Abstract
DNA methylation has been implicated in chromatin condensation and nuclear organization, especially at sites of constitutive heterochromatin. How this is mediated has not been clear. In this study, using mutant mouse embryonic stem cells completely lacking in DNA methylation, we show that DNA methylation affects nuclear organization and nucleosome structure but not chromatin compaction. In the absence of DNA methylation, there is increased nuclear clustering of pericentric heterochromatin and extensive changes in primary chromatin structure. Global levels of histone H3 methylation and acetylation are altered, and there is a decrease in the mobility of linker histones. However, the compaction of both bulk chromatin and heterochromatin, as assayed by nuclease digestion and sucrose gradient sedimentation, is unaltered by the loss of DNA methylation. This study shows how the complete loss of a major epigenetic mark can have an impact on unexpected levels of chromatin structure and nuclear organization and provides evidence for a novel link between DNA methylation and linker histones in the regulation of chromatin structure.
Introduction
DNA methylation is generally thought to silence gene expression and reduce transcriptional noise by compacting chromatin structure. However, how this is brought about has not been systematically investigated. The impact of DNA methylation on histone modifications is well established in plants (Tariq and Paszkowski, 2004) but not in mammalian cells. In human cells, ablation of the DNA methyltransferase DNMT1 leads to a partial reduction of DNA methylation, predominantly at repetitive sequences. This is reported to result in a depletion of di- (H3K9me2) and trimethylation (H3K9me3) at H3K9 and a concomitant increase in H3K9 acetylation (H3K9ac; Espada et al., 2004). Indirect reduction of DNA methylation at pericentromeric heterochromatin by loss of the chromatin remodeling protein Lsh also results in increased levels of histone acetylation (Huang et al., 2004). In the mouse, little change in H3K9 methylation was found in Dnmt1 − and Dnmt3 − embryonic stem (ES) cells (Dnmt3a −/−/Dnmt3b −/− ES cells), which have a 50% reduction in levels of DNA methylation (Martens et al., 2005). Because of the remaining levels of DNA methylation in all of the aforementioned studies, it has not been clear what the absolute relationship between DNA methylation and histone modifications is in mammalian cells.
There has been recent interest in links between linker histones and DNA methylation. Reduction of linker histone levels in vivo can give rise to altered DNA methylation at specific genomic sites (Fan et al., 2005). On the other hand, there is conflicting evidence about whether DNA methylation influences linker histone binding in chromatin. Unmethylated CpG islands appear to be depleted of H1 (Tazi and Bird, 1990), and H1-containing nucleosomes contain 80% of the 5′-methylcytosine (Ball et al., 1983), suggesting that linker histones may prefer to bind to methylated DNA. This is confirmed by some in vitro binding experiments (Levine et al., 1993; McArthur and Thomas, 1996). However, other studies suggest that DNA methylation does not affect H1 binding to nucleosomes (Campoy et al., 1995; Nightingale and Wolffe, 1995; Hashimshony et al., 2003).
Experimentally or genetically induced alterations in levels of DNA methylation have implicated this epigenetic modification in mammalian higher order chromatin condensation and nuclear organization, especially at sites of constitutive heterochromatin that have the highest concentration of DNA methylation (Schmid et al., 1984; Brown et al., 1995; Karymov et al., 2001; Gisselsson et al., 2005; Ma et al., 2005). Chromocenter clustering during differentiation has been recently attributed to increasing levels of DNA methylation and methyl-CpG–binding proteins (Brero et al., 2005).
To investigate the consequences of eliminating CpG methylation on multiple levels of mammalian chromatin structure and nuclear organization, we used mouse ES cells that lack both Dnmt3a and b (Dnmt3a −/−/Dnmt3b −/− but herein called Dnmt3 −; Okano et al., 1999). Chromatin structure in these cells was analyzed by biochemical, biophysical, and cytological assays and compared with that from wild-type (WT) cells. The absence of DNA methylation altered chromatin structure at the level of the nucleosome and at the level of nuclear organization. There is a genome-wide decrease in H3K9me2, an increase in histone acetylation, and an increased clustering of chromocenters in mouse ES cells that are devoid of DNA methylation. However, contrary to expectations, micrococcal nuclease (Mnase) digestion and sucrose gradient sedimentation analyses indicate that the compaction of the chromatin in general and of heterochromatin in particular is not affected. Instead, there is a surprising decrease in the mobility of linker histones in the absence of DNA methylation. These studies highlight the complex interplay between DNA methylation and chromatin structure and the need to assess the effects of epigenetic modifications at multiple levels of chromatin and nuclear organization.
Results
Complete loss of DNA methylation in the absence of Dnmt3a and b
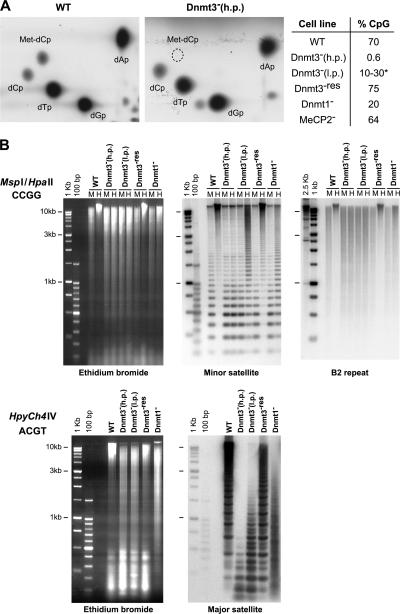
At the time of their establishment, Dnmt3 − cells show a partial loss of CpG methylation (Okano et al., 1999), but, after prolonged passage in culture, virtually no (0.6%) CpG methylation remains (Fig. 1 A; Jackson et al., 2004). This probably reflects the failure of Dnmt1 to efficiently maintain methylation (Chen et al., 2003). Southern blotting shows that methylation is similarly lost at euchromatic (B2 repeat) and heterochromatic (minor and major satellite) parts of the genome (Fig. 1 B). In contrast, MeCP2 − ES cells (Tate et al., 1996) and Dnmt3 − cells transfected with a Dnmt3b transgene (Dnmt3 −res; Jackson et al., 2004) show near normal levels of DNA methylation.
Figure 1.
DNA methylation levels in mutant ES cells. (A) Nearest neighbor analysis of CpG methylation in WT and Dnmt3 − ES cells. The table summarizes the percentage of CpGs that are methylated in WT, high passage (h.p.) Dnmt3 −, low passage (l.p.) Dnmt3 −, Dnmt3 −res, Dnmt1 −, and MeCP2 − cells. The asterisk indicates dependence on passage number. (B) Analysis of DNA methylation by Southern blotting in WT, high passage Dnmt3 −, low passage Dnmt3 −, Dnmt3 −res, and Dnmt1 − cells. B2 and minor satellite repeats were analyzed on MspI- (M) and HpaII (H)-digested DNAs. Mouse major satellite was analyzed on DNAs digested with HpyCH4IV (Lehnertz et al., 2003). Size markers are1-kb and 100-bp ladders.
