37°C chamber were used for collecting 0.18–0.2-μm-distanced optical sections in the z axis.
Images at single focal planes were processed with a deconvolution algorithm (100×: Olympus_100X_140_10103.otf). Settings were “enhanced ratio,” with noise filtering set to medium and 10 deconvolution cycles. The number of z stacks collected was variable (between 6 and 14), depending on the height of the individual cell. Images were projected into one picture using softWoRx 3.5.0 (Applied Precision). Exposure times and settings for image processing (deconvolution) were constant for all samples to be compared within any given experiment. Images were opened in Photoshop CS (Adobe) and sized and placed in figures with Illustrator CS2 (Adobe).
Immunoblotting was performed as described previously (Fry et al., 1998; Mayor et al., 2000; Bahe et al., 2005). Primary antibodies were used at the following concentrations: 1 μg/ml rabbit anti–Cep164 affinity-purified IgG (R171) or corresponding preimmune serum (1:1,000), mouse anti–α-tubulin mAb (1:5,000; DM1A; Sigma-Aldrich), anti–polo-like kinase 1 mAb (1:5; Yamaguchi et al., 2005), anti-cyclin E mAb (1:5; HE-12; provided by J. Bartek, Danish Cancer Society, Copenhagen, Denmark). Secondary antibodies were HRP-conjugated goat anti–rabbit (1:7,000; Bio-Rad Laboratories) or anti–mouse (1:7,000; Bio-Rad Laboratories) IgGs.
siRNA experiments and PC growth assays
Proteins to be tested in the siRNA screen were depleted using siRNA duplex oligonucleotides (Qiagen and Dharmacon) targeting the sequences described in Table S1. Cep164 was efficiently depleted using two different siRNA duplex oligonucleotides (278 and 279). A duplex targeting luciferase (GL2; Elbashir et al., 2001) was used for control. RNA oligonucleotides were used at 20 μM, and cells were analyzed 48 or 72 h later.
hTERT-RPE1 cells (provided by L. Kohen, Universiätsklinikum Leipzig, Leipzig, Germany) were grown on acid-treated, sterilized glass coverslips and transfected for 48 h with different siRNA duplexes. PC formation was induced by continued culturing in serum-free medium for another 48 h. Before methanol fixation, cells were incubated for 45 min on ice to depolymerize the microtubules.
qRT-PCR
RNA for qRT-PCR analysis was prepared as described in the text. cDNA was synthesized from RNA samples using random hexamers and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. PCR reactions contained cDNA, Power SYBR Green Master Mix (Applied Biosystems) and 300 nM of forward and reverse primers. Primers were designed with Primer Express software (Applied Biosystems) and the amplified fragment corresponded to an exon–exon junction. qRT-PCR was performed in optical 384-well plates and fluorescence was quantified with a sequence detection system (Prism 7900 HT; Applied Biosystems). Samples were analyzed in triplicate and the raw data consisted of PCR cycle numbers required to reach a fluorescence threshold cycle. Raw fluorescence threshold cycle values were obtained using SDS 2.0 (Applied Biosystems). The relative expression level of target genes was normalized with geNorm software (Primer Design Ltd.; Vandesompele et al., 2002) using eukaryotic translation elongation factor a-1 and β-glucuronidase genes as references to determine the normalization factor. The thermal profile recommended by Applied Biosystems was used for amplification (50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min). To verify the specificity of amplification, a melting curve analysis was included according to the thermal profile suggested by the manufacturer (95°C for 15 s, 60°C for 15 s, and 95°C for 15 s). The generated data were analyzed with SDS 2.2 software.
Miscellaneous techniques
Human centrosomes were isolated from KE37 cells as described previously (Andersen et al., 2003).