Increased chromocenter clustering in the absence of DNA methylation
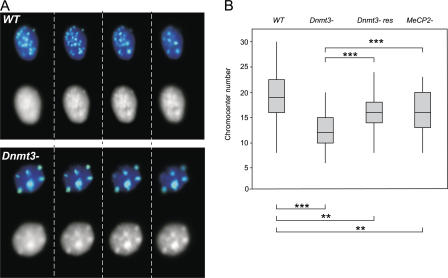
In the mouse nucleus, pericentric heterochromatin comprising of major satellite repeats tends to cluster into chromocenters. Recently, it has been suggested that increasing levels of DNA methylation contribute to progressive chromocenter clustering during differentiation and that this is mediated through methyl-CpG–binding proteins (Brero et al., 2005). If this is the case, there should be a reduction of chromocenter clustering in Dnmt3 − cells compared with WT. Using 3D FISH with a probe for major satellite, we analyzed the number of individual chromocenters visible in the nuclei of mutant ES cells and in their parental WT equivalents, J1 cells (Fig. 2 A). The number of chromocenters detected in Dnmt3 − ES cells (median = 12) was significantly less than in J1 WT cells (median = 19; P = 0.0000 in Mann-Whitney U analysis; n = 85; taken from three independent experiments; Fig. 2 B). This was verified by analysis of a subset of images by two independent investigators and was even apparent from the DAPI staining pattern in which fewer larger, brightly stained foci were visible in Dnmt3 − cells compared with WT. This was also confirmed using an independent WT ES cell line, CGR8. The median number of chromocenters in these cells (20) is not significantly different (P = 0.9) from that in the WT J1 cells and is significantly (P = 0.000) larger than the chromocenter number in Dnmt3 − cells.
Figure 2.
Chromocenter clustering in the nuclei of mutant ES cells. (A) Single-image planes taken at 1-μm intervals through (left to right) the z axis of WT (top) and Dnmt3 − (bottom) ES cells hybridized with a major satellite probe (green). Nuclei are counterstained with DAPI (blue in color image and shown in black and white below that). (B) Box plots showing the number of distinct chromocenters in the nuclei of WT, Dnmt3 −, Dnmt3 −res, and MeCP2 − ES cells. The lower and upper limits of the boxes denote the 25th and 75th percentiles, respectively, and the line in the box is the median. Statistical comparisons of chromocenter number was performed by Mann-Whitney U test (**, P = 0.001; ***, P = 0.0000).
These data indicate that the loss of DNA methylation leads to increased clustering of pericentric heterochromatin into a few large chromocenters in ES cells. Restoring levels of DNA methylation in the Dnmt3 −res cells increased the number of chromocenters (median = 16; Fig. 2 B). Chromocenter clustering in Dnmt3 − cells is not simply caused by the subsequent loss of methyl-CpG–binding proteins because the number of chromocenters in MeCP2 − cells (median = 16.0) is significantly larger (P = 0.0000) than in Dnmt3 − cells (Fig. 2).
Loss of DNA methylation does not affect heterochromatin compaction
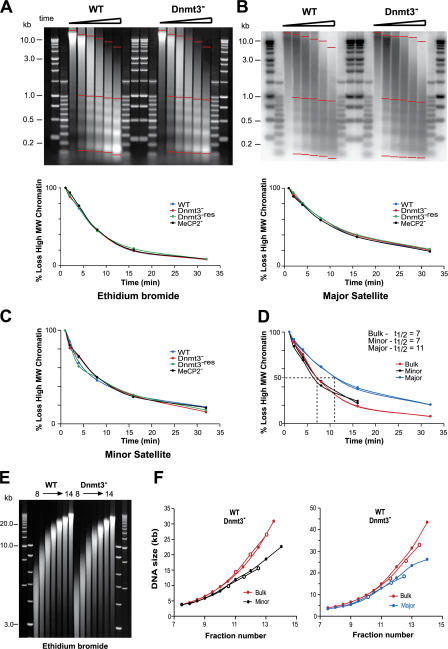
To investigate whether the altered nuclear organization of heterochromatin in DNA methylation–deficient ES cells is caused by a change in underlying secondary chromatin structures, we analyzed the Mnase sensitivity of chromatin from WT and mutant cells. All cell lines showed identical digestion kinetics of bulk chromatin (Fig. 3 A). Major satellite has a less accessible chromatin structure (i.e., is digested more slowly; t 1/2 = 11 min) than bulk chromatin or chromatin at minor satellite (t 1/2 = 7 min), which is independent of DNA methylation (Fig. 3, B–D). Nucleosome repeat length of bulk chromatin and major and minor satellites was also identical between cell lines. As previously shown, dinucleosomes at major satellite are refractory to Mnase digestion compared with those at minor satellite (Gilbert and Allan, 2001; Guenatri et al., 2004), but this is also unaffected by the absence of DNA methylation (unpublished data).
Figure 3.
Sensitivity and compaction of Mnase-digested chromatin from WT and mutant cells. (A and B) EtBr-stained gel (A) and Southern blot probed for major satellite (B) of DNAs prepared from nuclei of WT and Dnmt3 − cells after an Mnase digestion time course. Markers are 1-kb and 100-bp ladders. The percent loss of high molecular weight signal for bulk (A) or major satellite (B) DNAs, which was calculated by measuring the signal between the two top red bars and the total signal between the top and bottom red bars, is graphed below each gel. (C) Percent loss of high molecular weight signal for minor satellite. (D) Percent loss of high molecular weight material signal in WT and Dnmt3 − cells for bulk chromatin (EtBr; red) and minor (black) and major (blue) satellites. (E) EtBr-stained gel of DNA prepared from WT and Dnmt3 − chromatin fractionated on a sucrose gradient. (F) The DNA size of the peak EtBr signals (red) and the minor (black) or major satellite (blue) signals detected after the Southern blotting of E was measured for the WT (closed circles) and mutant (open circles) ES cell chromatins and plotted against fraction number (sedimentation rate).
A 16-kb region of silent repetitive chromatin adjacent to the chicken β-globin locus, which is resistant to Mnase digestion (Prioleau et al., 1999), has been shown to sediment through sucrose with a frictional coefficient consistent with a rodlike shape of approximately the dimensions expected of a compact 30-nm chromatin fiber (Ghirlando et al., 2004). Similarly, and consistent with the aforementioned Mnase sensitivity, the rate of sedimentation of mouse and human satellite DNAs in sucrose gradients suggests that they are packaged into 30-nm chromatin fibers that are more compact in shape than those from the bulk genome (Gilbert and Allan, 2001; Gilbert et al., 2004). Bulk chromatin fibers from WT and Dnmt3 − cells have identical sedimentation rates (Fig. 3, E and F). Moreover, major and minor satellite–containing chromatin fibers from Dnmt3 − ES cells still sediment more rapidly than equivalently sized bulk chromatin fragments from the same cells (Fig. 3 F), indicating that these satellite regions remain more compact than bulk chromatin even in the complete absence of DNA methylation.
Altered histone modifications in the absence of DNA methylation
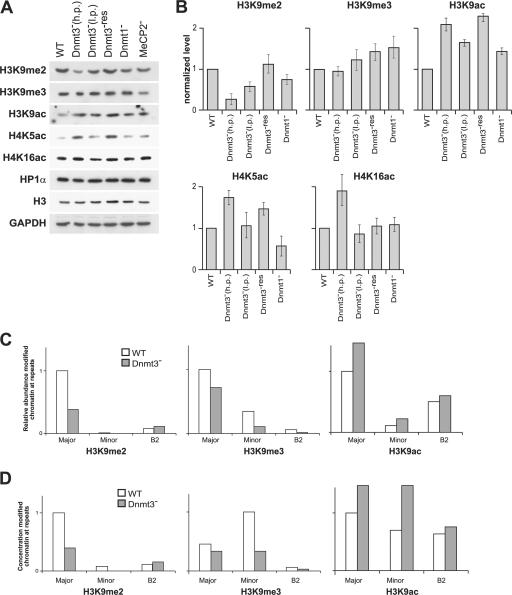
The surprising absence of any effect of DNA methylation on secondary chromatin structure suggests that the main influence might be on primary chromatin (nucleosome) structure. Indeed, Western blotting revealed that as DNA methylation disappeared in the Dnmt3 − cells, levels of H3K9me2 reduced concomitantly (Fig. 4, A and B). Adding back Dnmt3b (Dnmt3 −res ) restored H3K9me2 to WT levels. The loss of H3K9me2 is also not simply the result of the depletion of methyl-CpG–binding proteins because it is not seen in MeCP2 − cells.
Figure 4.
Histone methylation and acetylation levels in mutant ES cells. (A) Western blots of proteins from WT, Dnmt3 −, Dnmt3 −res, Dnmt1 −, and MeCP2 − ES cells incubated with antibodies that detect H3K9me2, H3K9me3, H3K9ac, H4K5ac, H4K16ac, HP1α, and H3. GAPDH is shown as a control. (B) Mean (±SEM [error bars]) levels of histone modifications detected in the different mutant ES cells normalized relative to GAPDH and WT (n = 3 or 4). (C and D) Abundance (C) and concentration (per kilobase; D) of H3K9me2, H3K9me3, and H3K9ac modifications detected by ChIP at different repeats in WT (white bars) and Dnmt3 − (gray bars) cells. h.p., high passage; l.p., low passage.
The loss of H3K9me2 in Dnmt3 − cells was paralleled by a progressive increase in the levels of H3K9ac. Acetylation levels at H4K5 and H4K16 are also elevated (Fig. 4, A and B). Although levels of H4K16ac and H4K5ac are completely or partially restored in Dnmt3 −res cells, levels of H3K9ac are not rescued in Dnmt3 −res cells (Fig. 4 B) or in Dnmt3 − cells rescued with Dnmt3a (not depicted). In human cells lacking Dnmt1, H3K9ac levels did return to those of WT when Dnmt1 was restored (Espada et al., 2004). However, in that case, the effects of DNA methylation and, therefore, presumably histone modification were concentrated in repetitive sequences. We suggest that in the Dnmt3 − ES cells, the increased H3K9ac levels are refractory to rescue by the reintroduction of Dnmt3a or b and that this may reflect a problem in retargeting some histone deacetylases. Because we found that both the sedimentation rate of chromatin fibers through sucrose (not depicted) and chromocenter nuclear organization (Fig. 2) from Dnmt3 −res cells are similar to WT, this indicates that histone acetylation does not affect secondary chromatin fiber structure or the nuclear organization of heterochromatin in these assays.
No decrease in global levels of H3K9me3 was detected in mutant cells by Western blotting. However, histone methylation marks are differentially distributed among tandem and interspersed repeats in the mouse genome (Martens et al., 2005). We used chromatin immunoprecipitation (ChIP) and Southern blotting of the immunoprecipitated material to investigate H3K9me2, H3K9me3, and H3K9ac in Dnmt3 − cells at minor and major satellite repeats and at the interspersed B2 repeat. Levels of H3K9me2 were almost undetectable at minor satellite even in WT, but, at the major satellite, there was a large (63%) reduction in H3K9me2 and an increase in H3K9ac (Fig. 4 C). Although most abundant at the major satellite (Gilbert et al., 2003; Guenatri et al., 2004; Martens et al., 2005), levels of H3K9me3 were reduced by 30% at all of the repeats in Dnmt3 − cells (Fig. 4 C).
Major satellite, minor satellite, and B2 comprise 3%, 0.45%, and 2.39% of the mouse genome, respectively (Waterston et al., 2002; Martens et al., 2005). Normalizing our ChIP data for repeat abundance showed that the concentration (per kilobase) of H3K9me2 is higher at major satellite than at minor satellite or B2 (Fig. 4 D). In contrast, the concentration of H3K9me3 is higher at minor satellite, although its total abundance is less. Loss of DNA methylation redistributes H3K9me3 so that its concentration at major and minor satellites is similar.