To monitor Cep164 levels during cell cycle progression, HeLaS3 cells were synchronized using double thymidine block or a thymidine–nocodazole treatment, followed by release into fresh medium. To analyze expression levels of CEP164 transcripts, total RNA was extracted from HeLaS3 cells at various cell cycle stages using an RNeasy Mini kit (QIAGEN). Transcript levels were determined by qRT-PCR. For the analysis of siRNA efficiency, total RNA was extracted from HeLa S3 cells treated for 72 h with siRNA oligonucleotide duplexes targeting individual centrosomal proteins (Table S1).
Online supplemental material
Table S1 indicates the siRNA oligonucleotide sequences used for protein depletion. Figs. S1 and S2 illustrate the localizations and dependencies of appendage proteins. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707181/DC1.
Supplementary Material
Acknowledgments
We thank Dr. L. Kohen for providing hTERT-RPE-1 cells, Dr. J. Salisbury for anti-centrin antibodies, and Dr. J. Bartek for anti–cyclin E antibodies. We also thank P. Descombes for carrying out qRT-PCR experiments, R. Malik for help with bioinformatics, and E. Nigg for excellent technical assistance. Finally, we thank C. Wilkinson for his help at the early stages of this study and Dr. Paul Grossfeld for helpful information about Jacobsen syndrome.
This work was supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft (grant SFB413).
Abbreviations used in this paper: ALMS, Alstrom syndrome; BBS, Bardet-Biedl syndrome; Cep, centrosomal protein; C-Nap1, centrosomal Nek2-associated protein 1; IF, immunofluorescence; IFT, intraflagellar transport; immuno-EM, immunogold electron microscopy; Odf, outer dense fiber; PC, primary cilium; qRT-PCR, quantitative real-time PCR.
References
- Andersen, J.S., C.J. Wilkinson, T. Mayor, P. Mortensen, E.A. Nigg, and M. Mann. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574. [DOI] [PubMed] [Google Scholar]
- Anderson, R.G. 1972. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J. Cell Biol. 54:246–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss, T., A.M. Maer, E. Koundakjian, A. Polyanovsky, T. Keil, S. Subramaniam, and C.S. Zuker. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117:527–539. [DOI] [PubMed] [Google Scholar]
- Baala, L., S. Audollent, J. Martinovic, C. Ozilou, M.C. Babron, S. Sivanandamoorthy, S. Saunier, R. Salomon, M. Gonzales, E. Rattenberry, et al. 2007. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am. J. Hum. Genet. 81:170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano, J.L., N. Mitsuma, P.L. Beales, and N. Katsanis. 2006. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7:125–148. [DOI] [PubMed] [Google Scholar]
- Bahe, S., Y.D. Stierhof, C.J. Wilkinson, F. Leiss, and E.A. Nigg. 2005. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 171:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson, J., and M. Wright. 2003. Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 15:96–104. [DOI] [PubMed] [Google Scholar]
- Blacque, O.E., E.A. Perens, K.A. Boroevich, P.N. Inglis, C. Li, A. Warner, J. Khattra, R.A. Holt, G. Ou, A.K. Mah, et al. 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15:935–941. [DOI] [PubMed] [Google Scholar]
- Bornens, M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14:25–34. [DOI] [PubMed] [Google Scholar]
- Boucher, C., and R. Sandford. 2004. Autosomal dominant polycystic kidney disease (ADPKD, MIM 173900, PKD1 and PKD2 genes, protein products known as polycystin-1 and polycystin-2). Eur. J. Hum. Genet. 12:347–354. [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing, V., M. Moudjou, D.J. Ferguson, S. Mucklow, Y. Belkaid, G. Milon, and P.R. Crocker. 1996. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J. Cell Sci. 109:179–190. [DOI] [PubMed] [Google Scholar]
- Chang, P., T.H. Giddings Jr., M. Winey, and T. Stearns. 2003. Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat. Cell Biol. 5:71–76. [DOI] [PubMed] [Google Scholar]