Altered binding of linker histones in the absence of DNA methylation
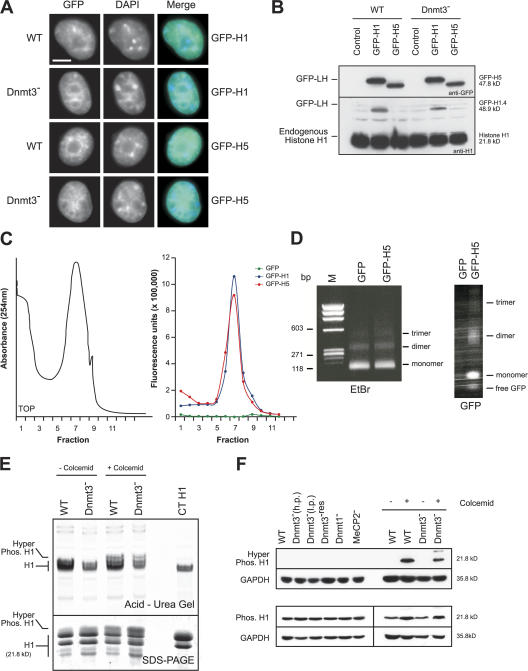
The binding of linker histones to nucleosomes in order to form the chromatosome is a fundamental aspect of dynamic chromatin structure. The precise sites of linker histone binding to the nucleosome remain in dispute, but there is evidence, mainly from in vitro analyses, arguing both for (Ball et al., 1983; McArthur and Thomas, 1996) and against (Campoy et al., 1995; Nightingale and Wolffe, 1995; Hashimshony et al., 2003) a role of DNA methylation in linker histone binding. To investigate this in vivo, we used FRAP to analyze the mobility of linker histones H1 (subtype H1.4) and H5, which were tagged with GFP at their N termini, in WT and Dnmt3 − ES cells. We chose this orientation to minimize any interference of GFP with the high-affinity C-terminal chromatin-binding domain of H1 (Hendzel et al., 2004). We were able to select for somatic cells stably transfected with these constructs but not undifferentiated ES cell lines. However, in transient transfections of ES cells, viable expressing cells were still visible 72 h after transfection, the tagged linker histones localized correctly in the nucleus, and Western blotting with GFP and H1 antibodies confirms that the levels of GFP–linker histone were low compared with endogenous H1 (Fig. 5, A and B). Fractionation of Mnase-digested chromatin from these cells on a sucrose step gradient showed that the GFP fluorescence cosediments with chromatin (Fig. 5 C). To confirm that GFP–linker histone incorporates into nucleosomes, soluble polynucleosomes, which were released from GFP- or GFP-H5–transfected COS cells with Mnase, were fractionated on a nucleoprotein gel, and GFP fluorescence was analyzed by scanning (Fig. 5 D). We conclude that GFP–linker histones are correctly incorporated into nucleosomal chromatin in transiently transfected ES cells but that expression of the exogenous linker histone is incompatible with the long-term propagation of undifferentiated ES cells.
Figure 5.
GFP-tagged linker histone characterization. (A) Distribution of GFP fluorescence (green) in WT or Dnmt3 − ES cells transiently transfected with GFP-H1 or -H5. DNA is counterstained with DAPI (blue). Bar, 5 μm. (B) Western blot of nuclei prepared from GFP-H1– or -H5–transfected WT and Dnmt3 − cells probed with an anti-GFP or -H1 antibody; the position of the GFP-tagged linker histone (GFP-LH) and endogenous H1 are indicated. Transfection efficiency is ∼30%. (C) Chromatin isolated from Dnmt3 − cells transfected with GFP–linker histone or free GFP and fractionated on a 10/ 50% sucrose step gradient. Chromatin was monitored by absorbance at 254 nm (left). GFP fluorescence was monitored across the fractions at 507 nm (right). (D) EtBr-stained 1% agarose gel of DNA purified from Mnase-digested chromatin from cells transfected with GFP or GFP-H5 (left). Chromatin was fractionated on a native nucleoprotein gel and scanned for GFP fluorescence (right). M, marker. (E) Coomassie-stained acid-urea (top) or SDS-PAGE (bottom) gels of perchloric acid–extracted linker histones from WT and Dnmt3 − cells. (F) Western blot with antibodies detecting phosphorylated or hyperphosphorylated H1 on protein extracts from WT and mutant ES cells in the presence or absence of colcemid treatment. GAPDH was used as a loading control.
Linker histone mobility has been shown to be influenced by phosphorylation (Hendzel et al., 2004). Perchloric acid–extracted linker histones from WT and Dnmt3 − cells were examined on an acid-urea gel that is able to discriminate proteins based on their phosphorylation state. Although the linker histone phosphorylation associated with mitosis (induced by colcemid treatment) was readily apparent, we did not detect any differences in the linker histone phosphorylation state between WT and Dnmt3 − interphase cells (Fig. 5 E). This was confirmed by Western blotting of the cells with antibodies that detect phosphorylated or hyperphosphorylated H1 (Fig. 5 F).
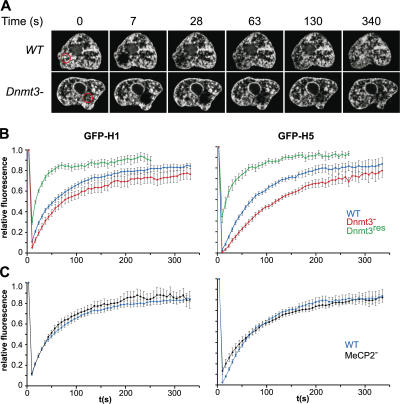
Linker histone mobility was then analyzed in ES cells 24 h after transfection by following the fluorescence recovery every 7 s (for a total of 340 s) after photobleaching (Fig. 6 A). The recovery kinetics of H1 in WT ES cells (t 1/2 = 50 s) is consistent with previous analyses in differentiated cells (Misteli et al., 2000; Hendzel et al., 2004). The t 1/2 for H5 was longer (70 s), which is consistent with the higher affinity of the arginine-rich H5 linker histone for the chromatin fiber (Thomas and Rees, 1983). Strikingly, compared with WT cells, the recovery kinetics of both H1 and H5 were slowed in Dnmt3 − cells (Fig. 6 B), with t 1/2 values increasing to 70 s for H1 and 100 s for H5 (Table I). In contrast, the t 1/2 for both H1 and H5 were considerably shorter (25–28 s) in Dnmt3 −res cells than in WT (Fig. 6 B). This suggests that the prolonged binding of linker histones to chromatin in Dnmt3 − cells is caused by the lack of DNA methylation and not by the elevated levels of histone acetylation because the latter persists in Dnmt3 −res cells (Fig. 4).
Figure 6.
Linker histone mobility in WT and mutant cells. (A) Representative confocal images of GFP-H1–transfected WT and Dnmt3 − cells during FRAP. The bleach area is marked by red circles, and the fluorescence recovery is monitored over time. (B) Relative fluorescence within the bleach ROI during FRAP of H1-GFP or -GFP expressed in WT (blue), Dnmt3 − (red), and Dnmt3 −res (green) cells. Graphs show mean values (±SEM [error bars]) for 10 cells at each time point. (C) Relative fluorescence (mean ± SEM) within the bleach ROI during FRAP of H1-GFP or -GFP expressed in WT (blue) and MeCP2 − (black) cells.
Table I.
Linker histone FRAP analysis
| Cell line | DNA methylation | H3K9me | H3K9ac | Linker histone t 1/2 |
|---|---|---|---|---|
| s | ||||
| WT | High | High | Low | 50 (H1) 70 (H5) |
| Dnmt3− | Low | Low | High | 70 (H1) 100 (H5) |
| Dnmt3−res | High | High | High | 27 (H1) 27 (H5) |
| MeCP2 | High | High | Low | 53 (H1) 64 (H5) |
The mobility of both H1 and H5 in MeCP2 − ES cells was indistinguishable from WT (Fig. 6 C and Table I). Therefore, the altered kinetics of linker histone binding in Dnmt3 − cells is likely caused by the lack of DNA methylation itself and not by the subsequent loss of this methyl-CpG–binding protein.
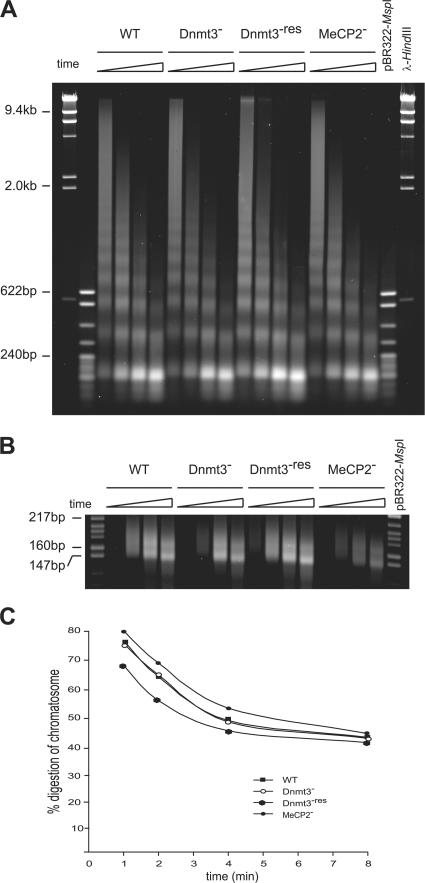
Linker histones retard Mnase digestion of the core particle, pausing digestion at 178 nucleotides before protection of the 168-bp chromatosome, which comprises the nucleosome core particle together with the DNA that is protected from digestion by linker histone binding to the dyad (Muyldermans et al., 1981). If the slowed mobility of linker histone in Dnmt3 − cells is caused by its altered binding to the nucleosome core particle, the rate of Mnase trimming of nucleosomes might change. However, the chromatosome was trimmed at the same rate in WT, Dnmt3 −, and other mutant cells (Fig. 7). Therefore, although the overall in vivo mobility of linker histone on chromatin was reduced in Dnmt3 − cells, its residence time at the nucleosome dyad of the chromatosome measured in vitro was not altered.
Figure 7.
Mnase trimming of chromatosomal particles. (A) EtBr-stained gel of DNA prepared from nuclei of WT and mutant ES cells after time course digestion with 360 U/ml Mnase. Markers are λ-HindIII and pBR322-MspI ladders. (B) High resolution agarose gel to distinguish between the nucleosome (146 bp) and chromatosome (168 bp) bands. (C) Loss of the 168-bp band with respect to the total DNA in the 146- and 168-bp bands.
Discussion
Using mutant mouse ES cells, we show that the absence of DNA methylation leads to altered nuclear organization with an increase in the level of chromocenter clustering (Fig. 2). This is reminiscent of the increased associations between juxtacentromeric heterochromatin that are seen in the nuclei of individuals with immunodeficiency centromeric instability facial anomalies (ICF) syndrome, which is caused by the mutation of DNMT3b and the consequent hypomethylation of satellite sequences (Gisselsson et al., 2005). Increased chromocenter clustering in Dnmt3 − cells is in contrast to the suggested role of DNA methylation and methyl-CpG–binding proteins in promoting chromocenter clustering during the terminal differentiation of mouse cells (Brero et al., 2005) and might indicate differences between cell types. Altered nuclear organization in the Dnmt3 − cells suggests that there is some underlying changes in chromatin structure when DNA methylation is absent. Indeed, we have identified such changes at two levels of primary chromatin structure: histone modifications and linker histone binding.
ES cells lacking DNA methylation have globally elevated levels of acetylation at H3K9, H4K5, and H4K16 (Fig. 4). Increased acetylation of H4K5 has been previously reported in these cells (Jackson et al., 2004). We also saw reduced levels of H3K9me2 in Dnmt3 − cells by both Western blotting and ChIP and a redistribution of histone methylation within satellite-containing heterochromatin compartments (Fig. 4). This contrasts with the unaltered histone methylation seen in a previous analysis of Dnmt3 − ES cells (Martens et al., 2005). However, in that analysis, the Dnmt3 − cells had substantial residual DNA methylation. Both Dnmts and methyl-CpG–binding proteins can physically associate with histone deacetylases (Rountree et al., 2000; Fuks et al., 2001) and histone methyltransferases (Fuks et al., 2003a,b; Lehnertz et al., 2003; Sarraf and Stancheva, 2004), so the altered histone modifications in cells that completely lack CpG methylation may reflect the loss of these interactions.
It has been suggested that histone modifications can directly affect secondary chromatin structures by, for example, altering nucleosome–DNA or nucleosome–nucleosome interactions and by neutralizing charge in the histone N-terminal tails (Wolffe and Hayes, 1999; Carruthers and Hansen, 2000; Wang et al., 2001). However, neither Mnase digestion kinetics nor sucrose gradient sedimentation (Fig. 3) revealed any evidence for a general decompaction of secondary chromatin structure in the absence of DNA methylation. Satellite sequences remain in a more compact structure than the bulk genome using these assays regardless of the state of DNA methylation.
Most unexpectedly, the loss of DNA methylation leads to the altered binding of linker histones. By FRAP with GFP-tagged H1 and H5, t 1/2 times for both linker histones were increased in Dnmt3 − cells compared with WT (Fig. 6 and Table I). This was not an indirect effect of the loss of a methylated CpG–binding protein because recovery times were normal in MeCP2 − cells (Fig. 6 C). The tighter binding is also not an indirect effect of elevated histone acetylation levels because in Dnmt3 −res cells, in which DNA methylation levels are restored but H3K9ac levels remain elevated compared with WT, t 1/2 times are dramatically shortened (Fig. 6 B). This is consistent with the increased mobility of linker histones found in trichostatin A–treated cells (Misteli et al., 2000). The recovery kinetics that we found for H1 in WT and mutant ES cells are similar to those previously reported in differentiated somatic cells (Misteli et al., 2000; Hendzel et al., 2004). We do not detect the fast recovery kinetics that have recently been reported for H1 in ES cells (Meshorer et al., 2006). This might be the result of differences in the linker subtypes or ES cells used or of differential behavior between C- and N-terminally tagged H1 (Hendzel et al., 2004). Because there was no change in the ability of linker histones to protect the chromatosome from trimming by Mnase in Dnmt3 − cells (Fig. 6), we suggest that the loss of DNA methylation does not alter the binding of linker histones to the dyad of the nucleosome but that it is binding to another site (perhaps linker DNA) that is enhanced (Zhou et al., 1998; Bharath et al., 2003). This is consistent with the two-site binding model proposed by Phair et al. (2004). The recent identification of a link between H1 depletion and altered DNA methylation at specific genomic regions (Fan et al., 2005) suggests that a better understanding of the mechanistic and regulatory interactions between these two key chromatin modulators is required.