- Collin, G.B., J.D. Marshall, A. Ikeda, W.V. So, I. Russell-Eggitt, P. Maffei, S. Beck, C.F. Boerkoel, N. Sicolo, M. Martin, et al. 2002. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat. Genet. 31:74–78. [DOI] [PubMed] [Google Scholar]
- Delgehyr, N., J. Sillibourne, and M. Bornens. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118:1565–1575. [DOI] [PubMed] [Google Scholar]
- den Hollander, A.I., R.K. Koenekoop, S. Yzer, I. Lopez, M.L. Arends, K.E. Voesenek, M.N. Zonneveld, T.M. Strom, T. Meitinger, H.G. Brunner, et al. 2006. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 79:556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S.K. 2003. Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii. Traffic. 4:443–451. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- Fry, A.M., T. Mayor, P. Meraldi, Y.D. Stierhof, K. Tanaka, and E.A. Nigg. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle–regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely, F., V.M. Draviam, and J.W. Raff. 2003. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman, A., E.E. Davis, and N. Katsanis. 2006. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat. Genet. 38:961–962. [DOI] [PubMed] [Google Scholar]
- Gromley, A., A. Jurczyk, J. Sillibourne, E. Halilovic, M. Mogensen, I. Groisman, M. Blomberg, and S. Doxsey. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161:535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfeld, P.D., T. Mattina, Z. Lai, R. Favier, K.L. Jones, F. Cotter, and C. Jones. 2004. The 11q terminal deletion disorder: a prospective study of 110 cases. Am. J. Med. Genet. A. 129:51–61. [DOI] [PubMed] [Google Scholar]
- Guarguaglini, G., P.I. Duncan, Y.D. Stierhof, T. Holmstrom, S. Duensing, and E.A. Nigg. 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell. 16:1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck, R., Y.D. Stierhof, C.J. Wilkinson, and E.A. Nigg. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. [DOI] [PubMed] [Google Scholar]
- Hearn, T., G.L. Renforth, C. Spalluto, N.A. Hanley, K. Piper, S. Brickwood, C. White, V. Connolly, J.F. Taylor, I. Russell-Eggitt, et al. 2002. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alstrom syndrome. Nat. Genet. 31:79–83. [DOI] [PubMed] [Google Scholar]
- Hildebrandt, F., and W. Zhou. 2007. Nephronophthisis-associated ciliopathies. J. Am. Soc. Nephrol. 18:1855–1871. [DOI] [PubMed] [Google Scholar]
- Ibañez-Tallon, I., N. Heintz, and H. Omran. 2003. To beat or not to beat: roles of cilia in development and disease. Hum. Mol. Genet. 12:R27–R35. [DOI] [PubMed] [Google Scholar]
- Ishikawa, H., A. Kubo, S. Tsukita, and S. Tsukita. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524. [DOI] [PubMed] [Google Scholar]
- Jurczyk, A., A. Gromley, S. Redick, J. San Agustin, G. Witman, G.J. Pazour, D.J. Peters, and S. Doxsey. 2004. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 166:637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn, J., J. Westendorf, M. Le Clech, R. Habedanck, Y.D. Stierhof, and E.A. Nigg. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202. [DOI] [PubMed] [Google Scholar]
- Kozminski, K.G., K.A. Johnson, P. Forscher, and J.L. Rosenbaum. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 90:5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, W., and E.A. Nigg. 1991. Mutations of p34cdc2 phosphorylation sites induce premature mitotic events in HeLa cells: evidence for a double block to p34cdc2 kinase activation in vertebrates. EMBO J. 10:3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., R. Vega, K. Nelms, N. Gekakis, C. Goodnow, P. McNamara, H. Wu, N.A. Hong, and R. Glynne. 2007. A role for Alström syndrome protein, alms1, in kidney ciliogenesis and cellular quiescence. PLoS Genet. 3:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, M.S., and R.D. Burk. 2006. Primary cilium formation requires von hippel-lindau gene function in renal-derived cells. Cancer Res. 66:6903–6907. [DOI] [PubMed] [Google Scholar]