Materials and methods
Cell culture and transfection
Mouse WT J1, Dnmt3−, Dnmt1 − (S/S allele) (gift from E. Li, Novartis Institutes for Biomedical Research, Cambridge, MA; Lei et al., 1996; Okano et al., 1999), Dnmt3 −res (Jackson et al., 2004), and MeCP2 − (gift from A. Bird, Wellcome Trust Centre for Cell Biology, University of Edinburgh, Edinburgh, UK; Tate et al., 1996) ES cells were cultured under standard conditions in the presence of leukemia inhibitory factor. Cells were transfected using LipofectAMINE 2000 (Invitrogen).
CpG methylation analysis, DNA digestion, and Southern hybridization
Nearest neighbor analysis was performed as described previously (Jackson et al., 2004). Genomic DNAs were digested with methylation-sensitive isoschisomer pairs (HpaII–MspI) or a methylation-sensitive enzyme (HpyCH4IV) and fractionated on a 0.7% agarose gel in Tris–phosphate buffer supplemented with ethidium bromide (EtBr). Gels were Southern blotted onto Hybond N (GE Healthcare) in 20× SSC and were probed for minor and major satellites and the B2 repeat (Gilbert and Allan, 2001).
3D FISH
ES cells were cultured on gelatine-coated microscope slides for 4 h before fixation in 4% PFA for 10 min and processed for 3D FISH as previously described (Mahy et al., 2002). Major satellite was detected by hybridization with digoxigenin-labeled pSAT (Lewis et al., 1992). After washing and detection, slides were counterstained with 50 ng/ml DAPI and mounted in Vectashield. Slides were examined with an epifluorescence microscope (Axioskop; Carl Zeiss MicroImaging, Inc.) equipped with a 100× NA 1.3 lens and a CCD camera (Micromax; Princeton Instruments). A Pifoc piezo-driven objective focusing device was used to capture images at 0.25-μm intervals through the z axis. Images were captured and analyzed using custom IPlab (BD Biosciences) scripts. The significance of differences between cell lines was assessed using a Mann-Whitney U nonparametric test.
Preparation and fractionation of nuclei and chromatin
Nuclei were prepared as previously described (Gilbert et al., 2003) but with a reduced concentration (0.05%) of NP-40 in nuclei buffer B. For Mnase sensitivity digests, the nuclei concentration was adjusted to 4 A260 in nuclei buffer R. 20 or 50 U/ml Mnase (Worthington) was added, and aliquots were removed into stop buffer (2% SDS, 200 μg/ml proteinase K, amd 10 mM EDTA) at various time intervals. Purified DNAs were fractionated on a 1% agarose gel in Tris-borate buffer in the presence of EtBr. To distinguish nucleosomal (146 bp) and chromatosome-protected (166 bp) DNA after MNase digestion, the nuclei concentration was adjusted to 4 A260 in nuclei buffer R and were digested with 180 or 360 U/ml Mnase. Purified DNAs were fractionated on a 4% (1% regular agarose and 3% 3:1 NuSieve agarose [Flowgen]) gel in Tris-borate buffer in the presence of EtBr. Gels were scanned on a phosphorimager (FLA5100; Fuji) equipped with a 532-nm laser and 575-nm bandpass filter and were transferred to Hybond N by Southern blotting if required. Soluble chromatin was prepared and fractionated on sucrose gradients as described previously (Gilbert and Allan, 2001; Gilbert et al., 2004). Gel and blot images were analyzed using the Aida software package version 3.52 (Raytek).
Linker histones were isolated by extracting whole cell lysates with 5% perchloric acid and subsequently by precipitation with acetone. The linker histones were analyzed on either a 15% (80:1 acrylamide/bis-acrylamide) acid-urea gel (Barratt et al., 1994) or a 18% (200:1 acrylamide/bis-acrylamide) SDS-PAGE gel (Thomas and Kornberg, 1978).
Western blotting and ChIP
Cells were lysed in 1× SDS sample buffer (62.5 mM Tris-HCl, pH 6.75, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, and bromophenol blue), sonicated, and ∼5 μg of total protein was fractionated on a 12% SDS polyacrylamide gel. Proteins were transferred to Hybond P by electroblotting, and membranes were probed with antibodies that detect the following: H3K9me2 (1:2,000; Upstate Biotechnology), H3K9me3 (1:2,000; provided by T. Jenuwein, Research Institute of Molecular Pathology, Vienna, Austria), H3K9ac (1:2,000; Upstate Biotechnology), H4K5ac (1:20,000; Upstate Biotechnology), H4K16ac (1:10,000; Upstate Biotechnology), HP1α (1:2,500; MAB3446; Chemicon), phosphorylated H1 (0.15 μg/ml; clone 12D11; Upstate Biotechnology), hyperphosphorylated H1 (1:500; Upstate Biotechnology), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:2,000, Abcam). Detection was performed by ECL (Pierce Chemical Co.).
ChIP was performed as described previously (Chambeyron and Bickmore, 2004) with antibodies recognizing the aforementioned H3K9me2, H3K9me3, and H3K9ac. Immunoprecipitated chromatin was dot blotted onto Hybond N+ (GE Healthcare). Membranes were probed with minor and major satellite repeats and the interspersed B2 repeat, and the blots were analyzed on a phosphorimager (FLA5100; Fuji).
H1 and H5 constructs
Chicken linker histone H5 cDNA (Krieg et al., 1982) was subcloned from pBR-H5 using a blunted NotI–XmnI fragment into the SmaI site of pUC18 to give pUCH5. The coding sequence was PCR amplified with primers 5′-GAAGATCTTCCGGAATGACGGAGAGCCTGGTC-3′ and 5′-GGCCGCTCGAGTTACTTCAGCTCACTTCTTCTTGGGCGATTT-3′. Human histone H1.4 (a gift from D. Doenecke, German Cancer Research Center, Heidelberg, Germany) genomic clone (GenBank/EMBL/DDBJ accession no. M60748) was PCR amplified using primers 5′-GAAGATCTTCCGGAATGTCCGAGACTGCGCCT-3′ and 5′-GGCCGCTCGAGTTACTTCAGCCTACTTTTTCTTGGCTGCCGC-3′. Both PCR fragments were digested with BglII–XhoI and cloned into a BglII–SmaI site of pDsRed1-C1 (CLONTECH Laboratories, Inc.). The linker histone–containing portion was removed using BspEI–BamHI and was cloned into EGFP-C1 at BspEI–BamHI. Constructs were checked by sequencing, by transient transfection, and by Western blotting for GFP.
To examine the nuclear distribution of the GFP fusion proteins, transfected cells were fixed in 4% PFA and visualized by fluorescence microscopy. To investigate the association of GFP–linker histone to chromatin, 3-kb nucleosomal fragments from transiently transfected cells were generated by Mnase digestion and fractionated on a 10/50% sucrose step gradient at 48 K for 105 min in a rotor (SW55; Beckman Coulter). Fractions were collected by upward displacement with continuous monitoring of the chromatin at 254 nm (Fig. 5 C). GFP fluorescence was measured in each fraction on a fluorometer (Envision; PerkinElmer) at 510 nm. Proteins were precipitated from the gradient fractions and analyzed by SDS-PAGE to confirm that the GFP–linker histone fusion protein was present. Binding of GFP-H5 to individual nucleosomes was investigated by isolating chromatin from COS7 cells transfected with GFP-H5. Soluble chromatin was dialysed overnight against TEP80 and were further digested with Mnase to prepare short oligonucleosomes. The chromatin fragments were fractionated on a 5% polyacrylamide gel in Tris-borate buffer at 4°C. The gel was analyzed for GFP fluorescence using a scanner (FLA2000; Fuji) equipped with a 479-nm laser and 520-nm bandpass filter. The EtBr-stained gel was scanned using a 479-nm laser and 580-nm bandpass filter.
FRAP
Transiently transfected ES cells were grown on 0.17-mm culture dishes (DeltaT; Bioptechs) and, 24 h after transfection, were mounted onto a heated stage (Bioptechs) on a confocal microscope (LSM510; Carl Zeiss MicroImaging, Inc.). An objective warmer (Bioptechs) was used to maintain a stable temperature of the medium in the culture dish. Cells expressing high levels of H1- or H5-GFP fusion protein (total cellular pixel intensity > 35,000) were excluded from analysis.
For FRAP, a 3-μm-diameter region of interest (ROI) of the nucleus in the midfocal plane was bleached with 10–15 iterations at 100% power with an argon laser at 6.1 mA. The pinhole size for the confocal was set at 1 Airy U. The time series software option was used to specify the appropriate time delay between rounds of 3D image stack capture. Images were captured with a 100× objective at 7-s intervals for a total of 340 s using 8% of laser power. Each image was processed by an interactive script (IPLAB version 3.6; Scanalytics) to correct for nuclear rotation and cell movement. Loss of fluorescence attributed to the imaging process alone was assessed from the sum of pixel intensities in the cell. The fluorescence intensity for each ROI over time was then normalized to this (Phair and Misteli, 2000).
Acknowledgments
We thank Heidi Sutherland for the initial immunofluorescence analysis of Dnmt3 − cells, Michael Whitehead for assistance with preliminary chromatin analysis, and Elizabeth Kerr for technical support. We are grateful to Thomas Jenuwein for the gift of antibody against H3K9me3 and to D. Doenecke for human H1 clones. Dnmt3- and MeCP2-null ES cells were gifts from En Li (Boston) and Adrian Bird (Edinburgh). We also thank Richard Meehan for his critical reading of the manuscript.
N. Gilbert is a Wellcome Trust Career Development Fellow, and W.A. Bickmore is a Centennial Fellow of the James S. McDonnell Foundation. This work was supported by the UK Medical Research Council and, in part, through the European Union FP6 Network of Excellence Epigenome (grant LSHG-CT-2004-503433).
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; ES, embryonic stem; EtBr, ethidium bromide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Mnase, micrococcal nuclease; ROI, region of interest; WT, wild type.
References
- Ball, D.J., D.S. Gross, and W.T. Garrard. 1983. 5-methylcytosine is localized in nucleosomes that contain histone H1. Proc. Natl. Acad. Sci. USA. 80:5490–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt, M.J., C.A. Hazzalin, E. Cano, and L.C. Mahadevan. 1994. Mitogen- stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc. Natl. Acad. Sci. USA. 91:4781–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharath, M.M., N.R. Chandra, and M.R. Rao. 2003. Molecular modeling of the chromatosome particle. Nucleic Acids Res. 31:4264–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brero, A., H.P. Easwaran, D. Nowak, I. Grunewald, T. Cremer, H. Leonhardt, and M.C. Cardoso. 2005. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 169:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.C., E. Grace, A.T. Sumner, A.T. Edmunds, and P.M. Ellis. 1995. ICF syndrome (immunodeficiency, centromeric instability and facial anomalies): investigation of heterochromatin abnormalities and review of clinical outcome. Hum. Genet. 96:411–416. [DOI] [PubMed] [Google Scholar]
- Campoy, F.J., R.R. Meehan, S. McKay, J. Nixon, and A. Bird. 1995. Binding of histone H1 to DNA is indifferent to methylation at CpG sequences. J. Biol. Chem. 270:26473–26481. [DOI] [PubMed] [Google Scholar]
- Carruthers, L.M., and J.C. Hansen. 2000. The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275:37285–37290. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., and W.A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T., Y. Ueda, J.E. Dodge, Z. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada, J., E. Ballestar, M.F. Fraga, A. Villar-Garea, A. Juarranz, J.C. Stockert, K.D. Robertson, F. Fuks, and M. Esteller. 2004. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 279:37175–37184. [DOI] [PubMed] [Google Scholar]
- Fan, Y., T. Nikitina, J. Zhao, T.J. Fleury, R. Bhattacharyya, E.E. Bouhassira, A. Stein, C.L. Woodcock, and A.I. Skoultchi. 2005. Histone h1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 123:1199–1212. [DOI] [PubMed] [Google Scholar]
- Fuks, F., W.A. Burgers, N. Godin, M. Kasai, and T. Kouzarides. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20:2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks, F., P.J. Hurd, R. Deplus, and T. Kouzarides. 2003. a. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31:2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks, F., P.J. Hurd, D. Wolf, X. Nan, A.P. Bird, and T. Kouzarides. 2003. b. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278:4035–4040. [DOI] [PubMed] [Google Scholar]