- Marshall, W.F., and J.L. Rosenbaum. 2000. How centrioles work: lessons from green yeast. Curr. Opin. Cell Biol. 12:119–125. [DOI] [PubMed] [Google Scholar]
- Mayor, T., Y.D. Stierhof, K. Tanaka, A.M. Fry, and E.A. Nigg. 2000. The centrosomal protein C-Nap1 is required for cell cycle–regulated centrosome cohesion. J. Cell Biol. 151:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikule, K., B. Delaval, P. Kaldis, A. Jurcyzk, P. Hergert, and S. Doxsey. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9:160–170. [DOI] [PubMed] [Google Scholar]
- Mogensen, M.M., A. Malik, M. Piel, V. Bouckson-Castaing, and M. Bornens. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013–3023. [DOI] [PubMed] [Google Scholar]
- Nachury, M.V., A.V. Loktev, Q. Zhang, C.J. Westlake, J. Peranen, A. Merdes, D.C. Slusarski, R.H. Scheller, J.F. Bazan, V.C. Sheffield, and P.K. Jackson. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. [DOI] [PubMed] [Google Scholar]
- Ostrowski, L.E., K. Blackburn, K.M. Radde, M.B. Moyer, D.M. Schlatzer, A. Moseley, and R.C. Boucher. 2002. A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics. 1:451–465. [DOI] [PubMed] [Google Scholar]
- Ou, Y.Y., G.J. Mack, M. Zhang, and J.B. Rattner. 2002. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 115:1825–1835. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., and G.B. Witman. 2003. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 15:105–110. [DOI] [PubMed] [Google Scholar]
- Piperno, G., and M.T. Fuller. 1985. Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetorius, H.A., and K.R. Spring. 2005. A physiological view of the primary cilium. Annu. Rev. Physiol. 67:515–529. [DOI] [PubMed] [Google Scholar]
- Pugacheva, E.N., S.A. Jablonski, T.R. Hartman, E.P. Henske, and E.A. Golemis. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 129:1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J.L., and G.B. Witman. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825. [DOI] [PubMed] [Google Scholar]
- Salisbury, J.L., K.M. Suino, R. Busby, and M. Springett. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12:1287–1292. [DOI] [PubMed] [Google Scholar]
- Satir, P., and S.T. Christensen. 2007. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69:377–400. [DOI] [PubMed] [Google Scholar]
- Schermer, B., C. Ghenoiu, M. Bartram, R.U. Muller, F. Kotsis, M. Hohne, W. Kuhn, M. Rapka, R. Nitschke, H. Zentgraf, et al. 2006. The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. J. Cell Biol. 175:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey, J.M. 2003. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 19:423–443. [DOI] [PubMed] [Google Scholar]
- Schultz, J., R.R. Copley, T. Doerks, C.P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla, V., and J.F. Reiter. 2006. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 313:629–633. [DOI] [PubMed] [Google Scholar]
- Thoma, C.R., I.J. Frew, C.R. Hoerner, M. Montani, H. Moch, and W. Krek. 2007. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat. Cell Biol. 9:588–595. [DOI] [PubMed] [Google Scholar]
- Tucker, R.W., A.B. Pardee, and K. Fujiwara. 1979. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 17:527–535. [DOI] [PubMed] [Google Scholar]
- Valente, E.M., J.L. Silhavy, F. Brancati, G. Barrano, S.R. Krishnaswami, M. Castori, M.A. Lancaster, E. Boltshauser, L. Boccone, L. Al-Gazali, et al. 2006. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 38:623–625. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar, E.K., and T. Stearns. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev, I.A., and Yu.S. Chentsov. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., H. Goto, T. Yokoyama, H. Silljé, A. Hanisch, A. Uldschmid, Y. Takai, T. Oguri, E.A. Nigg, and M. Inagaki. 2005. Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J. Cell Biol. 171:431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala, M.A., M.R. Knowles, and H. Omran. 2007. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 69:423–450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.