- Ghirlando, R., M.D. Litt, M.N. Prioleau, F. Recillas-Targa, and G. Felsenfeld. 2004. Physical properties of a genomic condensed chromatin fragment. J. Mol. Biol. 336:597–605. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., and J. Allan. 2001. Distinctive higher-order chromatin structure at mammalian centromeres. Proc. Natl. Acad. Sci. USA. 98:11949–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., S. Boyle, H. Sutherland, H. J.de Las, J. Allan, T. Jenuwein, and W.A. Bickmore. 2003. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 22:5540–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., S. Boyle, H. Fiegler, K. Woodfine, N.P. Carter, and W.A. Bickmore. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibres. Cell. 118:555–566. [DOI] [PubMed] [Google Scholar]
- Gisselsson, D., C. Shao, C.M. Tuck-Muller, S. Sogorovic, E. Palsson, D. Smeets, and M. Ehrlich. 2005. Interphase chromosomal abnormalities and mitotic missegregation of hypomethylated sequences in ICF syndrome cells. Chromosoma. 114:118–126. [DOI] [PubMed] [Google Scholar]
- Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony, T., J. Zhang, I. Keshet, M. Bustin, and H. Cedar. 2003. The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 34:187–192. [DOI] [PubMed] [Google Scholar]
- Hendzel, M.J., M.A. Lever, E. Crawford, and J.P. Th'ng. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028–20034. [DOI] [PubMed] [Google Scholar]
- Huang, J., T. Fan, Q. Yan, H. Zhu, S. Fox, H.J. Issaq, L. Best, L. Gangi, D. Munroe, and K. Muegge. 2004. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 32:5019–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M., A. Krassowska, N. Gilbert, T. Chevassut, L. Forrester, J. Ansell, and B. Ramsahoye. 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 24:8862–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karymov, M.A., M. Tomschik, S.H. Leuba, P. Caiafa, and J. Zlatanova. 2001. DNA methylation-dependent chromatin fiber compaction in vivo and in vitro: requirement for linker histone. FASEB J. 15:2631–2641. [DOI] [PubMed] [Google Scholar]
- Krieg, P.A., A.J. Robins, M.J. Gait, R.C. Titmas, and J.R. Wells. 1982. Chicken histone H5: selection of a cDNA recombinant using an extended synthetic primer. Nucleic Acids Res. 10:1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz, B., Y. Ueda, A.A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A.H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192–1200. [DOI] [PubMed] [Google Scholar]
- Lei, H., S.P. Oh, M. Okano, R. Juttermann, K.A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 122:3195–3205. [DOI] [PubMed] [Google Scholar]
- Levine, A., A. Yeivin, E. Ben Asher, Y. Aloni, and A. Razin. 1993. Histone H1-mediated inhibition of transcription initiation of methylated templates in vitro. J. Biol. Chem. 268:21754–21759. [PubMed] [Google Scholar]
- Lewis, J.D., R.R. Meehan, W.J. Henzel, I. Maurerfogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular-localization of a novel chromosomal protein that binds to methylated DNA. Cell. 69:905–914. [DOI] [PubMed] [Google Scholar]
- Ma, Y., S.B. Jacobs, L. Jackson-Grusby, M.A. Mastrangelo, J.A. Torres-Betancourt, R. Jaenisch, and T.P. Rasmussen. 2005. DNA CpG hypomethylation induces heterochromatin reorganization involving the histone variant macroH2A. J. Cell Sci. 118:1607–1616. [DOI] [PubMed] [Google Scholar]
- Mahy, N.L., P.E. Perry, S. Gilchrist, R.A. Baldock, and W.A. Bickmore. 2002. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, J.H., R.J. O'Sullivan, U. Braunschweig, S. Opravil, M. Radolf, P. Steinlein, and T. Jenuwein. 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur, M., and J.O. Thomas. 1996. A preference of histone H1 for methylated DNA. EMBO J. 15:1705–1714. [PMC free article] [PubMed] [Google Scholar]
- Meshorer, E., D. Yellajoshula, E. George, P.J. Scambler, D.T. Brown, and T. Misteli. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D.T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881. [DOI] [PubMed] [Google Scholar]
- Muyldermans, S., I. Lasters, L. Wyns, and R. Hamers. 1981. Protection of discrete DNA fragments by the complex H1-octamerhistones or H5- octamerhistones after micrococcal nuclease digestion. Nucleic Acids Res. 9:3671–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale, K., and A.P. Wolffe. 1995. Methylation at CpG sequences does not influence histone H1 binding to a nucleosome including a Xenopus borealis 5 S rRNA gene. J. Biol. Chem. 270:4197–4200. [DOI] [PubMed] [Google Scholar]
- Okano, M., D.W. Bell, D.A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99:247–257. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D.T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau, M.N., P. Nony, M. Simpson, and G. Felsenfeld. 1999. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 18:4035–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree, M.R., K.E. Bachman, and S.B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269–277. [DOI] [PubMed] [Google Scholar]
- Sarraf, S.A., and I. Stancheva. 2004. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 15:595–605. [DOI] [PubMed] [Google Scholar]
- Schmid, M., T. Haaf, and D. Grunert. 1984. 5-Azacytidine-induced undercondensations in human chromosomes. Hum. Genet. 67:257–263. [DOI] [PubMed] [Google Scholar]
- Tariq, M., and J. Paszkowski. 2004. DNA and histone methylation in plants. Trends Genet. 20:244–251. [DOI] [PubMed] [Google Scholar]
- Tate, P., W. Skarnes, and A. Bird. 1996. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat. Genet. 12:205–208. [DOI] [PubMed] [Google Scholar]
- Tazi, J., and A. Bird. 1990. Alternative chromatin structure at CpG islands. Cell. 60:909–920. [DOI] [PubMed] [Google Scholar]
- Thomas, J.O., and R.D. Kornberg. 1978. The study of histone–histone associations by chemical cross-linking. Methods Cell Biol. 18:429–440. [PubMed] [Google Scholar]
- Thomas, J.O., and C. Rees. 1983. Exchange of histones H1 and H5 between chromatin fragments. A preference of H5 for higher-order structures. Eur. J. Biochem. 134:109–115. [DOI] [PubMed] [Google Scholar]
- Wang, X., C. He, S.C. Moore, and J. Ausio. 2001. Effects of histone acetylation on the solubility and folding of the chromatin fiber. J. Biol. Chem. 276:12764–12768. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., K. Lindblad-Toh, E. Birney, J. Rogers, J.F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature. 420:520–562. [DOI] [PubMed] [Google Scholar]
- Wolffe, A.P., and J.J. Hayes. 1999. Chromatin disruption and modification. Nucleic Acids Res. 27:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y.B., S.E. Gerchman, V. Ramakrishnan, A. Travers, and S. Muyldermans. 1998. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 395:402–405. [DOI] [PubMed] [Google Scholar]